Abstract
The E4orf6 protein of group C adenovirus is an oncoprotein that, in association with the E1B 55-kDa protein and by E1B-independent means, promotes virus replication. An arginine-faced amphipathic α-helix in the E4orf6 protein is required for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein and to enhance replication of an E4 deletion virus. In this study, E4orf6 protein variants containing arginine substitutions in the amphipathic α-helix were analyzed. Two of the six arginine residues within the α-helix, arginine-241 and arginine-243, were critical for directing nuclear localization of the E1B 55-kDa protein. The four remaining arginine residues appear to provide a net positive charge for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein. The molecular determinants of the arginine-faced amphipathic α-helix that were required for the functional interaction between the E4orf6 and E1B 55-kDa proteins seen in the transfected cell differed from those required to support a productive infection. Several E4orf6 protein variants with arginine-to-glutamic acid substitutions that failed to direct nuclear localization of the E1B 55-kDa protein restored replication of an E4 deletion virus. Additionally, a variant containing an arginine-to-alanine substitution at position 243 that directed nuclear localization of the E1B 55-kDa protein failed to enhance virus replication. These results indicate that the ability of the E4orf6 protein to relocalize the E1B 55-kDa protein to the nucleus can be separated from the ability of the E4orf6 protein to support a productive infection.
Open reading frame 6 of early region 4 (E4orf6) of group C adenovirus (Ad) encodes a multifunctional protein that enhances viral replication (reviewed in reference 34) and acts as an oncoprotein (38, 40, 42). Ad mutants that lack the entire E4 region are severely defective for viral DNA replication and late viral protein synthesis (11, 26, 28). However, expression of the E4orf6 protein in trans largely corrects the growth defect of an E4 deletion virus (2, 31). The E4orf6 protein promotes the biogenesis and cytoplasmic accumulation of late viral mRNA, enhances stability of late viral RNA in the nucleus, and, as part of a complex with the E1B 55-kDa protein, promotes the nucleocytoplasmic transport of processed late viral mRNA to the exclusion of many host mRNAs (reviewed in reference 17).
It has been proposed that the E4orf6-E1B 55-kDa protein complex binds a component of the host nucleocytoplasmic transport system to achieve the selective transport of late viral mRNA (46). The E4orf6 protein, in cooperation with the E1A and E1B proteins of Ad, promotes the oncogenic transformation of baby rat kidney (BRK) cells (40, 42). The E4orf6-mediated transformation of BRK cells may stem from the ability of the E4orf6 protein to bind and inactivate tumor suppressor proteins such as p53 (16, 41) and p73 (27, 52), to bind and inactivate the cyclin A protein (25), or to increase the accumulation of mutations in the host cell genome (38, 43).
Several structural elements of the E4orf6 protein required for its function have been identified. A protein fragment containing the amino-terminal 58 amino acids of the E4orf6 protein binds both the E1B 55-kDa protein and the tumor suppressor p53 protein in vitro (16, 48). Additionally, it has been reported that the E4orf6 protein contains a cryptic leucine-rich nuclear export signal (15) needed for E4orf6-mediated degradation of p53 (41). However, the role of this cryptic nuclear export signal in late gene expression remains controversial (10, 18, 47, 54). Although E4orf6 proteins lacking the amino terminus or the cryptic nuclear export signal can cooperate with the E1A and E1B proteins to transform BRK cells, these cells are not as tumorogenic in nude mice as BRK cells transformed with the E1A and E1B proteins and the wild-type E4orf6 protein (41). E4orf6 variants with substitutions among several conserved cysteine and histidine residues fail to promote late viral gene expression, no longer coimmunoprecipitate with the E1B 55-kDa protein, fail to direct nuclear localization of the E1B 55-kDa protein, fail to destabilize the p53 protein, transform BRK cells with reduced efficiency, and produce transformed cells with diminished oncogenic potential in nude mice (9, 41). Boyer and Ketner have suggested that these conserved cysteine and histidine residues coordinate with two or more zinc ions to establish the proper tertiary structure of the E4orf6 protein (9).
A theoretical model for the structure of the E4orf6 protein was derived by Flint and associates (13). This model suggests the existence of a binuclear zinc coordination structure, as proposed by Boyer and Ketner (9). The molecular model of Flint and associates is also consistent with the existence of an arginine-faced amphipathic α-helix at the carboxy terminus of the E4orf6 protein. This structure appears to be required for many of the functions of the E4orf6 protein. E4orf6 variants that lack this structure or contain proline substitutions within the α-helix fail to promote virus replication (45). These E4orf6 variants also fail to direct the E1B 55-kDa protein to the cell nucleus after transfection.
The integrity of the arginine-faced amphipathic α-helix is required for E4orf6-mediated p53 degradation (41). An intact arginine-faced amphipathic α-helix is also required for the full oncogenic potential of the E4orf6 protein, as measured by the ability to transform BRK cells and to elicit an abnormal state of growth termed hypertransformation (41). Although these oncogenic functions may depend, in part, on the E1B 55-kDa protein, it is possible that the arginine-faced amphipathic α-helix of the E4orf6 protein binds to and alters the activity of cellular factors that control cell growth. For example, a motif within the amphipathic α-helix was suggested to mediate binding to cyclin A and augment expression of a transgene present on a recombinant adeno-associated virus (rAAV) (25). Because DNA-damaging agents promote transgene expression in rAAV-infected cells (1), it seems possible that the E4orf6 protein may promote rAAV transgene expression by disrupting the integrity of cellular DNA or perturbing signaling pathways associated with DNA damage (8, 43, 44).
Here we examine the molecular features of the arginine-faced amphipathic α-helix of the E4orf6 protein that are required to support a productive viral infection and to retain the rapidly shuttling E1B 55-kDa protein (18, 32) in the nucleus, which we interpret as a functional interaction between these two proteins. The hydrophilic face of this α-helix contains six arginine residues. Replacement of these arginine residues with similarly charged lysine residues had little impact on the ability to direct nuclear localization of the E1B 55-kDa protein. Two arginine residues, at positions 241 and 243, were found to be critical for a functional E4orf6-E1B 55-kDa protein interaction in cells. These residues lie on opposite sides of the α-helix. The other four arginine residues establish a positive charge in the central region of the α-helix. A net positive charge in this region appears to be required for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein.
Strikingly, the features of the arginine-faced amphipathic α-helix required for the functional interaction with the E1B 55-kDa protein in cells differ from those required for E4orf6 protein function during a productive Ad infection. Some E4orf6 variants that failed to retain the E1B 55-kDa protein in the nucleus provided wild-type function during productive Ad infection. However, an E4orf6 variant that successfully directed nuclear localization of the E1B 55-kDa protein expressed either by transfection or from the viral chromosome failed to supply E4orf6 protein function during Ad infection. These results indicate that the ability of the E4orf6 protein to relocalize the E1B 55-kDa protein to the nucleus can be separated from the ability of the E4orf6 protein to support a productive infection. The separation of these functions in the mutant proteins described here may reflect a diverse number of interactions that are mediated by the arginine-faced amphipathic α-helix of the E4orf6 protein for the many activities of this protein.
MATERIALS AND METHODS
Cell culture and viruses.
Cell culture media, cell culture supplements, and serum were obtained from Invitrogen/Life Technologies (Gaithersburg, Md.) through the Tissue Culture Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. HeLa and W162 cells (55) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% newborn calf serum as previously described (21).
