Abstract
A system of photoaffinity reagents for selective labeling of DNA polymerases in extracts has been examined. To create the photoreactive DNA probe in situ, DNA substrates containing a synthetic abasic site are incubated in mouse embryonic fibroblast (MEF) cellular extract in the presence of base-substituted arylazido derivatives of dNTPs. This results in synthesis of a photoreactive long patch base excision repair (BER) intermediate. The arylazido photoreactive group is then activated through energy transfer from the pyrene group of a dNTP analog (Pyr-dUTP), following 365 nm UV light exposure. Pyr-dUTP binds to the active site of DNA polymerases, and the pyrene group, when excited by 365 nm UV light, activates the nearby photoreactive group in the BER intermediate resulting in crosslinking of DNA-bound DNA polymerases. Under these conditions, various DNA binding proteins that are unable to bind Pyr-dUTP are not crosslinked to DNA. DNA polymerase β is the predominant crosslinked protein observed in the MEF extract. In contrast, several other DNA binding proteins are labeled under conditions of direct UV light activation of the photoreactive group at 312 nm. This study illustrates use of a new method of selective labeling of DNA polymerases in a crude cellular extract.
INTRODUCTION
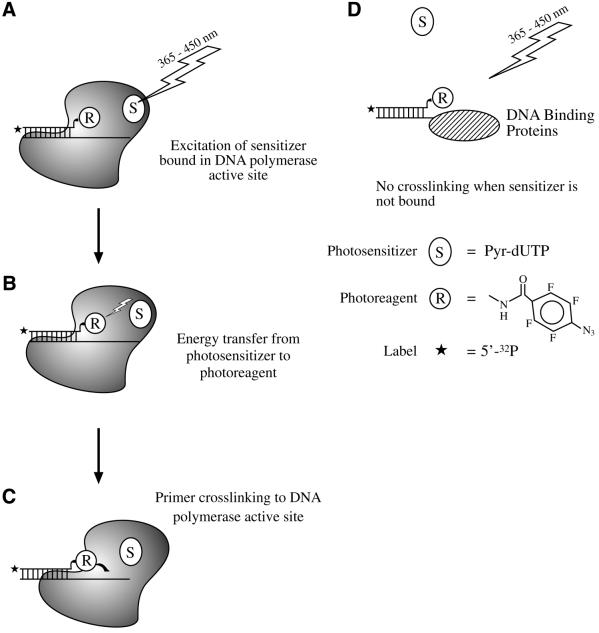
Photoaffinity labeling is a strategy for the study of macromolecular interactions in biochemical systems (1–3). We recently applied this approach to select base excision repair (BER) proteins from a crude cellular extract (4). We used a DNA photoaffinity probe formed in situ in mouse embryonic fibroblast (MEF) cellular extracts. With 312 nm UV light exposure of the extract-probe mixture, only six proteins were strongly labeled. Four of these proteins were identified as poly(ADP-ribose) polymerase-1 (PARP-1), flap endonuclease-1 (FEN-1), DNA polymerase β (β-pol) and apurinic/apyrimidinic endonuclease (APE) (4 and unpublished results). Yet, identification of proteins crosslinked to DNA in a cellular extract remains challenging. The amount of crosslinked product is often insufficient for identification by direct sequencing or mass spectroscopy. Therefore, protein identification mainly relies upon the use of specific antibodies. An alternative approach that could verify the nature of a crosslinked protein would be helpful, and one such alternative approach is through selective affinity labeling of proteins (5). Recently, a binary system was developed and used for selective photoaffinity labeling of DNA polymerases in a reconstituted system (6). The principle behind this technique is as follows. A radioactively labeled primer is extended by a DNA polymerase incorporating a dNMP bearing the arylazido group into the primer. This extended primer is capable of binding at the polymerase’s template-primer binding site. A base-substituted analog of dTTP bearing a pyrene group, 5-{N-[4-(1-pyrenyl)butylcarbonyl]-amino-trans-propenyl-1}-2′-deoxyuridine-5′-triphosphate (Pyr-dUTP), serves as photosensitizer. Pyr-dUTP binds to the enzyme’s dNTP-binding site, which is in close proximity to the template-primer binding site. The reaction conditions and template structure prohibit incorporation of the photosensitizer at the 3′ end of the primer. Next, if the reaction mixture is irradiated with 365 nm UV light, photoenergy is absorbed by the pyrene moiety of the photosensitizer and transferred to the nearby photoreagent, i.e. the arylazido group in the primer (Scheme 1A and B). The arylazido group does not directly absorb UV light energy at this wavelength. However, as a result of energy transfer from the excited pyrene, the arylazido group undergoes photodecomposition and forms a crosslink with the protein in or near the active site (Scheme 1C). The initial rate of photomodification and crosslinking is extremely sensitive to the distance between the sensitizer and photoreactive group because the efficiency of fluorescence resonance energy transfer is a function of the inverse sixth power of this distance. DNA binding proteins are crosslinked to the photoreactive template-primer DNA only if they have a Pyr-dUTP binding site that is close to the photoreactive primer; thus, DNA binding proteins that interact with DNA, but not with Pyr-dUTP, will not undergo crosslinking (Scheme 1D). It was anticipated that this approach could be useful for selective labeling of DNA polymerases in multicomponent systems, such as cellular extracts.
