Abstract
We identified UIC-94003, a nonpeptidic human immunodeficiency virus (HIV) protease inhibitor (PI), containing 3(R),3a(S),6a(R)-bis-tetrahydrofuranyl urethane (bis-THF) and a sulfonamide isostere, which is extremely potent against a wide spectrum of HIV (50% inhibitory concentration, 0.0003 to 0.0005 μM). UIC-94003 was also potent against multi-PI-resistant HIV-1 strains isolated from patients who had no response to any existing antiviral regimens after having received a variety of antiviral agents (50% inhibitory concentration, 0.0005 to 0.0055 μM). Upon selection of HIV-1 in the presence of UIC-94003, mutants carrying a novel active-site mutation, A28S, in the presence of L10F, M46I, I50V, A71V, and N88D appeared. Modeling analysis revealed that the close contact of UIC-94003 with the main chains of the protease active-site amino acids (Asp29 and Asp30) differed from that of other PIs and may be important for its potency and wide-spectrum activity against a variety of drug-resistant HIV-1 variants. Thus, introduction of inhibitor interactions with the main chains of key amino acids and seeking a unique inhibitor-enzyme contact profile should provide a framework for developing novel PIs for treating patients harboring multi-PI-resistant HIV-1.
Combination therapy or highly active antiretroviral therapy (HAART) using two or more reverse transcriptase inhibitors (RTIs) and protease inhibitors (PIs) has dramatically improved the quality of life and survival of patients infected with human immunodeficiency virus type 1 (HIV-1) (22). However, the ability to provide effective long-term antiretroviral therapy for HIV-1 infection has become a complex issue because 40 to 50% of those who initially achieve favorable viral suppression to undetectable levels experience treatment failure (13, 37). Moreover, 10 to 40% of antiviral therapy-naive individuals infected with HIV-1 have persistent viral replication (plasma HIV RNA >500 copies/ml) under HAART (14, 17, 32), possibly due to transmission of drug-resistant HIV-1 variants (35). In addition, it is evident that with these anti-HIV drugs, only partial immunologic reconstitution is attained in patients with advanced HIV-1 infection.
From an aspect of development of new anti-HIV-1 therapeutics, we have faced formidable challenges different from those in the design of the first-line drugs, invoking thoughts about selection pressure mechanisms in addition to the conventional issues of potency, pharmacology, safety, and mechanism of drug action (5). Indeed, HIV-1 can, without doubt, develop resistance to any existing anti-HIV-1 therapeutic. It is of note that the very features that contribute to the specificity and efficacy of RTIs and PIs provide the virus with a strategy to mount resistance (6, 22), and it seems inevitable that this resistance issue will remain problematic for years to come.
A number of studies indicate that cross-resistance is a major obstacle inherent to most PIs (4, 16, 30). This observation should not be a surprise because all the inhibitors were designed to bind wild-type enzyme. Mutations have been found in every subsite of HIV protease; however, not every subsite residue has been found to mutate for a particular drug (6, 7). In this respect, novel PIs targeting the critical sites on the enzyme other than sites with which the currently available PIs interact should be active against HIV-1 variants resistant to existing PIs.
We recently described the properties of UIC-94003, also referred to as TMC-126, a femtomolar PI with potent antiviral activity that was discovered using biochemical resistance and fitness profiling methods (J. W. Erickson et al., 2001, 8th Conference on Retroviruses and Opportunistic Infections, abstr. 12; J. W. Erickson et al., unpublished data). In this study, we describe the antiviral profile of UIC-94003 against a wide spectrum of HIV, as well as against a variety of multi-PI-resistant clinical strains in vitro. We also describe the selection of a novel protease mutant that leads to UIC-94003 resistance and the characterization of its virological properties and susceptibilities to other PIs.
MATERIALS AND METHODS
Patients.
Patients with HIV-1 infection having a viral burden >50,000 RNA copies/ml by the bDNA assay (Chiron Diagnostics, Markham, United Kingdom) were enrolled into a randomized clinical study of amprenavir (Glaxo Wellcome, Stevenage, United Kingdom) and abacavir (Glaxo Wellcome) (8). These patients were put on the study under an investigational new drug application by the Critical Care Medicine Department, Clinical Center, National Institutes of Health. For the present study, eight patients were randomly chosen from among the enrollees who had failed the amprenavir plus abacavir therapy.
Antiviral agents.
UIC-94003 was designed and synthesized as described elsewhere (10). 3′-Azido-2′,3′-dideoxythymidine (zidovudine) and 2′,3′-dideoxyinosine (didanosine) were purchased from Sigma (St. Louis, Mo.) and Calbiochem (La Jolla, Calif.), respectively. 3′-Thiacytidine (lamivudine) was a kind gift from R. F. Schinazi (Atlanta, Ga.). Saquinavir and ritonavir were kindly provided by Roche Products Ltd. (Welwyn Garden City, United Kingdom) and Abbott Laboratories (Abbott Park, Ill.), respectively. Amprenavir was a kind gift from Glaxo-Wellcome, Research Triangle, N.C. Nelfinavir and indinavir were kindly provided by Japan Energy Inc, Tokyo.
Cells and viruses.
MT-2 cells were grown in an RPMI 1640-based culture medium supplemented with 15% fetal calf serum (FCS; HyClone Laboratories, Logan, Utah) plus 50 U of penicillin and 50 μg of streptomycin per ml. The following HIV-1 viruses were used for a drug susceptibility assay: HIV-1LAI (2), HIV-1Ba-L (9), HIV-2EHO (29), HIV-1ERS104pre, a clinical HIV-1 strain isolated from a drug-naive patient with AIDS (31), and eight HIV-1 clinical isolates which were originally isolated from heavily pretreated patients described above, which were genotypically and phenotypically characterized as multi-PI-resistant HIV-1s (8, 39).
Drug susceptibility assay.
The sensitivities of HIV-1LAI, HIV-1Ba-L, HIV-2EHO, and the primary HIV-1 isolates against various drugs were determined as previously described with minor modifications (21, 27, 31, 33). Briefly, MT-2 cells (2 × 104/ml) were exposed to 100 50% tissue culture infective doses (TCID50s) of HIV-1LAI, HIV-1Ba-L, or HIV-2EHO in the presence of various concentrations of drugs in 96-well microculture plates and incubated at 37°C for 7 days. After 100 μl of the medium was removed from each well, 10 μl of 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (7.5 mg/ml) in phosphate-buffered saline (PBS) was added to each well in the plate, followed by incubation at 37°C for 2 h. After incubation, to dissolve the formazan crystals, 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 was added to each well, and the optical density was measured in a microplate reader (model 3550; Bio-Rad). All assays were performed in duplicate (standard deviation [SD], <25%).
