Abstract
A technique for genome-wide detection of differences in the integration site positions of interspersed repeats in related genomes (DiffIR) is described. The technique is based on a whole- genome selective PCR amplification of the repeats’ flanking regions followed by a differential hybridization screening of the arrayed library of the selected amplicons. The technique was successfully applied to the comparison of the integration sites in the human and chimpanzee genomes, allowing us to discover 11 new human-specific integrations of human endogenous retrovirus, K family (HML-2) long terminal repeats.
INTRODUCTION
Progress in the complete genome sequencing of different organisms has reawakened interest in the functional and evolutionary role of transposable elements, which were shown to constitute up to 47% of the sequenced human genome (1), much greater than believed earlier. The three most abundant retrotransposon groups represented by long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs) and long terminal repeat (LTR) retrotransposons comprise 13, 20 and 8%, respectively, of the genome.
Retroelements (REs) are now attracting much attention due to the suggestion that they carry crucial information about evolutionary events and might play an important role in evolution by causing genomic rearrangements, creating new genes and affecting the regulation of the existing genes. The ongoing integrations of REs and genomic rearrangements due to the recombinations between REs located in different genomic loci can cause various hereditary diseases (2–6) and can also be involved in cancer development (reviewed in 7–9).
RE insertions within the genome represent a new class of genetic polymorphic markers for the study of human evolution, population genetics and, possibly, for gene mapping (10–12). Being stable, they offer several advantages over other types of polymorphisms. In particular, the presence of an RE represents identity by descent, because the probability that two different REs were independently integrated into exactly the same site is negligible. The identification of young, recently integrated REs is of considerable interest for deeper understanding of primate genome evolution. In particular, the identification of human-specific integrations (HSIs) in the vicinity of human genes, especially those involved in embryo development, could reveal the REs that possibly played a role in the split of the human and other hominoid lineages. Polymorphic integrations in the human population would be of great help in disease gene mapping and for better understanding of interpopulation relationships.
Several groups of authors have described quite a number of polymorphic and human specific Alu and L1 (the most abundant and young subfamily of LINEs) integrations. However, no comprehensive genome-wide search for human-specific and polymorphic RE insertions has been done so far, though an attempt at such a screening for Alu and L1 was made (10,12,13). The rationale for the authors’ approach was that small genomic subsets of both Alu and L1 families amplified during recent evolutionary time (4–5 million years ago) should reveal considerable polymorphism in the human population as convincingly demonstrated for Ya5, Ya8 and Yb8 young subfamilies of Alu repeats (10) and for the Ta young subfamily of L1 elements (11,12). Therefore, a computer search for recently integrated REs in the human genomic DNA sequence databases could reveal the candidate polymorphic integrations. This approach has been used to identify recently integrated Ya5 and Yb8 Alu family members (10). 2640 Ya5 Alu and 1852 Yb8 Alu family members were found in the draft sequence of the human genome. Experimental analysis of 475 of these elements showed that ∼99% of them were restricted to the human genome and absent from the genomes of non-human primates. But only 106 of them represented insertion polymorphisms within the genomes of various human populations. These results demonstrate that such a computer search must be accompanied by experimental checking of which of the REs identified are indeed polymorphic. In addition, some highly polymorphic insertions can be absent from the available human genome sequence, which represents only a few haplotypes taken by chance for sequencing. Such a computer search is also impossible with non-sequenced genomes, in particular with non-human primate genomes.
The discussion above shows that efficient experimental techniques in the genome-wide search for polymorphic insertions would be very useful. These techniques would also be of help for identification of the RE HSIs with important evolutionary implications.
In this report we describe such a technique. The technique includes targeted simplification of the genomes being compared, by selective amplification of the sequences flanking REs, preparation of arrayed libraries of the amplicons, and subsequent differential hybridization of these libraries with the amplicons representing one or the other genome. Selective amplification allows us to obtain a portion of a complex genome small enough to be used directly as a hybridization probe to detect differences between two selectively amplified genomes. We demonstrate the applicability of the approach to the identification of differences between distributions of HERV LTRs in the human genome and the genomes of great apes.
The technique can be easily expanded to the microchip format.