Scheme 1. The model in (A) illustrates a polymerase (solid color) bound to a template-primer complex. The template, which is longer than the primer, has a single-stranded region. The 3′ end of the primer contains the photoreagent (R) covalently attached to the terminal base. The diagram also illustrates a photosensitizer (S) bound in the polymerase active site. Excitation of the photosensitizer with 365 nm UV light results in activation of the photoreagent, as shown in (B), and crosslinking to the polymerase (C). A DNA binding protein unable to bind the photosensitizer is not crosslinked by 365 nm UV light exposure (D). The 5′-32P radiolabel is illustrated by an asterisk.
BER is the major repair pathway for non-bulky lesions involving DNA bases, including the abasic or apurinic/apyrimidinic (AP) site arising through base loss. AP sites are one of the most common DNA lesions and they accumulate due to chemical base loss or DNA glycosylase action. Recent studies in higher eukaryotic cells indicate that AP sites are repaired by two alternative pathways: short patch or single-nucleotide BER, and long patch BER. The first step in each pathway is the incision of the phosphodiester backbone immediately 5′ to the AP site. This step is carried out by APE, and generates DNA ends with a 3′-hydroxyl and 5′-deoxyribose phosphate (dRP). In single-nucleotide BER, β-pol fills the single-nucleotide gap and excises the 5′-dRP group. In long patch BER, β-pol or another DNA polymerase (7) carries out limited strand displacement DNA synthesis. The displaced strand is incised by the structure-specific endonuclease FEN-1, and DNA ligase I or the DNA ligase III–x-ray cross complementing factor 1 (XRCC1) complex seals the phosphodiester backbone to complete the repair reaction.
This study uses selective labeling of DNA polymerases in mammalian cellular extracts to identify enzymes potentially involved in mammalian BER. A photoreactive BER intermediate was created in situ in a cellular extract. The endogenous DNA polymerases were challenged with the BER intermediate, a dNTP analog with a pyrene moiety as a photosensitizer, and lower energy UV light (365 nm). β-Pol was the predominant crosslinked extract protein observed using this ‘binary photolabeling system’; in contrast, several other proteins were crosslinked when the photosensitizer was omitted and higher energy UV light was used to activate the BER intermediate (4). These results indicate that this binary photolabeling system, making use of photoreactive BER intermediates and a photosensitive dNTP analog, is useful for selective labeling of BER DNA polymerases in a cellular extract.
MATERIALS AND METHODS
Materials
Rainbow colored molecular mass markers and [γ-32P]ATP were purchased from Amersham Biosciences. Synthetic oligodeoxyribonucleotides were obtained from Oligo Etc. (Wilsonville, OR). The photoreactive dCTP analog, exo- N-[β-(p-azidotetrafluorobenzamido)-ethyl]-2′-deoxycytidine-5′-triphosphate (FAB-dCTP), was synthesized as described by Wlassoff et al. (8). Synthesis of 5-{N-[4-(1-pyrenyl) butylcarbonyl]-amino-trans-propenyl-1}-2′-deoxyuridine-5′-triphosphate (Pyr-dUTP), 5-[N-(2,3,5,6-tetrafluoro-4-azidobenzoyl)-amino-trans-propenyl-1]-2′-deoxyuridine-5′-triphosphate (FAB-4-dUTP) and 5-{N-[N′-(2,3,5,6-tetrafluoro-4-azidobenzoyl)-4-aminobutyryl]-amino-trans-propenyl-1}-2′- deoxyuridine-5′-triphosphate (FAB-9-dUTP) was carried out as described previously (6,9,10). 5-[N-(2,3,5,6-tetrafluoro-4-azidobenzoyl)-amino-trans-propenyl-1]-2′,3′-dideoxyuridine-5′-triphosphate (lithium salt) (FAB-4-ddUTP) was synthesized according to a procedure described for the synthesis of a 5-substituted ddUTP derivative (11). Human β-pol, FEN-1 and APE were purified as described previously (12–14). FLAG-antibody specific agarose was purchased from Sigma (St Louis, MO).
Cells and extracts
Cells expressing a FLAG-epitope tagged β-pol were grown as described previously (15). Cellular extracts were prepared as described previously (4).