Two strains of adherent HeLa cells were used in this work. The “fast-growing” strain of HeLa cells (CCL2.2) was obtained from the American Type Culture Collection in the late 1980s. This strain exhibits the typical morphology and growth rate (doubling time, ≈20 h) of HeLa cells. Another strain of HeLa cells which had been propagated in the laboratory of T. Shenk is identified as a “slow-growing” strain of HeLa cells. The doubling time of this strain is approximately 36 h. This slow-growing strain of HeLa cells reaches confluence at a lower cell density than the fast-growing strain of HeLa cells, consistent with greater spread and a less cuboid morphology.
Ad type 5 (Ad5) strain dl309 served as the wild-type Ad5 used in these studies. dl309 lacks a portion of the E3 region which has been shown to be dispensable for growth in culture (29). The E4 deletion virus dl1014 was constructed by Bridge and Ketner and described previously (12). This virus is able to express only the orf4 protein from the E4 region. The wild-type virus, dl309, was propagated in 293 cells (24). and dl1014 was propagated in W162 cells (55). Virus stocks were prepared by sequential centrifugation through CsCl as described previously (21).
The recombinant vaccinia virus used to express the T7 RNA polymerase, vTF7.3, was created by Fuerst et al. (20). Expression of the E1B 55-kDa and E4orf6 genes from the T7 promoter was achieved as described previously (23). Briefly, 105 cells were infected with vTF7.3 in reduced-serum medium and transfected with 1 μg of plasmid DNA mixed with 3 μg of Fugene 6 per the manufacturer’s (Roche, Nutley, N.J.) recommendation. Cells were analyzed by immunofluorescence between 12 and 14 h after infection and transfection. The localizations of the E4orf6 and E1B 55-kDa proteins were identical to those observed in cells expressing these proteins from the major immediate-early promoter and enhancer of cytomegalovirus in noninfected cells.
For confocal laser scanning microscopy, HeLa cells were grown on glass cover slips, infected with dl309 (29) or dl1014 (12), and simultaneously transfected with plasmids expressing the E4orf6 variants under control of the cytomegalovirus immediate-early promoter. For these experiments, 105 cells were exposed to 1 μg of plasmid DNA, 3 μg of Fugene 6, and 106 PFU of Ad in a 2-ml volume of OptiMEM (Invitrogen/Life Technologies). After 6 h at 37°C, the virus and plasmid mixture was replaced with normal growth medium, and the cells were processed 14 h after infection.
Plasmids and site-directed mutagenesis.
The plasmids carrying the E4orf6 and E1B 55-kDa protein genes were described previously (23, 45). Most E4orf6 variants were created by site-directed mutagenesis using PCR (7, 49). Briefly, arginine codons were changed by performing PCR with a 5′ oligonucleotide primer containing the altered E4orf6 sequences and a 3′ primer corresponding to sequences beyond the 3′ end of the E4orf6 coding region. The resulting PCR product of approximately 150 bp was used with another oligonucleotide corresponding to sequences beyond the 5′ end of the E4orf6 coding region to perform a second PCR synthesis. The resulting 1-kbp fragment containing the intended mutation was subcloned into pGEM11z (Promega, Madison, Wis.) by standard means.
The R4K and R241,243,244,248A E4orf6 variants were created by using the R241P V250S variant described previously (45). Restriction digestion of R241P V250S cDNA with NruI and StuI removes a blunt-ended, 30-bp DNA fragment encoding amino acids 241 through 250 of the E4orf6 protein. A 30-base oligonucleotide encoding the desired changes and its complement were annealed and ligated into the digested R241P V250S cDNA to introduce the desired changes. The mutations in the E4orf6 gene were verified by restriction analysis and confirmed by automated DNA sequencing of approximately 600 to 800 bp of the construct through the DNA Sequencing and Gene Analysis Core Laboratory of Wake Forest University. A list of the mutagenic oligonucleotides with the diagnostic restriction sites used in this study will be provided upon request. For expression from an intrinsically active promoter, certain E4orf6 variant cDNAs were subcloned into the pCMV Neo-BamHI vector (3) by standard means.
Indirect immunofluorescence.
Indirect immunofluorescence and photomicroscopy of whole cells were conducted as previously described (46). Double-label immunofluorescence was performed with the mouse monoclonal antibody MAb3 (37), which is specific for the amino terminus of the E4orf6 protein, and rat monoclonal antibody 9C10 (Oncogene Science, Uniondale, N.Y.) (56), which is specific for the Ad5 E1B 55-kDa protein. The secondary antibodies used were multiple-label-qualified goat antibodies conjugated to fluorescein and Rhodamine Red-X (Jackson ImmunoResearch, West Grove, Pa.) or Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.).
Samples were examined with a Leitz Dialux 20 EB microscope fitted for epifluorescent illumination and photographed using TMax film developed to an exposure index of 1600 ASA (Eastman Kodak, Rochester, N.Y.). Some samples were analyzed by a Zeiss LSM 510 confocal laser scanning device fitted to an Axioplan 2 microscope. Alexa Fluor 488 and Rhodamine Red-X dyes were excited by krypton ion and helium-neon laser excitation, respectively. Single optical sections of approximately 1.5-μm depth at the level of the cytoplasm were recorded as TIFF files using the LSM 510 software.
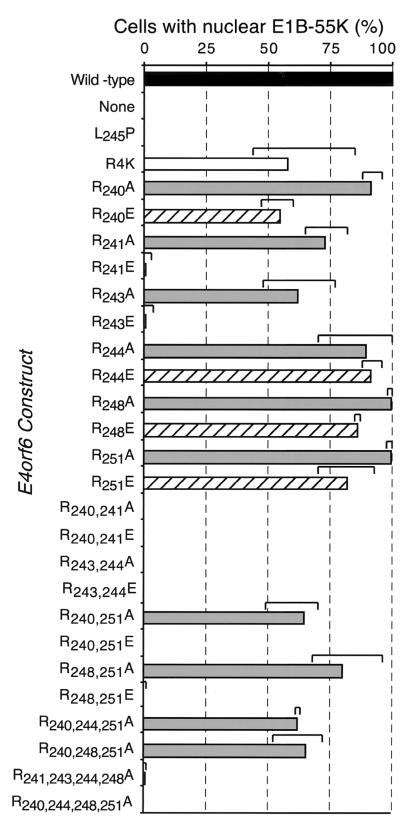
For the quantitative measurements shown in Fig. 5, a “double-blind” approach was used to quantify the degree of nuclear localization of the E1B 55-kDa protein. Appropriate mixtures of plasmids were prepared in randomly encoded tubes. A second investigator performed the infection or transfection using randomly labeled cultures of cells. The presence of both E4orf6 and E1B 55-kDa proteins was verified by double-label immunofluorescence as described above, and the localization of the E1B 55-kDa protein was scored as either cytoplasmic or nuclear as described in the text associated with Fig. 5. At least 100 cells were scored for each independent transfection.
FIG. 5.