MATERIALS AND METHODS
An aliquot of 500 ng of human and chimpanzee genomic DNA (brain and peripheral blood, respectively) in 50 µl was digested with 15 U of AluI restriction enzyme (Promega) at 37°C for 80 min. The digested DNAs were then precipitated with ethanol, dissolved in 5 µl of sterile water, and ligated to an excess adapter (2 µM) at 16°C overnight using T4 DNA ligase (Promega). To form the adapter, 20 µM A1A2 and A1 oligonucleotides (Table 1) were annealed in TM (10 mM Tris–HCl pH 7.8, 10 mM MgCl2) buffer. The ligation was terminated by incubation at 65°C for 5 min. The ligates were then separated from the excess primers by passing through a QIAquick DNA Purification Kit (Qiagen, USA). The 3′ ends of the DNA duplexes were extended with Klenow fragment of Escherichia coli DNA polymerase. The extended ligate was precipitated with ethanol and redissolved in 30 µl sterile water.
Table 1. Oligonucleotides/primers used for the library construction and analysis.
| A1A2 | TGTAGCGTGAAGACGACAGAAAGGGCGTGGTGCGGAGGGCGGT |
| A1 | TGTAGCGTGAAGACGACAGAA |
| A2 | AGGGCGTGGTGCGGAGGGCGGT |
| a2 | ACCGCCCTCC |
| A2ctG | AGGGCGTGGTGCGGAGGGCGGTCTG |
| A2ctA | AGGGCGTGGTGCGGAGGGCGGTCTA |
| A2ctT | AGGGCGTGGTGCGGAGGGCGGTCTT |
| A2ctC | AGGGCGTGGTGCGGAGGGCGGTCTC |
| T1 | TCCTCCGTATGCTGAACGCTGGTTCC |
| T2 | TCTCTAGACTTTGTCTCTGTGTC |
| T3 | CACAGGTGTGGAGGGGCAG |
Fractions of LTR-U5 flanking sequences were amplified using the PCR suppression method (14,15). A1 and T1 primers (Table 1) were used in the first round of PCR. All T primers (T1, T2 and T3 in Table 1) correspond to most conserved parts of the U5 LTR region. The PCR mixture contained 10 ng of the ligate in 25 µl of 1× PCR Buffer for Display Taq (Display System Biotech) containing 200 µM dNTPs, 0.4 µM of each primer, and 0.5 U of the Display Taq DNA polymerase (Display System Biotech). PCR was antibody hot started (TaqStart antibodies; Clontech) and processed in a thermal cycler (OmniGene Hybaid, tube control mode) as follows: 25 cycles of 94°C for 20 s, 65°C for 15 s and 72°C for 40 s. The PCR products were diluted 500-fold and re-amplified with T2 and A2 nested primers (Table 1) in the second antibody hot started PCR round (25 cycles of 94°C for 20 s, 68°C for 15 s and 72°C for 40 s).
The human-specific PCR products were cloned into a T-easy vector using a PCR products T-easy cloning kit (Promega). Randomly selected clones were arrayed in 96-well microtiter dishes with 150 µl of Luria–Bertrani broth medium supplemented with 100 µg/ml ampicillin and grown overnight on a shaker. Of the bacterial culture, 0.5 µl was used as a template for PCR with M13 for/rev primers in 96-well PCR plates. Aliquots of the PCR products were spotted and immobilized on nylon membranes (Hybond N; Amersham) according to the manufacturer’s protocol. Two identical membranes were prepared for each plate of the arrayed library.
Probes for differential hybridization were prepared by PCR using A2 and T3 primers (25 cycles, touchdown). One of the two identical membranes was then hybridized with the probe derived from the human genomic DNA, and the other with the probe derived from the chimpanzee genomic DNA. After labeling by random priming, the probes were diluted, mixed with competitive oligonucleotides (20 pmol each), denatured at 100°C for 3 min, chilled on ice and added to the hybridization mix. Competitive oligonucleotides corresponding to the identical parts of the probes and DNA immobilized on membranes (see Results) were added to minimize the background caused by cross-hybridization between these parts. Dot blot hybridization was carried out at 68°C.