Radioactive labeling of oligonucleotide primers
Dephosphorylated primers were 5′-[32P]-phosphorylated with T4 polynucleotide kinase as described by Sambrook et al. (16). Unreacted [γ-32P]ATP was removed by passing the mixture over a Nensorb-20 column using the manufacturer’s protocol.
Primer-template annealing
Lyophilized oligonucleotides were resuspended in 10 mM Tris–HCl pH 7.8 and 1 mM EDTA. Primer-templates were annealed by heating the primer with equimolar template at 90°C for 3 min followed by slow cooling to room temperature.
Photoreactive DNA synthesis
The reaction mixture (10 µl) contained 50 mM Tris–HCl, pH 7.8, 50 mM KCl, 10 mM MgCl2, 4 µl cellular extract prepared from β-pol null MEF cells expressing a FLAG-epitope tagged β-pol (4). The protein concentration of the cellular extract was 4–5 mg/ml. The reaction mixture (10 µl) reconstituted with purified BER proteins that contained 50 mM Tris–HCl, pH 7.8, 10 mM MgCl2, 50 mM KCl, 1 µM FEN-1, 1 µM β-pol and 0.7 µM APE. To produce photoreactive DNA2 or DNA5, 0.2 µM [32P]-labeled DNA1 or DNA4, respectively, were incubated with 10 µM FAB-dCTP for 30 min at 25°C in a standard reaction mixture. DNA4 has the same sequence as DNA1 except that it has a 4-nt flap in place of tetrahydrafuran at the nick. To produce photoreactive DNA3, 0.2 µM DNA1, 10 µM dCTP, 10 µM [α-32P]dGTP and 10 µM of one photoreactive dTTP analog (FAB-4-dUTP, FAB-9-dUTP or FAB-4-ddUTP) were added to the reaction mixture and incubated for 30 min (or the indicated time) at 25°C. The reaction was terminated by adding 10 µl 90% formamide, 50 mM EDTA, 0.1% bromophenol blue and xylene cyanol. The mixtures were heated for 3 min at 80°C and products were separated in 15% polyacryamide gel containing 8 M urea in 89 mM Tris–HCl, 89 mM boric acid and 2 mM EDTA, pH 8.8, and visualized by autoradiography.
Photochemical crosslinking
Reaction mixtures were as described above. After preincubation for 30 min at 25°C, the reaction mixtures were spotted onto Parafilm on ice. The samples were irradiated with UV light λmax = 312 nm (5–7 mJ) (‘direct’ crosslinking) or with UV light λmax = 365 nm (3 mJ) in the presence of 10–40 µM Pyr-dUTP. The UV light source was an UV-Stratalinker (Stratagene Cloning Systems). The control reaction mixture did not contain Pyr-dUTP. The photochemically crosslinked proteins were separated by 10% SDS–PAGE; dried gels were subjected to autoradiography.
Immunoprecipitation
Immunoprecipitation was as described previously (4) using M2-FLAG antibody conjugated to agarose. Briefly, the photoaffinity-labeled reaction mixture (200 µl) was mixed with 5 µg anti-FLAG M2 antibody conjugated to agarose, incubated with rotation for 4 h at 4°C and pelleted by centrifugation at 14 000 r.p.m. (15 s). The supernatant fraction was discarded and the protein-bound agarose complex was washed four times with extract buffer. After a final wash, the buffer was removed and the protein agarose suspension was mixed with 50 µl SDS-gel loading buffer and incubated in a boiling water bath for 5 min.