E4orf6 variants bearing arginine replacement mutations within the amphipathic α-helix retain the E1B 55-kDa protein (55K) in the nucleus less effectively than the wild-type E4orf6 protein. Expression of the E4orf6-related proteins and the E1B 55-kDa protein was established as described in the legend to Fig. 2 but in a double-blind fashion as described in Materials and Methods. Expression of the E4orf6 protein variant (indicated on the left) was established, and the localization of the E1B 55-kDa protein was determined. Approximately 100 cells expressing each E4orf6 variant and the E1B 55-kDa protein were evaluated in each of four independent transfections. The localization of the E1B 55-kDa protein was scored as nuclear or cytoplasmic as indicated in the legend to Fig. 4. Each bar represents the average fraction of cells containing predominantly nuclear E1B 55-kDa protein. The brackets above each bar indicate the minimum and maximum values measured among four experiments. The solid black bar represents the value measured for the wild-type E4orf6 protein, the solid white bar represents the R4K variant, the gray bars represent arginine-to-alanine replacement variants, and the hatched bars represent arginine-to-glutamic acid variants.
Protein expression.
Replicate cultures of infected and transfected cells were collected, suspended in urea sample buffer (7.5 M urea, 50 mM Tris [pH 6.8], 1% sodium dodecyl sulfate [SDS], 50 mM dithiothreitol, 5% β-mercaptoethanol, 0.05% bromophenol blue), sonicated, and heated for 10 min at 65°C. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose, and the E4orf6-related proteins and the E1B 55-kDa protein were visualized by immunoblotting with MAb3 (37) and 2A6 (50), respectively, a secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch), and chemiluminescence detection (Pierce, Rockford, Ill.).
Sequence analysis and molecular modeling.
Sequence analysis and secondary structure prediction were performed with the suite of programs available as the Wisconsin Package version 10.1 (Genetics Computer Group, Madison, Wis.) Molecular models of the amphipathic α-helix were created and analyzed with the Sybyl suite of programs (Tripos Associates, Inc., St. Louis, Mo.). The Composer module was used to assemble peptide models corresponding to residues 239 through 255 of the E4orf6 protein and related variants. The amino and carboxy termini of the model were blocked with neutral blocking groups, and all hydrogen atoms and unpaired electrons were added. Atomic charges were assigned from the Amber 95 model provided by Tripos Associates, Inc., and the α-carbons were constrained to the spacing of a standard α-helix.
Electrostatic effects were introduced with a dielectric constant that varied with interatomic distance, and the model was energetically minimized by a reiterative process until the energy change per step was less than 0.02 kcal/mol. The Dynamics module was used to identify energetically reasonable configurations for the amino acid side chains by simulating exposure to 300 K for 8 ps. The initial velocities of the atoms in the model were randomized between repeated simulations to sample the variety of allowable configurations. This repeated analysis with different initial conditions confirmed that the simulation parameters allowed all nonconstrained atoms to reach dynamic equilibrium. The resulting structures were again minimized before using the MOLCAD module to project the electrostatic potential onto the solvent-accessible surface of the peptide models.
Complementation analysis.
HeLa cells were infected with dl309 (29) or dl1014 (12) and simultaneously transfected with the E4orf6 variant cDNA constructs analyzed in Fig. 7. For these experiments, 4 × 105 cells in a 65-mm culture dish were exposed to 1 μg of plasmid DNA, 3 μg of Fugene 6, and 4 × 106 PFU of virus in a 2-ml volume of OptiMEM (Invitrogen/Life Technologies). After 6 h at 37°C, the virus and plasmid mixture was replaced with normal growth medium. Detailed methods for Ad plaque assays have been described elsewhere (30). In brief, virus was harvested from the infected and transfected HeLa cells by multiple cycles of freezing and thawing. The cell lysates were clarified by centrifugation and serially diluted for infection of W162 cells (55) grown in six-well tissue culture dishes for plaque assays. Typically, valid data were collected from three dilutions in each series of dilutions. The virus yield was determined by linear regression and expressed as the number of plaques per milliliter of initial lysate.
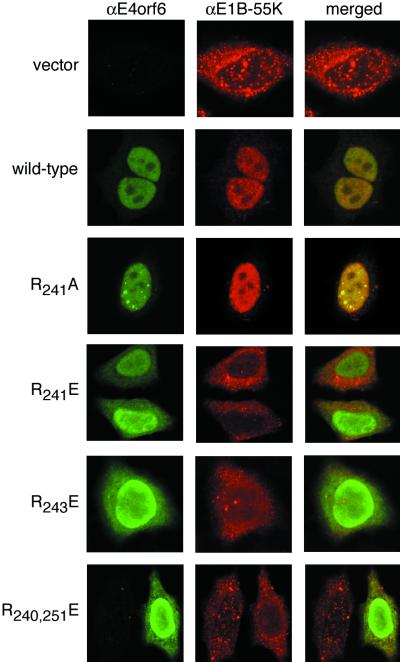
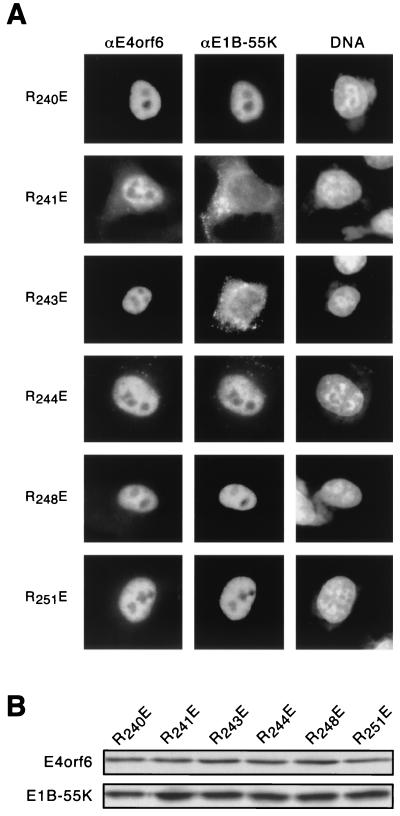
FIG. 7.
E4orf6 variants retain the E1B 55-kDa protein (55K) in the nucleus of Ad-infected cells with efficiency similar to that observed in noninfected, transfected cells. HeLa cells were infected with the E4 deletion virus dl1014 at 10 PFU per cell and simultaneously transfected with cDNAs expressing the E4orf6-related constructs indicated on the left. The cells were processed for immunofluorescence after 14 h. Representative confocal laser scanning images of approximately 1.5-μm depth were recorded at a plane of focus that includes the cytoplasm. Staining for the E4orf6 protein is seen in the left column (αE4orf6), and staining for the E1B 55-kDa protein is in the center column (αE1B-55K). The merged image (right column) shows the superposition of both signals as yellow.
Computer-aided graphics.
Film used to record chemiluminescence and immunofluorescence micrographs was scanned at 300 and 600 dpi, respectively, to produce a digitized image. Digital images were cropped using Photoshop 5.5 (Adobe Systems, Inc., San Jose, Calif.) and assembled with Canvas 5.1 and 7.0 (Deneba, Miami, Fla.) operating on a Macintosh microcomputer. Images produced by the Sybyl suite of programs were generated as 16-bit RGB files, transported to a Macintosh microcomputer as 8-bit RGB files, assigned a new color mapping scheme with Photoshop 5.5, and saved as 8-bit CMYK images.
RESULTS
Arginine residues in the amphipathic α-helix between positions 239 and 251 form a positively charged surface-exposed domain.