Positive clones were sequenced by the dye termination procedure using an Applied Biosystems 373 automatic DNA sequencer. Homology searches against the GenBank database and mapping were done using the BLAST Web-server at NCBI (http://www.ncbi.nlm.nih.gov/BLAST) and the Human Draft Genome Browser (http://genome.ucsc.edu/goldenPath/hgTracks.html). Genomic sequences were taken to design primers for the human LTR flanks. These primers were used for genomic PCR with different primate DNA samples to verify the human specificity of integrations.
RESULTS
The differences in the integration sites of low and medium copy number interspersed repeats (DiffIR) technique was applied to the identification of the human endogenous retrovirus, K family (HERV-K) (HML-2) LTRs integration sites, which differ in the human and chimpanzee genomes. This family of LTRs comprises about 2000 members scattered throughout the human genome (16). Several human-specific integrations of the REs were identified by us (15,17) and others (16,18), suggesting that the family remained transpositionally active until recently, at least in the common ancestors of human and chimpanzee (16). However, before now no attempts had been made to perform a genome-wide screening for HSIs, most probably due to the lack of efficient screening techniques. The method we developed can be successfully used for comparison of various interspersed repeats integration sites within closely related genomes. The method is based on selective amplification of LTR-flanking genomic fragments followed by a differential hybridization screening of an arrayed library of the selected amplicons.
Selective amplification of LTR-flanking genomic fragments
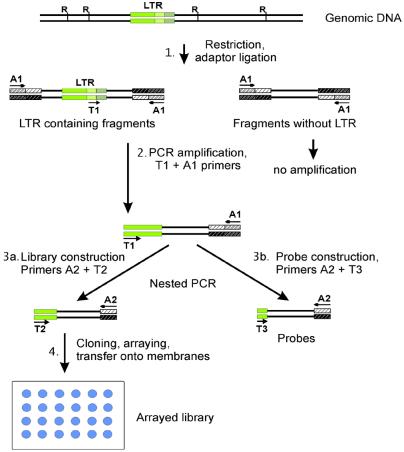
This stage, schematized in Figure 1, is needed to selectively simplify both the genomic DNAs being compared. Digestion of the genomic DNAs with a frequent cutter restriction enzyme (here AluI), and ligation of the restriction fragments to a pan-handle forming adapter (see Materials and Methods) results in the DNA restriction fragments tailed with inverted repeats at their termini (Stage 1). Therefore, the single-stranded fragments formed during the denaturation steps of PCR amplification are capable of forming stable intramolecular stem–loop structures (pan handle-like structures). PCR of the DNA fragments with such termini is suppressed when only one A1 primer targeted at the 5′ ends of the ligated adapter is used. In contrast, a pair of A1 primer and T1 primer targeted at the repeats within the single-stranded moiety of the stem–loop structure (19,20) can initiate DNA synthesis by DNA polymerase. The resulting amplified DNA will have different termini unable to form any stem–loop structures, and can be further efficiently amplified with A1+T1 primers (Stage 2).
Figure 1.
Stages of selective amplification of LTR flanking regions, and preparation of hybridization probes. Tentative positions of the restriction sites in the genomic DNA are indicated by Rs. LTRs are shown as green boxes. Positions of the LTR-targeted primers (T) and adapter primers (A) are indicated by arrows.
In the second (nested) round of PCR (Stage 3a and b) A2 and T2 primers were used to produce amplicons for the library construction (Stage 4), while A2 and T3 primers were taken to prepare hybridization probes. It should be remembered that the sequences of the T-primers correspond to the HERV-K LTR consensus sequence (15,21). The amplicons obtained were cloned and an arrayed library of human amplified LTR-flanking fragments was constructed.
2000 HERV-K (HML-2) LTR flanking fragments is a small enough number to allow the use of a reasonable size library for differential screening; about 6000 clones are necessary to detect a fragment with 95% probability. On the other hand, the complexity of the amplified fragments mixture is expected to be 250 (the average size of the fragments obtained due to AluI digestion) × 2000 = 5 × 105, that is ∼10% of the bacterial genome. The low complexity allows one to use this mixture directly as a hybridization probe for differential screening of the libraries to be compared. Even more complex probes such as the whole-genome bacterial DNAs are known to be successfully used for differential screening (22,23).