RESULTS
Synthesis of photoreactive BER intermediates
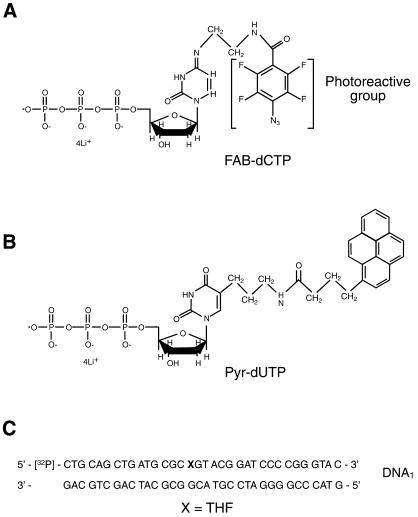
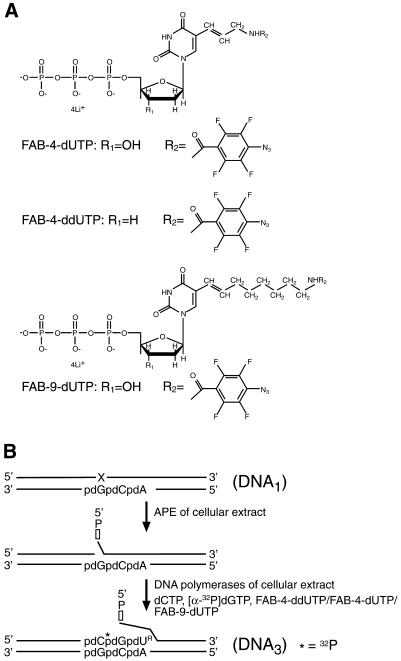
The structures of the photoreagents and dNTP analogs used in this study are shown in Figures 1 and 2. The photoreactive group was 2,3,5,6-tetrafluoro-4-azidobenzoyl (FAB) and the photoreactive dNTP analogs used were FAB-dCTP (Fig. 1A), FAB-4-dUTP, FAB-4-ddUTP and FAB-9-dUTP (Fig. 2A). The FAB group forms a crosslink with several protein side chain groups when exposed to UV irradiation (17) and can be activated by resonance energy or electron transfer from an excited pyrene residue, i.e. Pyr-dUTP (Fig. 1B), if the distance between the two groups is small (18). Any importance of size of the linker between the DNA base and the photoreactive group was tested to determine whether this affects the specificity of protein labeling. Results were compared using FAB-9-dUTP and FAB-4-dUTP. FAB-4-ddUTP was also used to introduce the photoreactive group at the 3′-end of the nascent DNA. This prevented elongation of 3′-photoreactive group-containing primers by endogeneous dNTPs in the cellular extract.
Figure 1.
Structures of photoreactive dNTP analogs and sequence of DNA1. (A) Structure of FAB-dCTP. (B) Structure of Pyr-dUTP. (C) Sequence of DNA1, with X representing the synthetic abasic site with tetrahydrofuran (THF).
Figure 2.
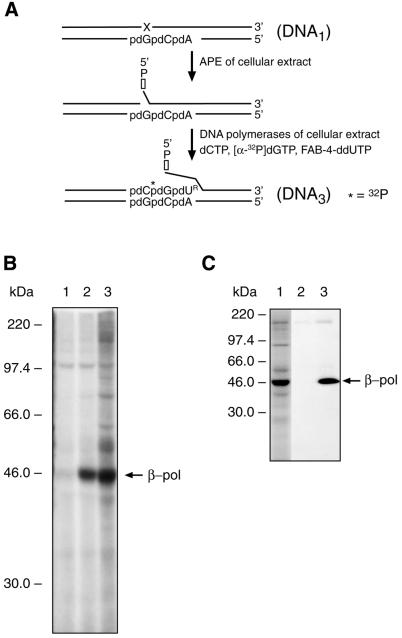
Structures of FAB-dUTP analogs and synthesis of photoreactive DNA3. (A) Structures of three FAB-dUTP analogs. (B) Schematic representation of photoreactive DNA3 synthesis in situ. The synthetic abasic site (X) in DNA1 is incised by APE; DNA polymerase of the MEF extract incorporates dCMP, [α-32P]dGMP or one of the photoreactive dUMP analogs (FAB-4-dUMP, FAB-4-ddUMP or FAB-9-dUMP) into the DNA. DNA3 is 32P-radiolabeled by incorporating dGMP (asterisk) and contained a 3′-end photoreactive dUMP (dUR) residue.
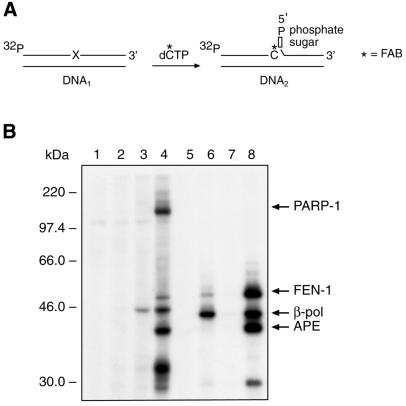
Figure 1C shows the structure of DNA1 containing a synthetic abasic site. This substrate can be repaired by long patch BER. Radiolabeled DNA1 was incubated in a MEF extract in the presence of FAB-dCTP. According to the scheme depicted in Figure 3A, one FAB-dCMP residue is introduced into DNA1 to produce DNA2. This was verified as shown in Figure 3B, lane 2. In the absence of FAB-dCTP, a product was detected with dCMP incorporated at the 3′ end of the primer (Fig. 3B, lane 1); this product was likely to have been formed using endogeneous dCTP, and had the same mobility as a product formed by incorporation of exogenous dCTP (Fig. 2B, lane 4). Addition of the photosensitizer, Pyr-dUTP, did not influence primer elongation in this reaction (Fig. 3B, lane 3).
Figure 3.