Using circular dichroism spectroscopy, we previously demonstrated that a peptide corresponding to amino acids 239 through 255 of the 294-residue E4orf6 protein can exist as an arginine-faced amphipathic α-helix (45). Variants of the E4orf6 protein that lack residues 241 through 250 or contain a proline in this region were defective. These mutant proteins failed to retain the E1B 55-kDa protein in the nucleus when expressed by transfection in noninfected cells, and these mutant proteins failed to promote replication of an E4 deletion virus when expressed by transfection in Ad-infected cells. These studies suggested that an intact α-helix in the E4orf6 protein, centered about residue 245, is required for a functional interaction between the E4orf6 and E1B 55-kDa proteins in the cell.
To aid in identifying features of this region that underlie the functional interaction between the E4orf6 and E1B 55-kDa proteins, we generated a molecular model of the α-helix. The space-filling model in Fig. 1 illustrates the 4.7 turns of the α-helix and a potential arrangement of residues 239 through 255 of the E4orf6 protein. In this model, the peptide backbone was constrained to a standard α-helix and the side chains were allowed to adopt an energetically favored configuration using molecular dynamics as described in Materials and Methods.
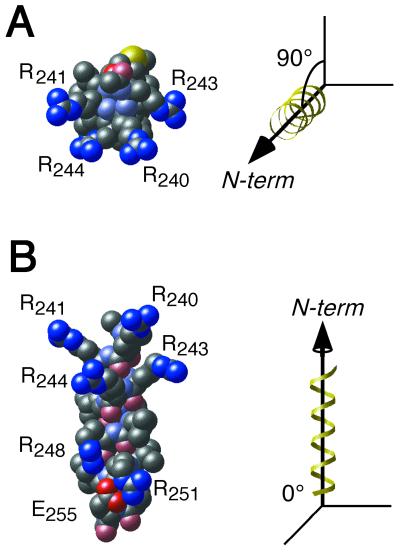
FIG. 1.
Representation of the arginine-faced amphipathic α-helix. Amino acids 239 through 255 of the E4orf6 protein were modeled as an α-helix. Side chains were allowed to adopt an energetically reasonable configuration using molecular dynamics as described in Materials and Methods. The identity of each atom is indicated by color: nitrogen, blue; carbon, gray; oxygen, red; sulfur, yellow. Hydrogen atoms are not shown. Atoms in the peptide backbone are rendered in a muted color. The charged residues that are visible are labeled near the side chain. (A) View down the helix axis from N terminus to C terminus. (B) View of the hydrophilic face.
The amphipathic nature of this structure is evident when viewed down the helix axis (Fig. 1A). Arginine-241 and arginine-243 occur at the extreme sides of the hydrophilic face. The only nonaliphatic residues in the hydrophobic backbone of the α-helix are threonine-242 and methionine-246. A likely distribution of the charged residues can be seen in the view of the α-helix shown in Fig. 1B. Arginine-240, arginine-244, and arginine-248 align on the hydrophilic face over three turns of the α-helix (Fig. 1B). In repeated modeling efforts, arginine-251 established a bond with the glutamic acid residue at position 255. The theoretical structure proposed for the E4orf6 protein by Brown et al. (13) suggests that the protein adopts an α-helical configuration beyond alanine-249. Thus, the pair of electronegative glutamic acid residues at positions 255 and 256 may interact with the guanidinium group of arginine-251.
It seems that the remaining five arginines are free to adopt a wide variety of configurations. This property would be consistent with their position on the solvent-exposed face of this amphipathic α-helix and would permit a significant degree of flexibility in binding diverse ligands. These arginine residues may form a loosely structured, positively charged region on the surface of the E4orf6 protein.
To determine whether the identity of these six arginine residues or the basic nature of these residues is critical for E4orf6-E1B 55-kDa protein interaction, an E4orf6 variant, R4K, bearing four lysine substitutions, was created and tested for a functional interaction with the E1B 55-kDa protein (Fig. 2A). The R4K and wild-type E4orf6 proteins were expressed with the E1B 55-kDa protein in HeLa cells using the vaccinia virus/T7 RNA polymerase infection-transfection expression system, and the localization of the proteins was determined. As reported previously, the E4orf6 protein is diffusely distributed throughout the nucleus and is excluded from the nucleoli (23, 45). Although the E1B 55-kDa protein shuttles between the cytoplasm and nucleus (18, 32), when expressed alone, this protein is found primarily in the cytoplasm (Fig. 2B, second row). However, the subcellular localization of the E1B 55-kDa protein is changed when expressed with the E4orf6 protein (23, 45). The E4orf6 protein retains the E1B 55-kDa protein in the cell nucleus. In most cells expressing both of these adenoviral proteins (Fig. 2B, third row), staining for the E1B 55-kDa protein appeared coincident with staining for the E4orf6 protein.
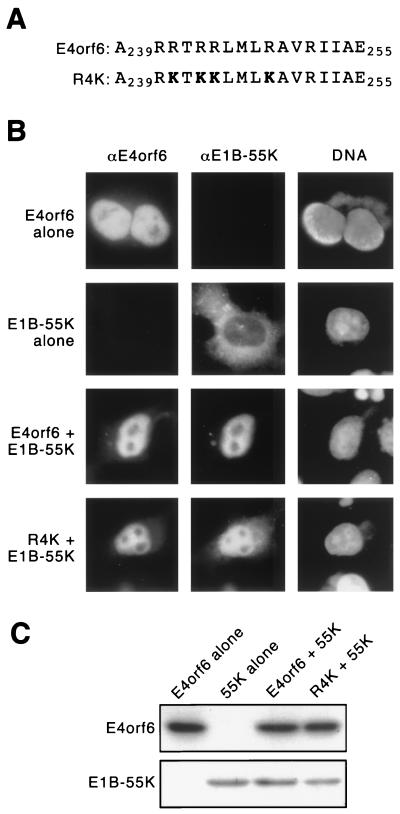
FIG. 2.
E4orf6 variant bearing arginine-to-lysine substitutions within the amphipathic α-helix retains the E1B 55-kDa protein (55K) in the nucleus after transfection. (A) The amino acid sequence of the amphipathic α-helix (residues 239 to 255) and a variant with lysine substitutions at positions 241, 243, 244, and 248. (B) HeLa cells were infected with the recombinant vaccinia virus vTF7.3 to establish expression of the T7 RNA polymerase and then transfected with cDNAs under control of the T7 promoter to express the E4orf6-related protein and the E1B 55-kDa protein. The transfected E4orf6 cDNAs are identified on the left. Ad proteins were visualized by double-label immunofluorescence 12 h after transfection. Representative cells are shown. E4orf6 proteins were visualized with the mouse monoclonal antibody MAb3 (37) (left column, αE4orf6), E1B 55-kDa protein was visualized with the rat monoclonal antibody 9C10 (56) (center column, αE1B-55K), and DNA was visualized with 4′,6′-diamidino-2-phenylindole (DAPI) (right column, DNA). (C) In parallel with the samples prepared for immunofluorescence, expression of the E4orf6 and E1B 55-kDa proteins was established by transfection of the cDNAs in vTF7.3-infected cells indicated above each lane. Total cell protein was isolated 12 h after transfection, separated by SDS-PAGE, and trans-ferred to a solid support. The E4orf6-related proteins and the E1B 55-kDa protein were visualized by immunoblotting with MAb3 and 2A6 (50), respectively. Only the portion of the membranes containing the E4orf6-related proteins and the E1B 55-kDa proteins is shown.