Therefore, we used human- and chimpanzee-specific amplicons as complex hybridization probes for differential screening of the human-specific library. The sequencing of 40 randomly chosen differential clones (see below) showed that all of them contained the expected U5 part of the LTR fragments. This result indicates that the specificity of the amplification (i.e. the numerical ratio of the LTR-containing amplification products to the total amplicons, both containing and lacking LTRs) is close to 100%. It is difficult to precisely estimate the amplification sensitivity, i.e. the portion of the total LTR flanks present in the genome that were actually amplified in our experiments. The sensitivity was evaluated in a special experiment. The genomic DNA purified from the hybrid cell line containing single human chromosome 21 was digested and amplified exactly as described above (see Materials and Methods). An analysis of the resulting amplicons by electrophoresis in a polyacrylamide gel revealed 14 fragments of different lengths. The number of the HERV-K (HML-2) LTRs on human chromosome 21 was previously estimated to be 16 (24). Therefore, the 14 fragments correspond to ∼87% sensitivity. The under-representation of the amplification products can be caused by many factors, in particular uneven distribution of the restriction nuclease recognition sites leading to inefficient amplification of longer fragments formed when the restriction site is far enough from the LTR. The sensitivity can be increased by using two restriction enzymes for the library and probes preparation.
Differential hybridization of the arrayed library of the human amplicons
The inserts of 864 clones of the arrayed library were PCR amplified with a universal pair of primers corresponding to the vector sequence. The PCR products were dotted in parallel on two sets of Hybond-N nylon membranes, one of which was hybridized with the human DNA-derived probe, and the other with the probe prepared from the chimpanzee DNA. The DNA fragments on the membranes and the fragments of hybridization probes contained the same sequences at their termini: 20 bp corresponding to A2 primer at the 3′ ends, and a stretch of ∼30 bp corresponding to the amplified LTR U5 region at their 5′ ends. The resulting unspecific hybridization was prevented by adding a 200-fold molar excess of unlabeled competitive double-stranded oligonucleotides.
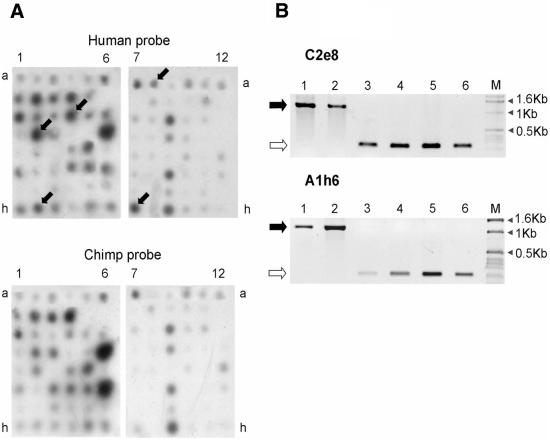
Figure 2A shows the results of the differential hybridization of the human and chimpanzee 32P-labeled probes with two sets of filters containing 864 dotted inserts of the arrayed library. In total, 40 DNA fragments that revealed strong hybridization signals with the human but not chimpanzee probe were identified. The corresponding clones were sequenced, and the sequences were then searched against human genome databases for homologs. Human specificity of the identified LTR insertions was further checked by phylogenetic analysis.
Figure 2.
(A) Examples of differential hybridization of the human arrayed library with the human (upper) and chimpanzee (lower) probes. Arrows indicate differential clones. (B) The results of PCR amplification with two individual LTR-containing loci in the human, chimpanzee and gorilla genomes. Black and white arrows to the left of the electropherograms indicate the predicted locations of the LTR-containing and LTR-free products, respectively. Lanes 1 and 2, amplification of genomic DNAs from two human individuals; lanes 3 and 4, chimpanzee (Pan troglodytes) genomic DNAs; lanes 5 and 6, gorilla (Gorilla gorilla) genomic DNAs; M, DNA fragments of a 1 kb ladder length marker (Gibco-BRL).