Phytohemagglutinin (PHA)-treated peripheral blood mononuclear cells (PBMC) (106/ml) were exposed to 50 TCID50s of each primary HIV-1 isolate in the presence or absence of various concentrations of drugs in 10-fold serial dilutions in 96-well microculture plates. All assays were performed in triplicate. The amounts of p24 antigen produced by the cells were determined on day 7 in culture using a commercially available radioimmunoassay kit. Drug concentrations which resulted in 50% inhibition (IC50) of p24 antigen production were determined by comparison with the p24 production level in drug-free control cell cultures as previously described (31, 33).
Generation of UIC-94003-resistant HIV-1 in vitro.
MT-2 cells (5 × 105) were exposed to 500 TCID50s of HIV-1NL4-3 (39) and cultured in the presence of UIC-94003 and amprenavir at an initial concentration of 0.0005 and 0.02 μM, respectively. Viral replication was monitored by observation of cytopathic effect in MT-2 cells. The culture supernatant was harvested on day 7 and used to infect fresh MT-2 cells for the next round of culture in the presence of increasing concentrations of each compound. When the virus began to propagate in the presence of the drug, the compound concentration was increased further. Proviral DNAs from the lysates of infected cells from several passages were subjected to nucleotide sequencing. This selection by UIC-94003 and amprenavir was carried out up to 2 μM and 10 μM, respectively.
Determination of nucleotide sequences.
The primers for the first PCR of the PR region were PR-5 (5′-GTT TCA ATT GTG GCA AAG AAG GGC-3′) and PR-12 (5′-CTC GTG ACA AAT TTC TAC TAA TGC-3′). The primers for the second PCR of the PR region were CS33 M (5′-TGT AAA ACG ACG GCC AGT AGG AAG GAC ACC AAA TGA AAG A-3′) and CS32 M (5′-CAG GAA ACA GCT ATG ACC ACT TTT GGG CCA TCC ATT CCT G-3′), which included the M13 forward and reverse sequences, respectively. The nucleotide sequence of each passaged HIV-1 variant was determined using cellular proviral DNA obtained from the infected MT-2 cells. The proviral DNA was prepared with InstaGene Matrix (Bio-Rad Laboratories, Hercules, Calif.).
The first PCR mixture consisted of 5 μl of the proviral DNA solution, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 0.01% gelatin, 0.2 mM deoxynucleoside triphosphates, 2.0 U of Taq DNA polymerase (Perkin Elmer, Foster, Calif.), and 12.5 pmol of each of the first PCR primers in a total volume of 50 μl. The PCR conditions employed were an initial 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, with a final 8-min extension at 72°C. The first PCR products (1 μl) were then amplified for 35 cycles, using the second PCR primers under the same reaction conditions. The second-round PCR products were purified with spin columns (PCR select III; 5 Prime 3 Prime, Boulder, Colo.) and were directly sequenced using both M13 forward and reverse dye-labeled primers on an ABI model 373 automated DNA sequencer.
Certain passaged HIV-1 variants were subjected to molecular cloning followed by sequence determination (39). The first-round PCR products (1 μl) of the PR region were used directly in the second round of PCR with primers KAPA-1 (5′-GCA GGG CCC CTA GGA AAA AGG GCT GTT GG-3′) and KSMA-2.1 (5′-CCA TCC CGG GCT TTA ATT TTA CTG GTA C-3′). The PCR conditions involved an initial 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, with a final 8-min extension at 72°C. The second-round PCR products were purified with spin columns and cloned directly (PCR-Script Amp cloning kit; Stratagene, La Jolla, Calif.). Molecular clones were sequenced as described above.
Molecular modeling study.
A structural modeling of HIV-1 protease complexed with UIC-94003 was performed by using the published crystal structures of HIV-1 protease complexed with amprenavir (Protein Data Bank code 1hpv) (20) and the protease complexed with various PIs containing bis-THF (12), followed by minimization of the active site in the Sybyl force field.
RESULTS
In vitro drug sensitivity of HIV-1 laboratory isolates.
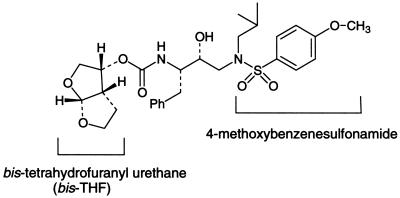
We designed, synthesized, and tested a variety of PIs containing a sulfonamide isostere for antiviral activity against HIV-1 in vitro (10, 11). Among them, UIC-94003, which has the same active isomer as amprenavir (Fig. 1), proved to be most potent against wild-type HIV. Further testing also revealed that UIC-94003 was potent against wild-type HIV protease and also against a wide spectrum of recombinant HIV protease mutants that were highly cross-resistant to one or more of the PIs used in first-line therapy (Erickson et al., abstract; Erickson et al., unpublished data). We initially tested UIC-94003 against HIV-1LAI and HIV-1Ba-L in PHA-PBMC and HIV-2EHO in MT-2 cells (Table 1). UIC-94003 was >10-fold more potent than any of the five currently available PIs against both HIV-1LAI and HIV-1Ba-L in PHA-PBMC (IC50, 0.0003 μM). As examined in MT-2 cells, the antiviral activity of ritonavir and amprenavir against HIV-2EHO was 9- and 13-fold less potent compared to that against HIV-1LAI, while UIC-94003 as well as indinavir, saquinavir, and nelfinavir were comparably active against both HIV-1LAI and HIV-2EHO (Table 1).
FIG. 1.
Structure of UIC-94003.
TABLE 1.