Like the E4orf6 protein, the R4K variant was found in the nucleus and retained a portion of the E1B 55-kDa protein in the nucleus of cells expressing both proteins. In contrast to the wild-type E4orf6 protein, the R4K protein appeared to be less efficient at retaining the E1B 55-kDa protein in the nucleus. In the example shown (Fig. 2B, fourth row), more cytoplasmic staining is evident for E1B 55-kDa protein in the presence of the R4K protein than in the presence of the wild-type E4orf6 protein (Fig. 2B, third row). Although the R4K protein retained at least a portion of the E1B 55-kDa protein in the nucleus of every cell examined, approximately 40% of these cells contained brighter staining for the E1B 55-kDa protein in the cytoplasm than in the nucleus. By contrast, a variant bearing alanine replacements at arginine-241, arginine-243, arginine-244, and arginine-248 failed to retain the E1B 55-kDa protein in the nucleus of cells expressing both proteins. Nonetheless, since expression of the R4K protein induced nuclear localization of at least a portion of the E1B 55-kDa protein in each cell and expression of the R241,243,244,248A variant did not, we conclude that the basic charge of the arginine residues within the amphipathic α-helix contributes to the functional interaction between the E4orf6 and E1B 55-kDa proteins seen in transfected cells.
The diminished ability of the R4K variant to retain the E1B 55-kDa protein in the nucleus is not due to reduced levels of the R4K protein. The steady-state level of the E4orf6, R4K, and E1B 55-kDa proteins in HeLa cells was determined by immunoblot. The amount of E4orf6 protein measured in cells expressing the E4orf6 protein alone or with the E1B 55-kDa protein was similar to the amount of R4K protein (Fig. 2C). Neither the E4orf6 protein nor the R4K variant affected the steady-state level of the E1B 55-kDa protein. An equivalent amount of E1B 55-kDa protein was detected in lysates derived from cells expressing the E1B 55-kDa protein alone, the E1B 55-kDa and E4orf6 proteins, or the R4K and E1B 55-kDa proteins.
Basic nature of arginines 241 and 243 is required for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein.
A collection of single arginine-to-glutamic acid substitution variants were created and tested for the ability to retain the E1B 55-kDa protein in the nucleus. Representative results are presented in Fig. 3A. In most cells expressing both the E1B 55-kDa protein and either the R240E, R244E, R248E, or R251E protein, a significant portion of the E1B 55-kDa protein was detected in the nucleus. However, variants of the E4orf6 protein with arginine-to-glutamic acid amino acid replacements at residues 241 or 243 failed to retain the E1B 55-kDa protein in the nucleus. Fewer than 1% of transfected cells expressing the E1B 55-kDa and either the R241E or R243E mutant protein contained the E1B 55-kDa protein in the nucleus. Furthermore, in this minority of cells, the E1B 55-kDa protein was uniformly distributed throughout the cell, reinforcing the defective nature of these mutant E4orf6 proteins.
FIG. 3.
E4orf6 variants with arginine-to-glutamic acid replacement substitutions at position 241 or 243 do not retain the E1B 55-kDa protein (55K) in the nucleus after transfection. (A) Expression of the E4orf6-related proteins (indicated on the left) and the E1B 55-kDa protein was established, and the localization of the Ad proteins was determined as described in the legend to Fig. 2. Representative images of a single cell from each transfection are presented, with the E4orf6 protein shown in the left column (αE4orf6), E1B 55-kDa protein in the center column (αE1B-55K), and DNA visualized with DAPI in the right column (DNA). (B) In parallel with the samples prepared for immunofluorescence, expression of the E4orf6 and E1B 55-kDa proteins was established by transfection of the cDNAs indicated above each lane, and the E4orf6-related proteins and the E1B 55-kDa protein were visualized by immunoblotting as described in the legend to Fig. 2.
However, like the R4K variant, the glutamic acid substitution variants were not as efficient as the wild-type E4orf6 protein at retaining the E1B 55-kDa protein in the nucleus. The E1B 55-kDa protein was predominantly nuclear in approximately 55% of the cells expressing both the E1B 55-kDa and R240E proteins. In cells expressing either the R244E, R248E, or R251E variant and the E1B 55-kDa protein, approximately 85% of the cells contained predominantly nuclear E1B 55-kDa protein.
The diminished ability of the R240E, R244E, R248E, and R251E variants to retain the E1B 55-kDa protein in the nucleus and the loss of this ability in the R241E and R243E variants cannot be attributed to a gross change in localization or to reduced levels of the variant E4orf6 proteins. The distribution of each of these E4orf6 variants in the vaccinia virus-infected cell was indistinguishable from that of the wild-type E4orf6 protein (compare E4orf6 protein localization in Fig. 2B with that in Fig. 3A). However, as noted previously (45), individual cells with high levels of E4orf6 protein contained some cytoplasmic staining for the E4orf6 protein. Immunoblot analysis revealed that these E4orf6-related proteins were expressed to similar levels (Fig. 3B). Furthermore, expression of these variants did not affect the level of E1B 55-kDa protein (Fig. 3B). Thus, the interaction of the E4orf6 and E1B 55-kDa proteins measured by this colocalization assay is ablated by the R241E and R243E mutations, reduced by the R240E mutation, and only slightly affected by the R244E, R248E, and R251E mutations.
All of the E4orf6 variants created in this study directed nuclear localization of the E1B 55-kDa protein with varying efficiency. The series of micrographs in Fig. 4 illustrate the various degrees to which the R240E mutant protein retained the E1B 55-kDa protein in the nucleus. More than half of the cells expressing both E1B 55-kDa and R240E proteins showed predominantly nuclear staining for the E1B 55-kDa protein (Fig. 4A and 4B). In slightly less than half of the cells, the E1B 55-kDa protein appeared to be uniformly distributed (Fig. 4C) or predominantly restricted to the cytoplasm (Fig. 4D and E). The variable efficiency with which the E4orf6 protein variants directed nuclear localization of the E1B 55-kDa protein formed the basis for the quantitative assay described below.
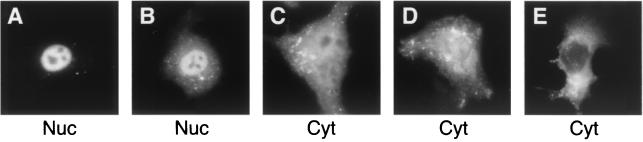
FIG. 4.
Extent of E1B 55-kDa nuclear localization in cells expressing the R240E protein varies from cell to cell. Expression of the R240E E4orf6 variant and the E1B 55-kDa protein was established and the localization of the E1B 55-kDa protein was determined as described in the legend to Fig. 2. The localization represented in A and B was seen in 55% of the cells and scored as nuclear (Nuc). The uniform distribution represented in C was seen in only 2% of the cells; the predominantly cytoplasmic localization seen in D and E was seen in 43% of the cells. Both the uniform and cytoplasmic distributions were scored as cytoplasmic (Cyt).