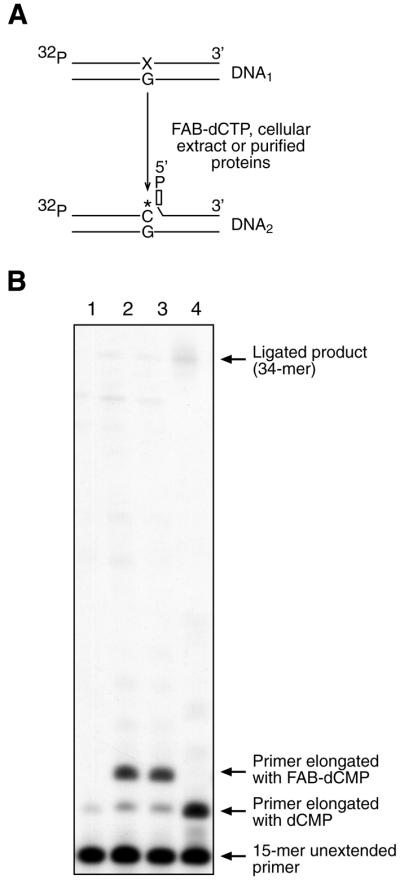
Synthesis of photoreactive DNA probe. (A) A scheme illustrating in situ conversion of radiolabeled DNA1 to the nicked DNA2 containing FAB-dCMP and deoxyribose phosphate at 3′ and 5′ margins, respectively, of the nick. (B) Photograph of an autoradiogram of a DNA sequencing gel showing primer extension with FAB-dCMP and dCMP by the MEF extract. DNA1 was incubated for 30 min at 25°C with MEF extract and products were analyzed as described in the Materials and Methods. Lane 1, DNA1 was incubated with MEF extract without FAB-dCTP; lane 2, DNA1 was incubated with MEF extract and FAB-dCTP; lane 3, DNA1 was incubated with MEF extract, FAB-dCTP and Pyr-dUTP; lane 4, DNA1 was incubated with MEF extract and dCTP (10 µM). The positions of the primer and the products are indicated.
DNA3 was prepared by incubating DNA1 with cellular extract in the presence of dCTP, [α-32P]dGTP and one of the photoreactive dTTP analogs (i.e. FAB-4-dUTP, FAB-9-dUTP or FAB-4-ddUTP). According to the scheme depicted in Figure 2B, the primer was elongated in the cellular extract, and dCMP, [α-32P]dGMP and one of the photoreactive dTMP analogs were incorporated at the 3′ end of the primer (data not shown). In the absence of a photoreactive analog, the major product of DNA synthesis was a 17mer oligonucleotide that corresponded to incorporation of two nucleotide residues. DNA synthesis of the 18mer extended primer, (i.e. DNA3) was not delayed when FAB-4-dUTP was replaced with FAB-4-ddUTP, suggesting that the endogeneous DNA polymerase utilized the ddNTP analog (data not shown). These results are consistent with the possibility that β-pol was the extract DNA polymerase detected by this assay, since β-pol has similar catalytic efficiency with dNTPs and ddNTPs (19).
Photocrosslinking of BER proteins in the mammalian cellular extract
Activation of the photoreactive group of DNA2 by 312 nm UV light caused several proteins in the MEF cellular extract to be crosslinked to the radiolabeled DNA (Fig. 4B, lane 4) (4). However, when the reaction mixture was supplemented with Pyr-dUTP and irradiated with 365 nm UV light, β-pol was the primary labeled protein (Fig. 4B, lane 3), and no protein labeling was observed in the absence of Pyr-dUTP with exposure to UV light at 365 nm (Fig. 4B, lane 2). These results indicate that Pyr-dUTP strongly enhances specificity of crosslinking of β-pol to the photoreactive BER intermediate, DNA2. Immunoprecipitation with antibodies against the FLAG-β-pol in the extract confirmed the identity of the labeled protein in lane 3, with electrophoretic mobility corresponding to ∼46 kDa (4).
Figure 4.
Photoaffinity labeling of BER proteins in MEF extract by a binary system of photoaffinity reagents, DNA2 and Pyr-dUTP. (A) A scheme illustrating synthesis of the BER intermediate DNA2 in cellular extract, as described in the Materials and Methods. (B) The reaction mixtures were incubated with DNA1 and then irradiated with 365 nm UV light. Absence of FAB-dCTP and Pyr-dUTP (lane 1); presence of FAB-dCTP (lane 2); presence of FAB-dCTP and Pyr-dUTP (lane 3). The reaction mixture was incubated with DNA1 and irradiated with 312 nm UV light: FAB-dCTP without Pyr-dUTP (lane 4). BER reaction was reconstituted with DNA1 and purified BER proteins, FEN-1, β-pol and APE (lanes 5–8). After incubation for 30 min at 25°C, the reaction mixture was irradiated with 365 nm UV light (lanes 5 and 6) or 312 nm UV light (lanes 7 and 8). Reaction was performed without Pyr-dUTP (lanes 5 and 7) or with Pyr-dUTP (lanes 6 and 8). The reaction products were separated by 10% SDS–PAGE followed by autoradiography. The positions of protein markers and crosslinked proteins are indicated on the left and right margins, respectively.