Sensitivities of HIV-1LAI, HIV-1Ba-L, and HIV-2EHO to various RTIs and PIsa
| Virus | Cells | Mean IC50 (μM) ± SDs
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RTIs
|
PIs
|
|||||||||
| AZT | ddI | 3TC | RTV | IDV | SQV | NFV | APV | UIC-94003 | ||
| HIV-1LAI | PBMC | 0.003 ± 0.0002 | 0.62 ± 0.02 | 0.021 ± 0.018 | 0.04 ± 0.008 | 0.015 ± 0.004 | 0.011 ± 0.005 | 0.009 ± 0.0003 | 0.017 ± 0.003 | 0.0003 ± 0.00009 |
| HIV-1Ba-L | PBMC | 0.011 ± 0.007 | 1.5 ± 1.1 | 0.054 ± 0.046 | 0.038 ± 0.02 | 0.017 ± 0.011 | 0.014 ± 0.01 | 0.003 ± 0.002 | 0.023 ± 0.009 | 0.0003 ± 0.00004 |
| HIV-1LAI | MT-2 | 0.024 ± 0.003 | 3.4 ± 0.2 | 0.66 ± 0.01 | 0.041 ± 0.005 | 0.019 ± 0.009 | 0.023 ± 0.002 | 0.005 ± 0.002 | 0.041 ± 0.01 | 0.0003 ± 0.0001 |
| HIV-2EHO | MT-2 | 0.003 ± 0.001 | 2.8 ± 0.6 | 0.4 ± 0.25 | 0.35 ± 0.025 | 0.01 ± 0.004 | 0.004 ± 0.0005 | 0.02 ± 0.01 | 0.53 ± 0.03 | 0.0005 ± 0.00007 |
Data shown represent mean values (with standard deviations) derived from the results of three independent experiments conducted in duplicate or triplicate. For PBMC, the IC50 were determined by employing PHA-PBMC exposed to each HIV-1 preparation (50 TCID50s per 105 PBMC) in the presence of each anti-HIV-1 agent and using the inhibition of p24Gag protein production as an endpoint on day 7 of culture. MT-2 cells (2 × 103) were exposed to 100 TCID50s of HIV-1LAI or HIV-2EHO and cultured in the presence of various concentrations of RTIs or PIs, and the IC50s were determined using the MTT assay on day 7 of culture. Abbreviations: AZT, zidovudine; ddI, didanosine; 3TC, lamivudine; RTV, ritonavir; IDV, indinavir; SQV, saquinavir; NFV, nelfinavir; APV, amprenavir.
In vitro activity of UIC-94003 against highly PI-resistant clinical HIV-1 strains.
HIV-1 was isolated from eight patients with AIDS who had failed 9 to 11 anti-HIV-1 drugs, propagated, titrated, and used as a source of infectious clinical HIV-1 strains as previously described (8, 39). These HIV-1 contained 9 to 14 amino acid substitutions in the protease-encoding region which have been reportedly associated with resistance against various PIs (ritonavir, indinavir, saquinavir, amprenavir, and nelfinavir) (4, 23, 25, 28, 36). Mutations seen in many of these isolates included L10I/R (8 of 8 isolates), M46I/L (7 of 8), I54V (5 of 8), L63P (8 of 8), A71V/T (7 of 8), V82A/T (8 of 8), and L90 M (6 of 8) (Table 2). As shown in Table 2, all strains had high-level resistance to ritonavir, indinavir, nelfinavir, and amprenavir (6- to >77-fold) compared to a wild-type clinical strain, HIV-1ERS104pre. With regard to saquinavir, four of the eight isolates were highly resistant (12- to 31-fold, with IC50 ranging from 0.1 to 0.3 μM), and the other four showed a moderately reduced sensitivity (3- or 4-fold increase in IC50 being ≈0.03 M). In contrast, UIC-94003 suppressed the replication of all eight isolates with IC50s ranging 0.0005 to 0.0055 μM (Table 2). While two strains (strains 1 and 7) were 6- and 8-fold less susceptible to UIC-94003, the IC50s remained extremely low, 0.004 and 0.0055 μM, respectively (Table 2).
TABLE 2.
Sensitivities of HIV-1 isolated from heavily drug-experienced individuals to PIs
| Virus | Amino acid substitutions in PR-encoding regiona | IC50b μM (fold change)
|
|||||
|---|---|---|---|---|---|---|---|
| RTV | IDV | SQV | NFV | APV | UIC-94003 | ||
| wild type | L63P | 0.044 (1) | 0.013 (1) | 0.010 (1) | 0.023 (1) | 0.025 (1) | 0.0007 (1) |
| 1 | L10I, K14R, L33I, M36I, M46I, F53L, K55R, I62V, L63P, A71V, G73S, V82A, L90M, I93L | >1 (>23) | >1 (>77) | 0.27 (27) | >1 (>43) | 0.27 (11) | 0.004 (6) |
| 2 | L10I, I15V, K20R, M36I, M46L, I54V, K55R, I62V, L63P, K70Q, V82A, L89M | >1 (>23) | 0.49 (38) | 0.037 (4) | 0.33 (14) | 0.28 (11) | 0.0013 (2) |
| 3 | L10I, I15V, E35D, N37E, K45R, I54V, L63P, A71V, V82T, L90M, I93L, C95F | >1 (>23) | 0.49 (38) | 0.036 (4) | >1 (>43) | 0.26 (10) | 0.001 (1) |
| 4 | L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, L90M | >1 (>23) | 0.21 (16) | 0.033 (3) | 0.09 (4) | 0.31 (12) | 0.0016 (2) |
| 5 | L10I, K43T, M46L, I54L, L63P, A71T, V82A, L90M, Q92K | >1 (>23) | >1 (>77) | 0.31 (31) | 0.41 (18) | 0.67 (27) | 0.0024 (3) |
| 6 | L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L | >1 (>23) | 0.30 (23) | 0.19 (19) | >1 (>43) | 0.16 (6) | 0.0005 (1) |
| 7 | L10I, L24I, L33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, V82A | >1 (>23) | >1 (>77) | 0.12 (12) | >1 (>43) | 0.49 (20) | 0.0055 (8) |
| 8 | L10R, N37D, M46I, I62V, L63P, A71V, G73S, V77I, V82T, L90M, I93L | >1 (>23) | 0.55 (42) | 0.042 (4) | >1 (>43) | 0.15 (6) | 0.001 (1) |
The amino acid sequence of each viral isolate was deduced from the nucleotide sequence and compared to the consensus B sequence cited from the Los Alamos data base. A clinical isolate, HIV-1ERS104pre (31), served as a source of wild-type HIV-1.