To determine whether the acquisition of a negative charge or the loss of a positive charge at the position normally occupied by arginine disrupted the E4orf6-E1B 55-kDa protein interaction, single arginine-to-alanine variants were tested for their ability to retain the E1B 55-kDa protein in the nucleus. The ability of these as well as all other mutant E4orf6 proteins to retain the E1B 55-kDa protein in the nucleus was quantified by a double-blind approach. Cells expressing both the E4orf6 variant and the E1B 55-kDa protein were evaluated for the relative staining intensity of the E1B 55-kDa protein in the nucleus and in the cytoplasm. Cells that contained stronger staining in the nucleus relative to the cytoplasm were scored as having a predominantly nuclear E1B 55-kDa protein. Each value plotted in Fig. 5 represents the average of four independent transfection-infections from two independent experiments. The ranges associated with each point indicate the minimum and maximum value of the four measurements.
The R240A variant retained the E1B 55-kDa protein in the nucleus of approximately 90% of cells expressing both proteins. This value was significantly greater than the 55% average measured for the related R240E variant. Moreover, the lowest percentage of cells containing nuclear E1B 55-kDa protein in the presence of the R240A variant (88%) exceeded the maximum value measured in the presence of the R240E variant (62%). Therefore, it seems likely that the acquisition of a negative charge at position 240 in the R240E variant rather than the loss of a positive charge at this position in the R240A variant reduced the strength of interaction with the E1B 55-kDa protein.
Because E4orf6 variants that contained glutamic acid in position 244, 248, or 251 behaved like the wild-type protein with respect to retaining the E1B 55-kDa protein in the nucleus, we expected the corresponding arginine-to-alanine variants to also function like the wild-type protein. This was indeed observed. Cells expressing the E1B 55-kDa protein and either the R244A, R248A, or R251A mutant protein contained predominantly nuclear E1B 55-kDa protein in 90 to 98% of the cells. By contrast, the R241A and R243A variants relocalized the E1B 55-kDa protein to the nucleus in 73 and 62% of the cells, respectively (Fig. 5). This intermediate value, although greater than that measured for the corresponding glutamic acid variants, is consistent with the notion that a positive charge at positions 241 and 243 is important for this function of the E4orf6 protein.
Further evidence in support of this idea can be derived from the ostensibly weaker interaction of the R241A and R243A proteins with the E1B 55-kDa protein seen in the slow-growing strain of HeLa cells. In this strain of HeLa cells, the R241A and R243A mutant proteins directed nuclear localization of the E1B 55-kDa protein in fewer than 20% of the cells, whereas the other alanine substitution mutant proteins did so to the same extent as in the fast-growing HeLa cell variant (data not shown).
The failure of the R241E and R243E variants and the diminished ability of the R241A and R243A variants to retain the E1B 55-kDa protein in the nucleus shows a requirement for a positively charged amino acid at positions 241 and 243 for this property of the E4orf6 protein. Since the R241A and R243A variants can retain at least a portion of the total E1B 55-kDa protein in the nucleus, it is possible that a neighboring arginine residue may partially compensate for the loss of arginine-241 or arginine-243. Therefore, double arginine-to-alanine variants were created and tested for their ability to retain the E1B 55-kDa protein in the nucleus. An E4orf6 variant bearing arginine-to-alanine substitutions at positions 240 and 241 did not relocalize any E1B 55-kDa protein to the nucleus. Similarly, the R243,244A variant was completely defective in its ability to retain the E1B 55-kDa protein in the nucleus. By contrast, two variants that preserved arginine-241 and arginine-243, R240,251A and R248,251A, retained the E1B 55-kDa protein in the nucleus of 70 and 82% of cells expressing both proteins, respectively. The properties of these double alanine substitution variants provide further support for the importance of residues 241 and 243 for the functional interaction between the E1B 55-kDa and E4orf6 proteins observed in cells.
Net positive charge among residues 240, 244, 248, and 251 is required for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein.
Under physiological conditions, the arginine residues of the amphipathic α-helix can form a positively charged surface. Arginine-241 and arginine-243 lie on opposite sides of the solvent-exposed face of this α-helix and are important for the functional interaction of the E4orf6 and E1B 55-kDa proteins observed in cells. Arginine-240, -244, -248, and -251 can form a colinear arrangement over nearly three turns of the α-helical segment to present a positively charged surface.
Variant E4orf6 proteins with multiple replacements of these four amino acids with either glutamic acid or alanine were tested for their ability to retain the E1B 55-kDa protein in the nucleus of cells transiently expressing both proteins. As discussed previously, E4orf6 variants containing two arginine-to-alanine replacement mutations (R240,251A and R248,251A) retained a portion of the E1B 55-kDa protein in the nucleus. By contrast, substituting glutamic acid for arginine at these positions (R240,251E and R248,251E) ablated the E4orf6-E1B 55-kDa protein interaction (Fig. 5). These double glutamic acid replacement variants appeared completely defective in their ability to retain the E1B 55-kDa protein in the nucleus. The distribution of the E1B 55-kDa protein in cells expressing any of the double arginine-to-glutamic acid E4orf6 variants resembled that observed in cells expressing the E1B 55-kDa protein alone.
Unlike the double arginine-to-alanine variants, the double arginine-to-glutamic acid variants have a net neutral charge among these four central residues. Therefore, to test the possibility that the functional E4orf6-E1B 55-kDa protein interaction in transfected cells requires a net positive charge among these amino acids, cDNAs were created to express E4orf6 proteins containing triple or quadruple arginine-to-alanine replacement mutations. When coexpressed with the E1B 55-kDa protein, the R240,244,251A protein directed the E1B 55-kDa protein to the nucleus in 60% of the cells. Similar results were observed in cells expressing the R240,248,251A variant and the E1B 55-kDa protein. By contrast, the quadruple mutant protein, R240,244,248,251A, failed to retain the E1B 55-kDa protein in the nucleus (Fig. 5). The localization of the E1B 55-kDa protein in these cells resembled that observed in cells expressing the E1B 55-kDa protein alone (Fig. 2). The quadruple alanine substitution variant R240,244,248,251A and the double glutamic acid substitution variants R240,251E and R248,251E have a net neutral charge among the four central residues. These results lead us to suggest that a positive surface on the hydrophilic face of the amphipathic α-helix is required for E4orf6-E1B 55-kDa protein interaction in transfected cells.
The overall charge of the amphipathic α-helix appeared to be important for the ability of the E4orf6 protein to retain the E1B 55-kDa protein in the cell nucleus. However, variants with a similar net charge differed in their ability to retain the E1B 55-kDa protein in the nucleus. To reveal a possible basis for these differences, molecular models of the amphipathic α-helix of the various E4orf6 mutant proteins were created, and the theoretical charge density of these models was determined (Fig. 6). The portion of the E4orf6 protein that was modeled was the same as that shown in Fig. 1 and included alanine-239 through glutamic acid-255.
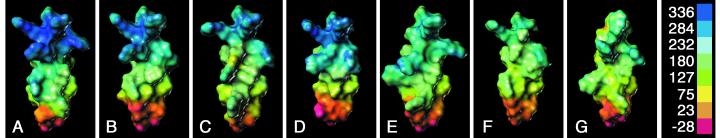
FIG. 6.