Similar experiments were peformed with purified BER proteins, β-pol, APE and FEN-1. For reference, when DNA2 was activated with 312 nm UV light, these three proteins were strongly radiolabeled (Fig. 4B, lane 8). When the reaction mixture was irradiated with 365 nm UV light in the presence of Pyr-dUTP, β-pol was the primary labeled protein along with minor labeling of FEN-1 (Fig. 4B, lane 6). This labeling was not observed in the absence of Pyr-dUTP (Fig. 4B, lane 5). These experiments indicate that β-pol was selectively crosslinked by photoreactive DNA2 in the MEF cellular extract or in the presence of purified BER proteins using the binary photoaffinity labeling system described.
Similar experiments were performed with photoreactive DNA3, a long patch BER intermediate (Fig. 5). The scheme by which this BER intermediate was prepared is shown in Figure 5A. In the presence of Pyr-dUTP, only β-pol was labeled (Fig. 5B, lane 2). However, in the absence of Pyr-dUTP and with UV light irradiation at 312 nm, many additional proteins in the extract were labeled (Fig. 5B, lane 3). One of the labeled proteins, i.e. migrating at ∼98 kDa, was observed without irradiation (Fig. 5B, lane 1) and was considered non-specific. Finally, similar results were obtained when FAB-9-dUTP was used, i.e. where the spacer carrying the photoreactive arylazido group was longer (data not shown). Immunoprecipitation experiments confirmed that the major labeled protein observed in the experiment (Fig. 5B, lane 2) was indeed FLAG-β-pol (Fig. 5C).
Figure 5.
Photoaffinity labeling of proteins in MEF extract using photoreactive long patch BER intermediate DNA3. (A) Photoreactive and radiolabeled DNA3 was produced in the MEF extract using DNA1 and FAB-4-ddUTP, as depicted in the scheme. (B) The reaction mixture containing FAB-4-ddUTP was irradiated with 365 nm UV light either without Pyr-dUTP (lane 1) or with Pyr-dUTP (lane 2); the reaction mixture in lane 3 was irradiated with 312 nm UV light after incubation with FAB-4-ddUTP and without Pyr-dUTP. (C) Immunoprecipitation of crosslinked products was performed with reaction mixture in lane 3 (B). Starting material (lane 1); reaction mixture immunoprecipitated with preimmune IgG (lane 2) or anti-FLAG IgG (lane 3). The UV-crosslinked products were separated by SDS–PAGE and visualized by autoradiography. The positions of crosslinked product and protein markers are indicated on the right and the left margins, respectively.
Another photoreactive long patch BER intermediate containing a flap structure was evaluated. The scheme by which this BER intermediate, DNA5, was prepared is shown in Figure 6A. β-Pol was the predominant DNA crosslinked protein in the MEF extract exposed to 365 nm UV light in the presence of Pyr-dUTP (Fig. 6B, lane 1); the crosslinked product at ∼98 kDa was considered non-specific, in view of the results in Figure 6B, lanes 2, 3 and 5. However, the labeling pattern with 312 nm UV light irradiation, without Pyr-dUTP (Fig. 6B, lane 4), was also selective for β-pol and was quite different from the pattern seen under these same conditions using DNA2 (compare Fig. 4, lane 4 with Fig. 6, lane 4). These results suggest that different proteins were involved in processing of these distinct BER intermediates, i.e. DNA2 and DNA5.
Figure 6.
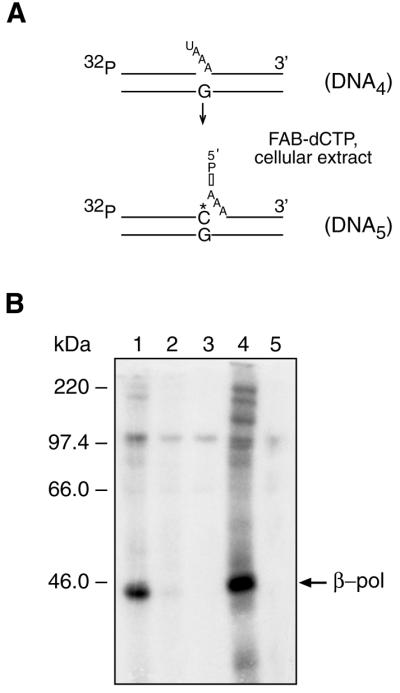
Photoaffinity labeling of proteins in MEF extract using photoreactive long patch BER intermediate DNA5. (A) Radiolabeled and photoreactive DNA5 was synthesized in the cellular extract starting with DNA4, as depicted in the scheme; the asterisk represents the position of FAB-dCMP. (B) After incubation, reaction mixtures were irradiated with UV light at 365 nm (lanes 1–3) or at 312 nm (lanes 4 and 5). Incubation was with (lane 1) and without Pyr-dUTP (lane 2), or without FAB-dCTP and Pyr-dUTP (lane 3). Reaction mixtures in lanes 4 and 5 were incubated with FAB-dCTP or without FAB-dCTP, respectively. UV-crosslinked products were separated by SDS–PAGE and visualized by autoradiography. The positions of crosslinked products and protein markers are indicated on the right and left margins, respectively.