The IC50 were determined by employing PHA-PBMC exposed to HIV-1 strains (50 TCID50s per 105 PBMC) in the presence of each anti-HIV-1 agent and using the inhibition of p24Gag protein production as an endpoint. All values were determined in triplicate, and those shown are representative of two or three separate experiments. Numbers in parentheses represent fold changes of IC50s against each isolate compared to IC50s against HIV-1wt. See Table 1, footnote a, for abbreviations.
Selection of UIC-94003-resistant HIV-1 in vitro.
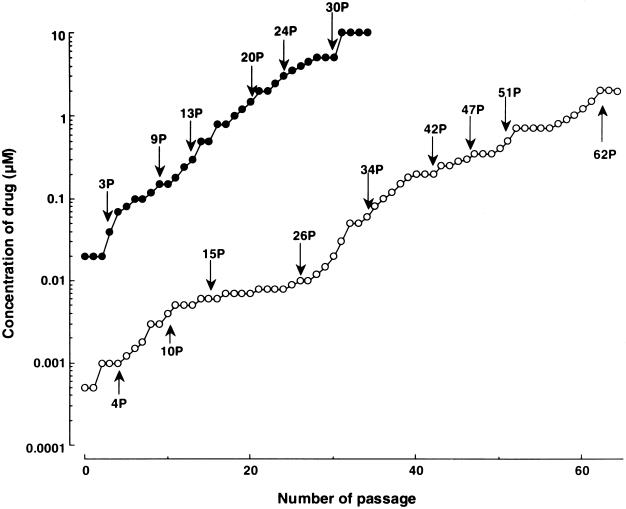
In order to select HIV-1 variants resistant to UIC-94003, a laboratory HIV-1 strain, HIV-1NL4-3, was propagated in MT-2 cells in the presence of increasing concentrations of amprenavir or UIC-94003 as previously described (39). The virus was initially exposed to 0.02 μM amprenavir and 0.0005 μM UIC-94003. The virus required 32 passages to acquire a 100-fold increase in the highest UIC-94003 concentration at which the virus could propagate, while it required only 21 passages for the virus to acquire the same level of resistance against amprenavir (Fig. 2). For the virus to acquire a 500-fold increase in UIC-94003 concentration, 46 passages were required, while 31 passages were required for viral acquisition of the same level of resistance against amprenavir (Fig. 2).
FIG. 2.
In vitro selection of HIV-1 variants resistant to UIC-94003 and amprenavir. A laboratory HIV-1 strain, HIV-1NL4-3, was passaged in the presence of increasing concentrations of UIC-94003 (○) or amprenavir (•) in MT-2 cells. The selection was carried out in a cell-free manner for a total of 34 to 64 passages with drug concentrations escalating from 0.0005 to 10 μM. Nucleotide sequences of proviral DNA were determined using cell lysates of HIV-1-infected MT-2 cells at the termination of each indicated passage.
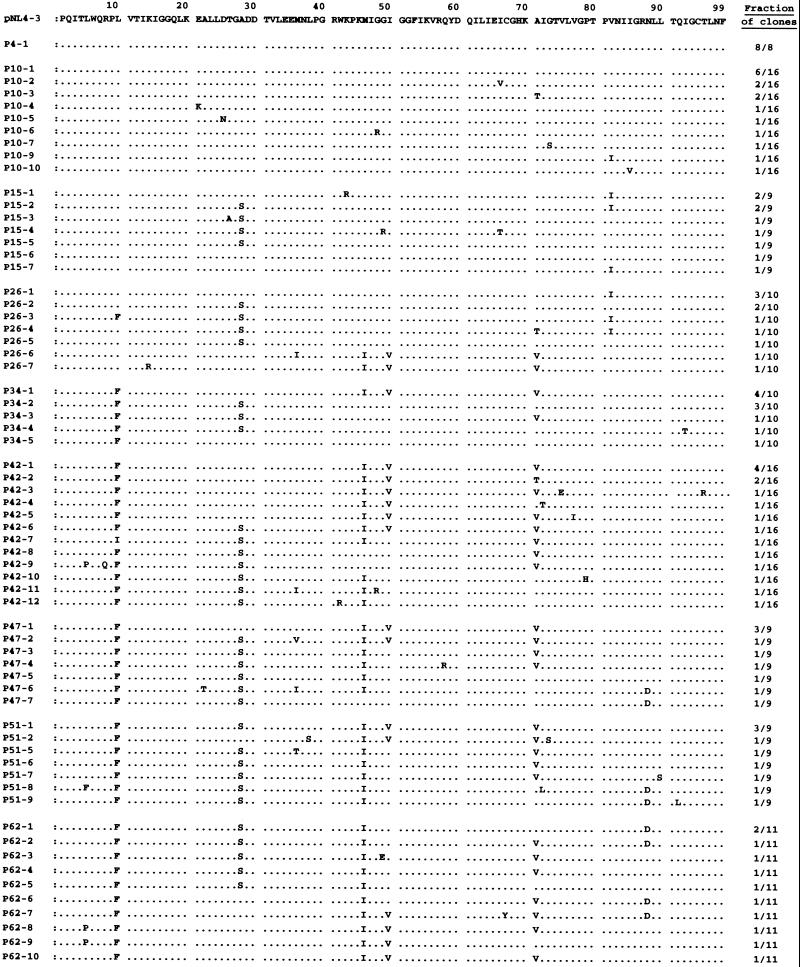
The protease-encoding region of the proviral DNA isolated from infected MT-2 cells was cloned and sequenced at passages 4, 10, 15, 26, 34, 42, 47, 51, and 62. Individual protease sequences and their frequency at each passage are depicted in Fig. 3, and six major substitutions (seen in HIV-1P62) are illustrated in Fig. 4. The wild-type protease gene sequence was seen in eight of eight clones derived from HIV-1NL4-3 at passage 4 (HIV-1UIC-P4). One active-site mutation V82I and one non-active-site mutation A71T emerged which were present in 1 and 2 of 16 clones of HIV-1UIC-P10, respectively. Val82 along with Ile84 represents an active-site residue whose side chains may be involved in making direct contact with inhibitor atoms (39), and the V82I substitution has been shown to be effective in conferring resistance when combined with a second active-site mutation such as V32I (19). However, the V82I mutation disappeared at passage 34 and beyond, possibly due to structural or biochemical constraints with the acquisition of another mutation(s).
FIG. 3.