Ability to retain the E1B 55-kDa protein in the nucleus is correlated with a more electropositive surface at the amino terminus of the amphipathic α-helix. Amino acids 239 through 255 of E4orf6 protein variants were modeled as an α-helix as described in the legend to Fig. 1. The solvent-accessible surface of the model peptide was calculated, and the electropositive potential of the molecule was projected onto this surface. Electropositive regions are mapped to deep blue and electronegative regions are mapped to red, as indicated by the scale (kilocalories per mole electron) on the right. The orientation of the α-helical peptides is the same as in Fig. 1B, where the amino terminus is at the top and the hydrophilic face is exposed. The models and associated value for nuclear E1B 55-kDa protein retention (from Fig. 5) are (A) wild-type E4orf6 protein, 100%; (B) R251A, 99%; (C) R244E, 91%; (D) R248,251A, 80%; (E) R241A, 72%; (F) R240,244,251A, 62%; and (G) R241E, 0.8%.
The orientation of the α-helices modeled in Fig. 6 is the same as that in Fig. 1B, where the amino terminus is at the top. These structures represent the solvent-accessible surface of the model peptide. The electropositive potential of the molecule was projected onto this surface and is indicated by the scale bar in Fig. 6, where the most positive regions are mapped to deep blue and the most negative regions are mapped to red. As expected, the positive charge of the multiple arginine residues in the wild-type structure (Fig. 6A) contributes to the overall positive nature (blue) of the amino-terminal portion of the α-helix. A possible interaction between arginine-251 and glutamic acid-255 diminishes the contribution of both residues to the potential, and that portion of the model is predicted to be nearly neutral (orange).
The structures in Fig. 6 are arranged in rank order with respect to the ability of the corresponding mutant E4orf6 protein to retain the E1B 55-kDa protein in the nucleus. This ability was reported in Fig. 5 as the fraction of cells expressing both proteins with predominantly nuclear E1B 55-kDa protein. These values are approximately 100, 99, 90, 80, 70, 60 and 1% for the proteins represented by Fig. 6A through G, respectively. This analysis reveals that the positive charge of the amino-terminal portion of the amphipathic α-helix is linked to the ability of the E4orf6 protein to retain the E1B 55-kDa protein in the nucleus. Although this pattern is not strictly observed, as can be seen by comparing Fig. 6C and D, this trend was noted for all 26 of the structures described in this work (data not shown).
We also noted that the resulting electropositive surface of the molecular model was not readily predicted from the amino acid changes. For example, although both R244E and R241E bear a single arginine-to-glutamic acid substitution in the amino terminus of the α-helix, the predicted surface charge changed considerably more in the R241E model (Fig. 6G) than in the R244E model (Fig. 6C). Collectively, these results suggest that the identity of the amino acids at positions 241 and 243 as well as the overall electropositive nature of the amino terminus of the amphipathic α-helix of the E4orf6 protein govern the functional interaction of the E4orf6 and E1B 55-kDa proteins, as defined by colocalization in the nucleus during transient expression.
E4orf6 variants exhibit a similar ability to retain the E1B 55-kDa protein in the nucleus of Ad-infected cells as in transfected cells.
Representative E4orf6 protein variants were tested for their ability to retain the E1B 55-kDa protein in the nucleus during an Ad infection. HeLa cells were infected with the E4 deletion virus dl1014 and simultaneously transfected with the appropriate E4orf6 expression vector. When the E1B 55-kDa protein was expressed from the viral chromosome in the absence of any E4 products other than orf4, the E1B 55-kDa protein was predominantly cytoplasmic. The limited amount of E1B 55-kDa protein found in the nucleus appeared to occur as irregularly shaped aggregates (Fig. 7, vector).
The E4orf6 variants that retained the E1B 55-kDa protein in the nucleus of cells after transfection of both Ad genes also retained the E1B 55-kDa protein in the nucleus of dl1014-infected cells. The wild-type and R241A E4orf6 proteins were diffusely distributed through the nucleus and excluded from the nucleoli. In addition, some cells showed an accumulation of E4orf6 and E1B 55-kDa proteins at what appeared to be viral replication centers (Fig. 7, R241A) (46).
The E4orf6 variants that failed to retain the E1B 55-kDa protein in the nucleus after transfection largely failed to retain the E1B 55-kDa protein in the Ad-infected cells. The examples shown in Fig. 7 (R241E, R243E, and R240,251E) are representative of cells with no more E1B 55-kDa protein in the nucleus than vector-transfected cells. However, the extent to which the E1B 55-kDa protein remained excluded from the nucleus varied among cells within a sample, as was observed in cells expressing both proteins by transfection (see Fig. 4).
Curiously, whereas both the R241E and R243E proteins retained the E1B 55-kDa protein in the nucleus of fewer than 1% of the vaccinia virus-infected cells, approximately 10% of the dl1014-infected cells expressing the R241E or R243E variant showed significant E1B 55-kDa nuclear localization (data not shown). In addition, the intranuclear distribution of the E4orf6 protein variants that failed to retain the E1B 55-kDa protein in the nucleus differed from that observed in non-Ad-infected cells. In the Ad-infected cells, the glutamic acid substitution variants were concentrated towards the periphery of the nucleus and were not excluded from nucleoli (Fig. 7, R241E, R243E, and R240,251E). Nonetheless, each E4orf6 variant analyzed in Ad-infected cells showed a similar ability to direct E1B 55-kDa protein nuclear localization whether the E1B protein was expressed by transfection or from the viral chromosome.
Ability of the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein is not strictly linked to its ability to promote virus replication.
Several reports have suggested that physical and functional interaction of the E4orf6 and E1B 55-kDa proteins is important for late viral gene expression (9, 54) and intranuclear localization of the E1B 55-kDa protein during a viral infection (46) and to correct the growth defect of an E4 deletion virus (45). To determine if the limited ability of the mutant E4orf6 proteins to relocalize the E1B 55-kDa protein correlated with reduced function during Ad infection, 11 of the mutant E4orf6 proteins were tested for their ability to correct the growth defect of an E4 deletion virus, dl1014. Surprisingly, the results described below demonstrate that the ability of the E4orf6 protein to retain the E1B 55-kDa protein in the nucleus is not strictly correlated with the ability of the protein to promote virus growth.
To measure the extent to which the mutant E4orf6 proteins correct the growth of the E4 deletion virus, both the fast- and slow-growing strain of HeLa cells were infected with dl1014 and simultaneously transfected with the appropriate E4orf6 cDNA expression construct. After 2 days, the cells and growth medium were collected, and the amount of virus produced was quantified by plaque assay. The phenotypically wild-type virus dl309 replicated to levels approximately 300-fold (Fig. 8A) or 3,000-fold (Fig. 8B) above that of the E4 deletion virus dl1014. Expression of the wild-type E4orf6 protein did not affect replication of the wild-type virus (Fig. 8).
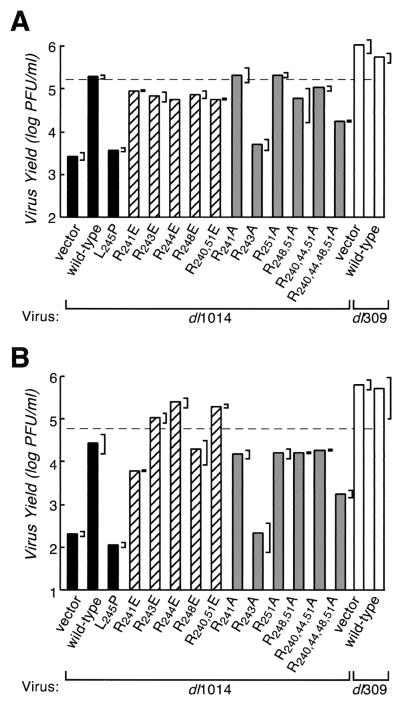
FIG. 8.