As mentioned above, all these 40 clones contained the corresponding LTR U5 part and an attached genomic region. Among all sequences of the clones, seven were shown by two clones each, and one sequence by three clones. Thus, 40 positive clones represented 31 individual sequences. Two LTR-flanking sequences had no homologs in the databases. The other 29 clones had highly homologous (97–100%; Table 2) genomic counterparts and were mapped within the human genome. The divergence as compared with the homologs in the GenBank database can be explained by single nucleotide polymorphism between different individuals or by sequencing errors. Another four clones could not be analyzed due to their integration into clusters of frequent low-divergence repeats, making the primer design for selective amplification of the corresponding genomic regions impossible. Seven sequences were reported previously: five of the LTRs were described as human specific (16,24–26), whereas two others were also detected in different hominoid genomes. The other 18 flanking sequences were phylogenetically analyzed to confirm their human specificity. For this purpose PCR primers corresponding to both 5′ and 3′ flanking sequences of each of the LTRs were designed. These primer pairs provided the amplification of either LTR-containing fragments or shorter fragments consisting of just the joined flanking sequences, depending on whether the LTR is present in the corresponding genome. Such pairs for each of the 18 loci were used in a PCR assay of the human and ape genomic DNAs (15,17). The amplification product of a locus with an LTR insert should be ∼1000 bp longer than that derived from an orthologous but LTR-free locus. Although we failed to obtain efficient PCR amplification for three loci, the analysis of the remaining 15 loci showed that 11 of them (Table 2) were human specific, whereas four were also found in the genomes of apes. In total, 16 of 22 analyzed LTRs were found to be human specific. Thus, the specificity of the differential hybridization was close to 73%. One of the reasons for false positive signals might be interspecies RFLP in the LTR-flanking regions due to less efficient PCR amplification of longer fragments. Another reason could be inefficient priming of some of the chimpanzee LTRs due to the mutations hampering or preventing human-specific primer binding. The results demonstrate that to achieve the most complete representation of differences between two genomes one has to take into account their divergence and use various sets of primers and restriction endonucleases.
Table 2. New LTR HSIs identified.
| Clone |
Accession numbera |
Identity (%)b |
Chromosome |
Subfamilyc |
|
|---|---|---|---|---|---|
| Identified HSIs | Homologs | ||||
| A1h6 | AF493416 | AC091895 | 98 | 5 | II-L4 |
| A3c4 | AF493417 | AL162723 | 97 | X | II-L2 |
| A4a12 | AF493418 | AC015525 | 98 | 3 | II-L4 |
| A4c1 | AF493419 | AC074019 | 99 | 2 | II-T |
| A4d1 | AF493420 | AL359644 | 99 | 9 | II-L4 |
| C2d10 | AF493421 | AC009132 | 97 | 16 | II-L4 |
| C2e8 | AF493422 | AC018539 | 98 | 11 | II-L4 |
| G1d12 | AF493423 | AC019155 | 99 | 7 | II-L4 |
| G1f5 | AF493424 | AC006035 | 99 | 7 | II-L4 |
| G2h11 | AF493425 | AP002513 | 98 | 11 | II-L4 |
| A1c10 | AF493426 | AL596188 | 100 | 6 | II-L4 |
aAccession numbers for the sequences found in the present work and submitted to GenBank are listed in the first column; accession numbers for homologs found in the GenBank database and used for primer design are listed in the second column.
bPercent identity between sequences found in the present work and the corresponding homologs.
cAccording to the classification reported in Lebedev et al. (15).
DISCUSSION
Eleven new human-specific LTR integrations were found in this work (Table 2) and 40 LTR HSIs have been revealed previously, whereas the total number of LTR HSIs in the human genome was estimated to be ∼70 (26). Although this number is most probably an underestimate, it seems unlikely that the real number exceeds 100. Analysis of more representative libraries could certainly reveal more HSIs.