Sequence analysis of the protease-encoding region of HIV-1 passaged in the presence of UIC-94003. The amino acid sequences of protease deduced from nucleotide sequences of the protease-encoding region of HIV-1 clones determined at nine different passages are illustrated. The fraction of clones examined is indicated on the right. The amino acid sequence of protease of a wild-type pNL4-3 clone is shown as a reference. Identity with this sequence at individual amino acid positions is indicated (dots).
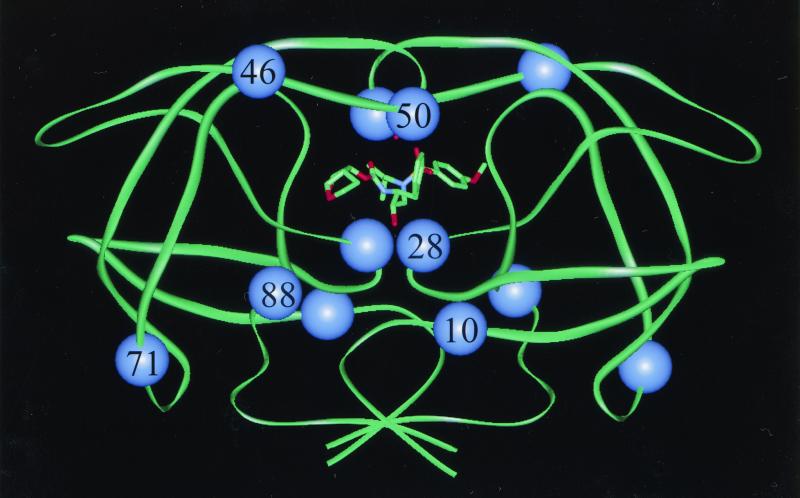
FIG. 4.
Backbone diagram of the dimeric HIV-1 protease. A model of UIC-94003 bound in the active site is shown in stick lines. The six residues which are commonly mutated during in vitro selection with UIC-94003 are indicated by spheres. Two of these residues lie within the active site (residues 28 and 50), while the other four residues lie outside the active site (residues 10, 46, 71, and 88).
An A28S mutation at the active site of the enzyme was seen early (at passage 15) in five of nine clones of HIV-1UIC-P15, and it was subsequently seen consistently at frequencies of >50% (except 44% for HIV-1P42), suggesting that this mutation was critical in conferring resistance to UIC-94003. A substitution at residue 50 from Ile to Val, seen in HIV-1 resistant to amprenavir (8, 28), was also identified at passage 26 and beyond. It appeared that the I50V mutation brought about a significant change in protease, because the virus started to replicate relatively rapidly in the presence of UIC-94003 following the emergence of I50V (Fig. 2). It is noteworthy that these two active-site mutations, A28S and I50V, did not coexist in any clones except in three clones throughout the selection (Fig. 3).
The L10F mutation also occurred in the presence of UIC-94003 in 1 of 10 clones at passage 26 and was subsequently seen in virtually all the clones examined. Leu10 is located distal to the active site of the enzyme, and the L10F mutation might act in concert with active-site mutations by compensating for a possible functional deficit caused by the latter. The M46I substitution was present in 2 of 10 clones at passage 26, in 4 of 10 clones at passage 34, and in virtually all subsequent clones (Fig. 3). Met46 along with Ile47 is located on the flap region of the enzyme. In this regard, I47V substitution reportedly emerges with viral resistance to amprenavir (24), but was not seen in the present study in either UIC-94003- or amprenavir-resistant HIV-1.
A commonly observed non-active-site mutation, A71V, also appeared in 2 of 10 clones at passage 26 and was seen predominantly beyond passage 34. Another non-active-site mutation, N88D, often selected in vivo with nelfinavir (26), was present in 2 of 9 clones at passage 52 and 5 of 11 clones at passage 62.
Sensitivity of UIC-94003-selected HIV-1 to various PIs.
Viruses at passage 62 with 2 μM UIC-94003 (HIV-1UIC-P62) and those at passage 30 with 5 μM amprenavir (HIV-1APV-P30) were titrated, and their susceptibilities to UIC-94003, amprenavir, and several other PIs were determined (Table 3). HIV-1UIC-P62 was highly resistant to amprenavir and UIC-94003 (IC50s were 20- and 70-fold greater than that of HIV-1NL4-3, respectively). However, HIV-1UIC-P62 was as sensitive to ritonavir and saquinavir as the parental HIV-1NL4-3, while it was moderately resistant to indinavir and nelfinavir (7- and 5-fold increases in IC50s, respectively). In contrast, HIV-1APV-P30 was highly resistant to all PIs tested except saquinavir (Table 3). Taken together, the present data suggest that UIC-94003 has significant advantages compared to amprenavir in terms of the emergence of drug resistance: (i) viral acquisition of resistance to UIC-94003 is substantially delayed (Fig. 2); (ii) UIC-94003 resistant HIV remains sensitive to all PIs except amprenavir; and (iii) in contrast, amprenavir-resistant virus is highly cross-resistant to all PIs except saquinavir (Table 3).
TABLE 3.
Amino acid substitutions in PR and sensitivities of drug-resistant HIV-1 strains to PIsa
| Virus | Amino acid substitutions | IC50, μM (fold change)
|
|||||
|---|---|---|---|---|---|---|---|
| RTV | IDV | APV | SQV | NFV | UIC-94003 | ||
| HIV-1NL4-3 | 0.038 (1) | 0.011 (1) | 0.042 (1) | 0.019 (1) | 0.023 (1) | 0.0003 (1) | |
| HIV-1UIC-P62 | L10F, A28S, M46I, I50V, A71V, N88D | 0.055 (1) | 0.08 (7) | 0.83 (20) | 0.01 (1) | 0.11 (5) | 0.021 (70) |
| HIV-1APV-P30 | L10F, V32I, M46I, I54M, A71V, I84V | >1.0 (>26) | 0.32 (30) | >1.0 (>25) | 0.035 (2) | >1.0 (43) | 0.029 (100) |
MT-2 cells (2 × 103) were exposed to HIV-1NL4-3, HIV-1UIC-P62, or HIV-1APV-P30 (all 100 TCID50s) and cultured in the presence of various drug concentrations. The IC50s were determined on day 7 of culture in the MTT assay. All values were determined in duplicate, and those shown are representive of two or three independent experiments. The numbers in parentheses represent fold changes compared to HIV-1NL4-3 (wild type). See Table 1, footnote a, for abbreviations.