DISCUSSION
X-ray crystallography, nuclear magnetic resonance and other physical biochemical methods can be highly informative concerning a protein’s structure and function. These methods, however, are generally restricted to highly purified samples; thus, it is difficult to apply these techniques to multicomponent reconstituted systems or to complex cellular extracts. Affinity labeling has been a useful method for studying DNA replication proteins (20–23) and DNA repair proteins (4) in nuclear and cellular extracts. This approach is also advantageous for analysis of interactions between DNA and proteins in cellular extracts. Approaches that increase the selectivity of such affinity labeling methods could eventually lead to broader use of affinity labeling in multicomponent systems.
Previous studies demonstrated that the selectivity of affinity labeling can be increased by differential labeling (24), catalytically competent labeling (25) or protein mediated energy transfer photolabeling (26). The data presented here show that a binary system for photoaffinity labeling, involving the combination of 365 nm UV light, BER intermediates with a photoreactive arylazido groups and the sensitizer Pyr-dUTP, can be highly selective for labeling of DNA polymerase in the MEF cellular extract. Thus, in MEF extracts, and with a labeling system based on BER intermediates, β-pol was the main crosslinking target. Selective β-pol labeling was probably a consequence of both binding of the Pyr-dUTP sensitizer to the polymerase’s dNTP-binding pocket and the polymerase’s affinity for the BER intermediate used. Other DNA binding proteins, such as PARP-1 and APE, were not crosslinked under these conditions, even though these proteins can interact with the photoreactive BER intermediate, DNA2, and are photocrosslinked when exposed to 312 nm UV light in the absence of the photosensitizer (Fig. 4, lane 4) (4).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Igor Safronov for providing FAB-dCTP, Dr Miriam Sander for assistance with the manuscript and Ms Jennifer Myers for editing the manuscript. This work was supported in part by NATO Collaborative Linkage Grant N978233 and by the grants of Russian Funds for Basic Research 02-04-48404 and 00-04-49309.
REFERENCES
- 1.Brunner J., (1993) New photolabeling and crosslinking methods. Annu. Rev. Biochem., 62, 483–514. [DOI] [PubMed] [Google Scholar]
- 2.Knorre D.G., and Godovikova,T.S. (1998) Photoaffinity labeling as an approach to study supramolecular nucleoprotein complexes. FEBS Lett., 433, 9–14. [DOI] [PubMed] [Google Scholar]
- 3.Meisenheimer K.M., and Koch,T.H. (1997) Photocross-linking of nucleic acids to associated proteins. Crit. Rev. Biochem. Mol. Biol., 32, 101–140. [DOI] [PubMed] [Google Scholar]
- 4.Lavrik O.I., Prasad,R., Sobol,R.W., Horton,J.K., Ackerman,E.J. and Wilson,S.H. (2001) Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J. Biol. Chem., 276, 25541–25548. [DOI] [PubMed] [Google Scholar]
- 5.Grachev M.A., Mustaev,A.A. and Zaychikov,E.F. (1996) Highly selective affinity labeling of RNA polymerases. In Kurganov,V.I., Nagradova,N.K. and Lavrik,O.I. (eds), Chemical Modification of Enzymes. Nova Sciences Publishers, Inc., New York, pp. 309–346.