The DiffIR technique described here can be used to compare the distribution of the integration sites of any low and medium copy number repetitive elements in related genomes. Such a comparison will be useful for analysis of integration peculiarities of certain subgroups of the important human genome constituents, such as HERVs, SINEs and LINEs. Although the latter two families of retroposons are extremely abundant in the human genome and represented by about 1.6 × 106 and 8.7 × 105 individual elements (1), respectively, each family consists of many less abundant subfamilies amplified during various evolutionary periods. Each subfamily can be analyzed separately with the DiffIR technique. The most recent subfamilies of L1 and Alu repeats are comprised of tens to hundreds of members (10,11). These youngest retrotransposons are of special interest because some of their insertions are still not fixed in the human species and provide useful polymorphic loci that allow identification of complex rearrangements that occurred during the evolution. The DiffIR technique is also applicable to any family of repeats providing that their number does not exceed several thousand copies per haploid genome. These might include conservative gene families, such as zinc finger encoding genes, certain families of simple tandem repeats, e.g. triple nucleotide repeats involved in some hereditary diseases, as well as evolutionarily young segmental genomic duplications characterized by high sequence identity and different distribution in related primate genomes (1). The technique can also be used for the detection of insertion/deletion polymorphisms in the human genome.
The specificity of the technique when applied to the human and chimpanzee genomes was close to 100%, at rather low (∼90%) sensitivity however. The low sensitivity is most probably explained by the divergence of the human and chimpanzee genomes. The sensitivity is supposed to be increased by using two restriction endonucleases that should reduce the effect of the restriction site polymorphism. Also, the negative effect of the divergence in the primer target sites can be reduced by using additional PCR cycles. We believe that this problem will be much less important when comparing individual human genomes because their identity is much higher than between the human and any non-human genomes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Drs Potapov and Skaptsova for oligonucleotide synthesis and Dr Glotov for helpful discussion. The research was partially supported by RFBR 02-04-48614 and INTAS-01-0759 grants.
DDBJ/EMBL/GenBank accession nos AF493416–AF493426
REFERENCES
- 1.International, Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Schwahn U., Lenzner,S., Dong,J., Feil,S., Hinzmann,B., van Duijnhoven,G., Kirschner,R., Hemberger,M., Bergen,A.A., Rosenberg,T. et al. (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nature Genet., 19, 327–332. [DOI] [PubMed] [Google Scholar]
- 3.Meischl C., Boer,M., Ahlin,A. and Roos,D. (2000) A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet., 8, 697–703. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian H.H. Jr, (2000) L1 retrotransposons shape the mammalian genome. Science, 289, 1152–1153. [DOI] [PubMed] [Google Scholar]
- 5.Brooks E.M., Branda,R.F., Nicklas,J.A. and O’Neill,J.P. (2001) Molecular description of three macro-deletions and an Alu-Alu recombination-mediated duplication in the HPRT gene in four patients with Lesch-Nyhan disease. Mutat. Res., 476, 43–54. [DOI] [PubMed] [Google Scholar]
- 6.Saikawa Y., Kaneda,H., Yue,L., Shimura,S., Toma,T., Kasahara,Y., Yachie,A. and Koizumi,S. (2000) Structural evidence of genomic exon-deletion mediated by Alu-Alu recombination in a human case with heme oxygenase-1 deficiency. Hum. Mutat., 16, 178–179. [DOI] [PubMed] [Google Scholar]
- 7.Lower R., (1999) The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol., 7, 350–356. [DOI] [PubMed] [Google Scholar]
- 8.Deininger P.L., and Batzer,M.A. (1999) Alu repeats and human disease. Mol. Genet. Metab., 67, 183–193. [DOI] [PubMed] [Google Scholar]