Molecular modeling of interaction of HIV-1 protease and UIC-94003.
A structural model of HIV-1 protease complexed with UIC-94003 was prepared based on the crystal structures of HIV-1 protease complexed with amprenavir (20) and with compound 49 (12). These two structures were chosen because UIC-94003 is closely related in structure to amprenavir, except for the presence of the P2 bis-THF moiety, which is contained in compound 49.
Figure 4 shows an optimized molecular model of the protease-UIC-94003 complex and illustrates the locations of amino acid residues where substitutions were identified in HIV-1P62. Ile50 is located on the internal surface of the flap of HIV-1 protease, from where its aliphatic side chain atoms make van der Waals contacts with inhibitor atoms (20, 38). Ala28 is located between the conserved catalytic triad, Asp25-Thr26-Gly27, and two major substrate and inhibitor binding residues, Asp29 and Asp30, but is not itself directly involved in binding. Met46 is located on the external surface of the flap of HIV protease. Ile10, Asn88, and Ala71 are located far from the active site of the enzyme. These mutations presumably improve the fitness of active-site-containing mutants by exerting long-range effects on inhibitor or substrate binding or by some other, unknown mechanism (6, 7).
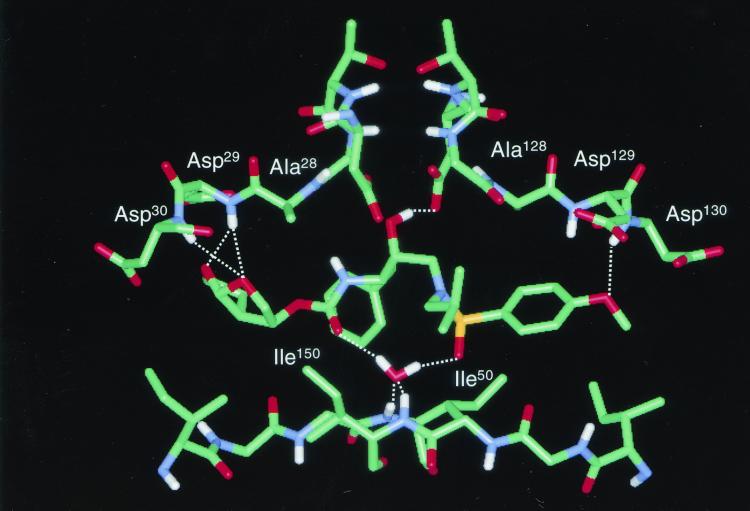
The model also showed that, using the bound conformation of amprenavir for UIC-94003, the two oxygen atoms of the bis-THF group of UIC-94003 could be positioned to form hydrogen bonds with the main chain amides of Asp29 and Asp30 in a manner similar to that observed previously for compound 49 (Fig. 5). In the model, UIC-94003 does not make any interactions with Ala28 (Fig. 5). This is consistent with the structure of the amprenavir-protease complex, in which Ala28 also does not play a direct role in binding.
FIG. 5.
Modeling of UIC-94003 bound in the active site of HIV-1 protease. Hydrogen bonds are represented as dotted lines.
DISCUSSION
UIC-94003 suppresses a wide spectrum of HIV-1, HIV-2, and various clinical HIV-1 strains at extremely low concentrations and over a very narrow spread of IC50s ranging from 0.0003 to 0.0055 μM (Tables 1 and 2). In contrast to other PIs, the presence of multiple, multidrug resistance-conferring mutations in HIV protease appears to have little if any effect on the antiviral potency of UIC-94003.
The pattern of drug resistance observed for PIs can be understood in light of the crystallographic data, and the pattern of amino acid substitutions conferring drug resistance confirms the particular binding modes of PIs to the enzyme (6, 7). The high potency of UIC-94003 relative to amprenavir is consistent with previous studies that have shown an increase in potency when a THF group is replaced by a fused-ring bis-THF moiety (10, 12). The increased potency appears to stem from the ability of the two conformationally constrained ring oxygen atoms in the bis-THF group to form hydrogen bonds with the main chain amide hydrogen atoms of Asp29 and Asp30 in the S2 subsite (Fig. 5). Since the main chain atoms cannot mutate, these interactions may be important for UIC-94003’s broad spectrum of activity against multidrug-resistant variants. The D30N mutation is a primary resistance mutation for nelfinavir, which forms a hydrogen bond with the side chain of Asp30 (25, 26). The D30N mutation has no effect on UIC-94003 binding (Erickson et al., unpublished). Consistent with these observations, exposure of HIV-1 to UIC-94003 does not select mutations at codon 30.
It is noteworthy that no amino acid mutations were seen (except at early passages during the selection of HIV-1 with UIC-94003) at the two active-site residues Val82 and Ile84, whose side chains are involved in making direct contacts with all PIs (6, 38). Mutations at these two active sites are commonly seen in HIV-1 resistant to various PIs. In particular, mutations at Val82 are highly effective at conferring resistance (4, 23) and, when combined with a second active-site mutation, such as V32I (19), can result in widespread biochemical cross-resistance to PIs (16, 39). The Ile84 residue, along with the symmetry-related I84′, makes interactions across S2/S1′ and S1/S2′ subsites, respectively (38), and the I84V mutation has been observed in clinical resistance with various PIs, including ritonavir (15), indinavir (3), saquinavir (36), and amprenavir (28). Our observation that HIV-1 propagated in the presence of UIC-94003 did not attain stable mutations at Val82 and did not acquire those at I84V (except for one clone) indicates that the acquisition of resistance to this PI by wild-type HIV-1 may require different evolutionary pathways than have been observed heretofore for most PIs.
Consistent with the above hypothesis, HIV-1 grown in the presence of increasing concentrations of UIC-94003 acquired the unique A28S mutation in the active site of the enzyme. The A28S mutation at the active site of the enzyme was seen at passage 15 and subsequently seen at frequencies of >50%, but never reaching 100%. After the appearance of the I50V substitution, another active-site mutation, during the selection with UIC-94003, the virus started to replicate relatively rapidly in the presence of UIC-94003 (Fig. 2). It is not clear why these two active-site mutations, A28S and I50V, did not coexist in nearly all clones throughout the selection (Fig. 3). However, these two mutations may produce unusual constraints on HIV protease activity.