- 6.Kolpashchikov D.M., Rechkunova,N.I., Dobrikov,M.I., Khodyreva,S.N., Lebedeva,N.A. and Lavrik,O.I. (1999) Sensitized photomodification of mammalian DNA polymerase β. A new approach for highly selective affinity labeling of polymerases. FEBS Lett., 448, 141–144. [DOI] [PubMed] [Google Scholar]
- 7.Frosina G., Fortini,P., Rossi,O., Carrozzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 8.Wlassoff W.A., Dobrikov,M.I., Safronov,I.V., Dudko,R.Y., Bogachev,V.S., Kandaurova,V.V., Shishkin,G.V., Dymshits,G.M. and Lavrik,O.I. (1995) Synthesis and characterization of (d)NTP derivatives substituted with residues of different photoreagents. Bioconjug. Chem., 6, 352–360. [DOI] [PubMed] [Google Scholar]
- 9.Kolpashchikov D.M., Pestryakov,P.E., Wlassoff,W.A., Khodyreva,S.N. and Lavrik,O.I. (2000) Study of interaction of human replication factor A with DNA using new photoreactive analogs of dTTP. Biochemistry (Mosc.), 65, 160–163. [PubMed] [Google Scholar]
- 10.Rechkunova N.I., Kolpashchikov,D.M., Lebedeva,N.A., Petruseva,I.O., Dobrikov,M.I., Degtyarev,S.K. and Lavrik,O.I. (2000) Highly selective affinity labeling of DNA-polymerase from Thermus thermophilus B35 by a binary system of photoreactive agents. Biochemistry (Mosc), 65, 244–249. [PubMed] [Google Scholar]
- 11.Kolpashchikov D.M., Aleksandrova,L.A., Zakirova,N.F., Khodyreva,S.N. and Lavrik,O.I. (2000) A photoreactive analogue of 2′,3′-dideoxyuridine 5′-triphosphate: preparation and use for photoaffinity modification of human replication protein A. Russian J. Bioorg. Chem., 26, 151–155. [PubMed] [Google Scholar]
- 12.Beard W.A., and Wilson,S.H. (1995) Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol., 262, 98–107. [DOI] [PubMed] [Google Scholar]
- 13.Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 14.Strauss P.R., Beard,W.A., Patterson,T.A. and Wilson,S.H. (1997) Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs–Haldane mechanism. J. Biol. Chem., 272, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 15.Sobol R.W., Prasad,R., Evenski,A., Baker,A., Yang,X.P., Horton,J.K. and Wilson,S.H. (2000) The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature, 405, 807–810. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Schnapp K.A., Poe,R., Leyva,E., Soundararajan,N. and Platz,M.S. (1993) Exploratory photochemistry of fluorinated aryl azides. Implications for the design of photoaffinity labeling reagents. Bioconjug. Chem., 4, 172–177. [DOI] [PubMed] [Google Scholar]
- 18.Dobrikov M.I., Gaidamakov,S.A., Koshkin,A.A., Luk’ianchuk,N.P., Shishkin,G.V. and Vlasov,V.V. (1995) Directed chemical modification of DNA fragments by perfluorarylazide derivatives of oligonucleotides in the presence of oligonucleotides bearing photosensitized pyrene groups. Dokl. Akad. Nauk, 344, 122–125. [PubMed] [Google Scholar]
- 19.Winters T.A., Russell,P.S., Kohli,M., Dar,M.E., Neumann,R.D. and Jorgensen,T.J. (1999) Determination of human DNA polymerase utilization for the repair of a model ionizing radiation-induced DNA strand break lesion in a defined vector substrate. Nucleic Acids Res., 27, 2423–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doerhoefer S., Khodyreva,S., Safronov,I.V., Wlassoff,W.A., Anarbaev,R., Lavrik,O.I. and Holler,E. (1998) Molecular constituents of the replication apparatus in the plasmodium of Physarum polycephalum: identification by photoaffinity labelling. Microbiology, 144, 3181–3193. [DOI] [PubMed] [Google Scholar]
- 21.Mass G., Nethanel,T. and Kaufmann,G. (1998) The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell. Biol., 18, 6399–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mass G., Wold,M.S., Lavrik,O.I., Nethanel,T. and Kaufmann,G. (2001) Replication protein A modulates its interface with the primed DNA template during RNA-DNA primer elongation in replicating SV40 chromosomes. Nucleic Acids Res., 29, 3892–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlotkin T., Kaufmann,G., Jiang,Y., Lee,M.Y., Uitto,L., Syvaoja,J., Dornreiter,I., Fanning,E. and Nethanel,T. (1996) DNA polymerase ε may be dispensable for SV40- but not cellular- DNA replication. EMBO J., 15, 2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips A.T., (1977) Differential labeling: a general technique for selective modification of binding sites. Methods Enzymol., 46, 59–69. [DOI] [PubMed] [Google Scholar]
- 25.Grachev M.A., Kolocheva,T.I., Lukhtanov,E.A. and Mustaev,A.A. (1987) Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur. J. Biochem., 163, 113–121. [DOI] [PubMed] [Google Scholar]
- 26.Goeldner M.P., and Hirth,C.G. (1980) Specific photoaffinity labeling induced by energy transfer: application to irreversible inhibition of acetylcholinesterase. Proc. Natl Acad. Sci. USA, 77, 6439–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]