- 9.Moran J.V., (1999) Murine A type retroviruses promote high levels of gene expression in embryonal carcinoma cells. Genetica, 107, 39–51. [DOI] [PubMed] [Google Scholar]
- 10.Carroll M.L., Roy-Engel,A.M., Nguyen,S.V., Salem,A.H., Vogel,E., Vincent,B., Myers,J., Ahmad,Z., Nguyen,L., Sammarco,M. et al. (2001) Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol., 311, 17–40. [DOI] [PubMed] [Google Scholar]
- 11.Sheen F.M., Sherry,S.T., Risch,G.M., Robichaux,M., Nasidze,I., Stoneking,M., Batzer,M.A. and Swergold,G.D. (2000) Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res., 10, 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boissinot S., Chevret,P. and Furano,A.V. (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol., 17, 915–928. [DOI] [PubMed] [Google Scholar]
- 13.Roy A.M., Carroll,M.L., Kass,D.H., Nguyen,S.V., Salem,A.H., Batzer,M.A. and Deininger,P.L. (1999) Recently integrated human Alu repeats: finding needles in the haystack. Genetica, 107, 149–161. [PubMed] [Google Scholar]
- 14.Diatchenko L., Lau,Y.F., Campbell,A.P., Chenchik,A., Moqadam,F., Huang,B., Lukyanov,S., Lukyanov,K., Gurskaya,N., Sverdlov,E.D. et al. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebedev Y., Belonovich,O., Zybrova,N., Khil,P., Kurdyukov,S., Vinogradova,T., Hunsmann,G. and Sverdlov,E. (2000) Differences in HERV-K LTR insertions in orthologous loci of human and great apes. Gene, 247, 265–277. [DOI] [PubMed] [Google Scholar]
- 16.Medstrand P., and Mager,D.L. (1998) Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol., 72, 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sverdlov E.D., (2000) Retroviruses and primate evolution. Bioessays, 22, 161–171. [DOI] [PubMed] [Google Scholar]
- 18.Turner G., Barbulescu,M., Su,M., Jensen-Seaman,M.I., Kidd,K.K. and Lenz,J. (2001) Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol., 11, 1531–1535. [DOI] [PubMed] [Google Scholar]
- 19.Lavrentieva I., Broude,N.E., Lebedev,Y., Gottesman,I.I., Lukyanov,S.A., Smith,C.L. and Sverdlov,E.D. (1999) High polymorphism level of genomic sequences flanking insertion sites of human endogenous retroviral long terminal repeats. FEBS Lett., 443, 341–347. [DOI] [PubMed] [Google Scholar]
- 20.Diatchenko L., Lukyanov,S., Lau,Y.F. and Siebert,P.D. (1999) Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol., 303, 349–380. [DOI] [PubMed] [Google Scholar]
- 21.Lavrentieva I., Khil,P., Vinogradova,T., Akhmedov,A., Lapuk,A., Shakhova,O., Lebedev,Y., Monastyrskaya,G. and Sverdlov,E.D. (1998) Subfamilies and nearest-neighbour dendrogram for the LTRs of human endogenous retroviruses HERV-K mapped on human chromosome 19: physical neighbourhood does not correlate with identity level. Hum. Genet., 102, 107–116. [DOI] [PubMed] [Google Scholar]
- 22.Bogush M.L., Velikodvorskaya,T.V., Lebedev,Y.B., Nikolaev,L.G., Lukyanov,S.A., Fradkov,A.F., Pliyev,B.K., Boichenko,M.N., Usatova,G.N., Vorobiev,A.A., Andersen,G.L. and Sverdlov,E.D. (1999) Identification and localization of differences between Escherichia coli and Salmonella typhimurium genomes by suppressive subtractive hybridization. Mol. Gen. Genet., 262, 721–729. [DOI] [PubMed] [Google Scholar]
- 23.Liang X., Pham,X.Q., Olson,M.V. and Lory,S. (2001) Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol., 183, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurdyukov S.G., Lebedev,Y.B., Artamonova,I.I., Gorodentseva,T.N., Batrak,A.V., Mamedov,I.Z., Azhikina,T.L., Legchilina,S.P., Efimenko,I.G., Gardiner,K. and Sverdlov,E.D. (2001) Full-sized HERV-K (HML-2) human endogenous retroviral LTR sequences on human chromosome 21: map locations and evolutionary history. Gene, 273, 51–61. [DOI] [PubMed] [Google Scholar]
- 25.Barbulescu M., Turner,G., Seaman,M.I., Deinard,A.S., Kidd,K.K. and Lenz,J. (1999) Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol., 9, 861–868. [DOI] [PubMed] [Google Scholar]
- 26.Buzdin A., Khodosevich,K., Mamedov,I., Vinogradova,T., Lebedev,Y., Hunsmann,G. and Sverdlov,E. (2002) A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics, 79, 413–422. [DOI] [PubMed] [Google Scholar]