Infectious clones containing either A28S or I50V in the pNL4-3 background replicated poorly compared to HIV-1wt in MT-2 cells (data not shown). In a previous biochemical study, the A28S mutation in HIV protease caused a more than 1,500-fold decrease in kcat/Km values for peptide substrates (18). These results suggest that A28S represents a critical mutation for UIC-94003 resistance but also confers a replication disadvantage on the virus. One can assume that, under greater UIC-94003 pressure, HIV-1 acquired another mutation (I50V) in the protease-encoding region and that a virus containing both mutations, A28S and I50V, had a replication disadvantage.
When HIV-1NL4-3 was propagated in the presence of increasing concentrations of UIC-94003 or amprenavir, the appearance of HIV-1 highly resistant to UIC-94003 seemed to be delayed compared to that of HIV-1 highly resistant to amprenavir (Fig. 2). However, it should be noted that the population size of HIV-1 in a culture is relatively small and the appearance of mutations can be affected by stochastic phenomena; i.e., rates of appearance of mutations in culture may not be reliable. It should be noted, however, that when we selected HIV-1LAI and HIV-1NL4-3/I84V against JE-2147 and JE-533, HIV-1 resistant to JE-533 appeared faster than that against JE-2147 reproducibly (39). Nevertheless, to address the issue of rates of mutation appearance, comparative studies on clinical application of these two PIs are needed.
Analysis of the model shows that UIC-94003 does not make interactions with either the backbone or the side chain atoms of Ala28. The A28S mutation will result in a slightly larger and more polar side chain. The reduction in potency with UIC-94003 could be due to steric hindrance with the larger side chain or, more likely, to unfavorable solvation energy effects during binding. The biochemical and crystallographic basis of the unusual ability of this PI to retain potency against multidrug-resistant HIV strains is described in a separate paper (Erickson et al., unpublished).
We also asked whether UIC-94003 could exert its activity in the presence of high concentrations of FCS, α1-acid glycoprotein (AAG), or human serum albumin (HSA). When we asked whether the activity of UIC-94003 and amprenavir against a wild-type clinical HIV-1 isolate, HIV-1ERS104pre, was altered in the presence of FCS in PHA-PBMC, the IC50s of UIC-94003 and amprenavir were 2- to 5-fold and 3-fold greater in the presence of 50% FCS, respectively, than in the presence of 15% FCS. The IC50s of UIC-94003 in the presence of 10 μM AAG and 300 μM HSA were 2- to 4-fold and 2-fold greater, respectively, than in the absence of AAG or HSA. On the other hand, the IC50s of amprenavir in the presence of 10 μM AAG and 300 μM HSA were 6- to 17-fold or 2-fold greater, respectively, than in the absence of AAG or HSA. These results suggested that the binding of UIC-94003 in human plasma is mainly attributable to AAG binding. However, the reduction in the antiviral activity of UIC-94003 through AAG binding is limited compared to the cases of other PIs, including amprenavir. It is noteworthy that UIC-94003, even in the presence of 10 μM AAG, is highly potent against HIV-1, with IC50s of 0.0002 to 0.0008 μM.
Taken together, the present data suggest that UIC-94003 has at least four advantages: (i) it exerts potent activity against a wide spectrum of drug-resistant HIV-1 variants, presumably due to its interaction with the main chains of the active-site amino acids Asp29 and Asp30; (ii) its unique contact with HIV-1 protease is different from that of other PIs; (iii) the viral acquisition of resistance is substantially delayed; and (iv) at least several PIs remain active in vitro against the virus selected in vitro with UIC-94003 (Table 3). These properties of UIC-94003 may provide a new framework for developing PIs with the bis-THF and a sulfonamide isostere. Further development of UIC-94003 for treating patients harboring multi-PI-resistant HIV-1 is warranted. A potent analog of UIC-94003, TMC-114, with a similar resistance profile, is currently undergoing clinical trials (Erickson et al., abstract).
Acknowledgments
We thank Pope Kosalaraksa and Takamasa Ueno for helpful discussions.
This work was supported in part by a grant from the National Institutes of Health (GM 53386, to A.K.G.), a grant from a Research for the Future Program of Japan Society for the Promotion of Science (JSPS-RFTF 97L00705, to K.Y. and H.M.), a Grant-in-Aid for Scientific Research (Priority Areas to H.M. and Encouragement of Young Scientists to K.Y.) from the Ministry of Education, Culture, Sports, Science, and Technology (Monbu-Kagakusho) of Japan (K.Y. and H.M.), and a Grant for Promotion of AIDS Research from the Ministry of Health, Labor and Welfare (Kosei-Rodosho) of Japan (K.Y. and H.M.).
REFERENCES
- 1.Baldwin, E. T., T. N. Bhat, S. Gulnik, B. Liu, I. A. Topol, Y. Kiso, T. Mimoto, H. Mitsuya, and J. W. Erickson. 1995. Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine. Structure 3:581–590. [DOI] [PubMed] [Google Scholar]
- 2.Clavel, F., D. Guetard, F. Brun-Vezinet, S. Chamaret, M. Rey, M. Santos-Ferreira, A. Laurent, C. Dauguet, C. Katlama, C. Rouzious, D. Klatzmann, J. Champalimaud, and L. Montagnier. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343–346. [DOI] [PubMed] [Google Scholar]
- 3.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569–571. [DOI] [PubMed] [Google Scholar]
- 5.De Clercq, E. 1997. In search of a selective antiviral chemotherapy. Clin. Microbiol. Rev. 10:674–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson, J. W., and S. K. Burt. 1996. Structural mechanisms of HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 36:545–571. [DOI] [PubMed] [Google Scholar]
- 7.Erickson, J. W., S. V. Gulnik, and M. Markowitz. 1999. Protease inhibitors: resistance, cross-resistance, fitness and the choice of initial and salvage therapies. AIDS 13:S189–204. [PubMed] [Google Scholar]
- 8.Falloon, J., S. Piscitelli, S. Vogel, B. Sadler, H. Mitsuya, M. F. Kavlick, K. Yoshimura, M. Rogers, S. LaFon, D. J. Manion, H. C. Lane, and H. Masur. 2000. Combination therapy with amprenavir, abacavir, and efavirenz in human immunodeficiency virus (HIV)-infected patients failing a protease-inhibitor regimen: pharmacokinetic Drug interactions and antiviral activity. Clin. Infect. Dis. 30:313–318. [DOI] [PubMed] [Google Scholar]
- 9.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, and R. C. Gallo. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, A. K., J. F. Kincaid, W. Cho, D. E. Walters, K. Krishnan, K. A. Hussain, Y. Koo, H. Cho, C. Rudall, L. Holland, and J. Buthod. 1998. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg. Med. Chem. Lett. 8:687–690. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, A. K., K. Krishnan, D. E. Walters, W. Cho, H. Cho, Y. Koo, J. Trevino, L. Holland, and J. Buthod. 1998. Structure based design: novel spirocyclic ethers as nonpeptidal P2-ligands for HIV protease inhibitors. Bioorg. Med. Chem. Lett. 8:979–982. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh, A. K., W. J. Thompson, P. M. Fitzgerald, J. C. Culberson, M. G. Axel, S. P. McKee, J. R. Huff, and P. S. Anderson. 1994. Structure-based design of HIV-1 protease inhibitors: replacement of two amides and a 10 pi-aromatic system by a fused bis-tetrahydrofuran. J. Med. Chem. 37:2506–2508. [DOI] [PubMed] [Google Scholar]
- 13.Grabar, S., C. Pradier, E. Le Corfec, R. Lancar, C. Allavena, M. Bentata, P. Berlureau, C. Dupont, P. Fabbro-Peray, I. Poizot-Martin, and D. Costagliola. 2000. Factors associated with clinical and virological failure in patients receiving a triple therapy including a protease inhibitor. AIDS 14:141–149. [DOI] [PubMed] [Google Scholar]
- 14.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734–739. [DOI] [PubMed] [Google Scholar]
- 15.Gulnik, S., J. W. Erickson, and D. Xie. 2000. HIV protease: enzyme function and drug resistance. Vitam. Horm. 58:213–256. [DOI] [PubMed] [Google Scholar]
- 16.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282–9287. [DOI] [PubMed] [Google Scholar]
- 17.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl for the AIDS Clinical Trials Group 320 Study Team. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725–733. [DOI] [PubMed] [Google Scholar]
- 18.Hong, L., J. A. Hartsuck, S. Foundling, J. Ermolieff, and J. Tang. 1998. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci. 7:300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, A. H., S. F. Michael, R. S. Wehbie, M. F. Knigge, D. A. Paul, L. Everitt, D. J. Kempf, D. W. Norbeck, J. W. Erickson, and R. Swanstrom. 1994. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc. Natl. Acad. Sci. USA 91:5597–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, E. E., C. T. Baker, M. D. Dwyer, M. A. Murcko, B. G. Rao, R. D. Tung, and M. A. Navia. 1995. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 117:1181. [Google Scholar]
- 21.Kodama, E., S. Shigeta, T. Suzuki, and E. De Clercq. 1996. Application of a gastric cancer cell line (MKN-28) for anti-adenovirus screening useing the MTT method. Antiviral Res. 31:159–164. [DOI] [PubMed] [Google Scholar]
- 22.Mitsuya, H., and J. Erickson. 1999. Discovery and development of antiretroviral therapeutics for HIV infection, p.751–780. In T. C. Merigan, J. G. Bartlet, and D. Bolognesi (ed.), Textbook of AIDS medicine. Williams & Wilkins, Baltimore, Md.
- 23.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760–766. [DOI] [PubMed] [Google Scholar]
- 24.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. J. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patick, A., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40:292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimitric assay for teh detection of anti-HIV compounds. J. Virol. Methods 20:309–321. [DOI] [PubMed] [Google Scholar]
- 28.Pazhanisamy, S., J. A. Partaledis, B. G. Rao, and D. J. Livingston. 1998. In vitro selection and characterization of VX-478 resistant HIV-1 variants. Adv. Exp. Med. Biol. 436:75–83. [DOI] [PubMed] [Google Scholar]
- 29.Rey-Cuille, M., J. Galabru, A. Laurent-Crawford, B. Krust, L. Montagnier, and A. Hovanessian. 1994. HIV-2 EHO isolate has a divergent envelope gene and induces single cell killing by apoptosis. Virology 202:471–476. [DOI] [PubMed] [Google Scholar]
- 30.Sardana, V. V., A. J. Schlabach, P. Graham, B. L. Bush, J. H. Condra, J. C. Culberson, L. Gotlib, D. J. Graham, N. E. Kohl, R. L. LaFemina, C. L. Schneider, B. S. Wolanski, J. A. Wolfgang, and E. A. Emini. 1994. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino acid substitutions in the enzyme’s substrate binding site. Biochemistry 33:2004–2010. [DOI] [PubMed] [Google Scholar]
- 31.Shirasaka, T., M. F. Kavlick, T. Ueno, W. Y. Gao, E. Kojima, M. L. Alcaide, S. Chokekijchai, B. M. Roy, E. Arnold, R. Yarchoan, and H. Mitsuya. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. USA 92:2398–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, and N. M. Ruiz for the Study 006 Team. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N. Engl. J. Med. 341:1865–1873. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, M., R. V. Srinivas, T. Ueno, M. F. Kavlick, F. K. Hui, A. Fridland, J. S. Driscoll, and H. Mitsuya. 1997. In vitro induction of human immunodeficiency virus type 1 variants resistant to 2′-β-fluoro-2′,3′-dideoxyadenosine. Antimicrob. Agents Chemother. 41:1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wainberg, M. A., and G. Friedland. 1998. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 279:1977–1983. [DOI] [PubMed] [Google Scholar]
- 36.Winters, M., J. Schapiro, J. Lawrence, and T. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wit, F. W., R. van Leeuwen, G. J. Weverling, S. Jurriaans, K. Nauta, R. Steingrover, J. Schuijtemaker, X. Eyssen, D. Fortuin, M. Weeda, F. de Wolf, P. Reiss, S. A. Danner, and J. M. Lange. 1999. Outcome and predictors of failure of highly active antiretroviral therapy: one-year follow-up of a cohort of human immunodeficiency virus type 1-infected persons. J. Infect. Dis. 179:790–798. [DOI] [PubMed] [Google Scholar]
- 38.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543–585. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: A dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. USA 96:8675–8680. [DOI] [PMC free article] [PubMed] [Google Scholar]