Ability of the E4orf6 protein to retain the E1B 55-kDa protein in the nucleus is neither necessary nor sufficient to correct the growth defect of an E4 deletion virus. A fast-growing strain of HeLa cells (A) and a slow-growing strain of HeLa cells (B) were infected with an E4 deletion virus lacking all E4 open reading frames except orf4 (dl1014) or a phenotypically wild-type virus (dl309) at 10 PFU per cell and simultaneously transfected with cDNAs expressing the E4orf6-related constructs listed below each bar. The E4orf6-related proteins were expressed under the control of the major immediate-early promoter of cytomegalovirus. Progeny virus was harvested after 48 h and quantified by plaque assay. A representative experiment (of three) showing the average amount of virus obtained from two independent infections is shown. The range of virus recovered in the two independent infections is indicated by the brackets on the right.
As reported previously (45), cells infected with dl1014 and simultaneously transfected with a wild-type E4orf6 expression vector produced over 200-fold more virus than cells transfected with an empty vector or a vector expressing the defective L245P E4orf6 mutant protein (Fig. 8 and data not shown). The transfection efficiency was determined to be 18% ± 2% and 13% ± 2% (average of six determinations ± standard deviation [SD]) for the cells analyzed in Fig. 8A and B, respectively. Therefore, the expected virus yield after transfection of a construct that fully restored E4 activity was 18 and 13% of the wild-type virus yield in the respective cell lines. The dashed lines in Fig. 8 indicate these values. To within a factor of 3, transfection of the wild-type E4orf6 expression vector restored the growth of the mutant virus to these levels.
The virus yield from cells infected with dl1014 and transfected with plasmids encoding the glutamic acid substitution variants R241E, R243E, R244E, R248E, and R240,251E was nearly the same (2- to 3-fold reduced) as that of dl1014-infected cells transfected with the wild-type E4orf6 construct (Fig. 8), with one exception. The exception, the R241E variant, nonetheless enhanced replication of the E4 mutant virus by nearly 100-fold in the slow-growing HeLa cell strain (Fig. 8B). We note that the extent to which the R248E variant supported dl1014 virus growth in this study is in good agreement with the results of others (47, 54).
We expected the R244E and R248E variants to promote virus growth because these E4orf6 variants behaved like the wild-type protein with respect to altering E1B 55-kDa protein localization. However, three of the glutamic acid substitution variants, R241E, R243E, and R240,251E, failed to retain the E1B 55-kDa protein in the nucleus, and their nearly wild-type function in this complementation assay was unexpected. Thus, it appears that ability to promote virus growth can be dissociated from the property of directing E1B 55-kDa protein to the nucleus.
The alanine substitution variants R251A, R248,251A, and R240,244,251A behaved as expected, based on their ability to direct nuclear localization of the E1B 55-kDa protein. Each of these variants retained the E1B 55-kDa protein in a majority of cells transiently expressing both proteins (Fig. 5), and each of these variants corrected the growth defect of dl1014 nearly as well as the wild-type E4orf6 protein (Fig. 8).
However, the behavior of other alanine substitution variants provided further support for the idea that virus growth enhancement can be dissociated from the property of E1B 55-kDa protein relocalization. The quadruple alanine substitution variant R240,244,248,251A failed to retain any E1B 55-kDa protein in the nucleus of over 400 cells evaluated that transiently expressed both proteins (Fig. 5). This E4orf6 variant restored the growth of dl1014 to 7 to 10% of the level associated with the wild-type E4orf6 protein. These values represent as much as a 300-fold increase over the yield of virus from infected cells transfected with the empty vector (Fig. 8B). Therefore, this E4orf6 variant has an intermediate ability to correct the growth defect of dl1014.
Cells infected with dl1014 and simultaneously transfected with the R241A expression construct produced approximately the same amount of virus as dl1014-infected cells transfected with the wild-type E4orf6 construct. This was measured in both the fast-growing HeLa cell strain (Fig. 8A), in which the R241A protein directed nuclear retention of the E1B 55-kDa protein in 73% of the cells (Fig. 5), and the slow-growing HeLa cell strain (Fig. 8B), in which only 16% of transfected cells contained predominantly nuclear E1B 55-kDa protein (data not shown). Thus, for these alanine substitution variants, the inability or reduced ability to direct nuclear retention of the E1B 55-kDa protein after transfection was not reflected in the ability of this variant to correct the growth defect of an E4 deletion virus.
In contrast to the nearly wild-type function of the R241A protein during a virus infection, the R243A protein was defective with respect to virus growth even though, by morphological criteria, the R243A variant resembled the R241A variant. The R243A variant directed nuclear retention of the E1B 55-kDa protein in 62% of the fast-growing HeLa cells (Fig. 5) and in 17% of the slow-growing HeLa cells (data not shown). However, both strains of HeLa cells infected with dl1014 and simultaneously transfected with the R243A expression construct failed to produce significantly more virus than dl1014-infected cells transfected with an empty plasmid vector (Fig. 8A and 8B). Thus, although preserving some ability to retain the E1B 55-kDa protein in the nucleus after transfection, the R243A protein appears to be unable to correct the growth defect of an E4 deletion virus. Together, these results lead us to conclude that the ability to direct nuclear localization of the E1B 55-kDa protein is neither sufficient nor necessary to complement the growth of the E4 deletion virus, dl1014, under the conditions of these assays.
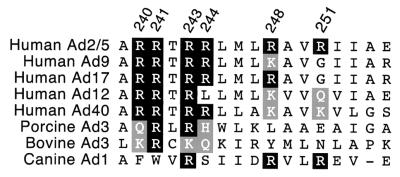
Key features of the E4orf6 amphipathic α-helix are conserved among adenoviruses.
To determine if the critical features of the arginine-faced amphipathic α-helix of the E4orf6 protein are conserved among different adenoviruses, we compared the predicted sequences of five human Ad E4orf6 proteins and three nonhuman Ad E4 proteins that are similar to the human Ad E4orf6 protein. The secondary-structure prediction algorithm of Chou and Fasman (14) confirmed that each protein could adopt an α-helical conformation at the carboxy terminus. These proteins exhibited overall identity to the Ad2 and Ad5 E4orf6 protein ranging from 62% (human Ad17) through 24% (bovine Ad3). The fraction of identical amino acids in the predicted α-helix region (Ad2 and Ad5 residues 239 through 255) also varied in a similar manner, from 88% (human Ad17) to 6% (bovine Ad3).
Alignment of these sequences reveals that the critical features of the amphipathic α-helix identified in this study may be conserved among this group of adenoviruses. In the alignment shown in Fig. 9, conserved arginine residues are shaded black. Basic amino acids found at the same position are shaded gray, and divergent amino acids are not shaded. With the exception of the canine Ad3 protein, each of these E4orf6 homologues contains a positively charged amino acid at the position equivalent to arginine-241 and arginine-243. All of these proteins preserve a net positive charge on the residues (240, 244, 248, and 251) that comprise the central face of the α-helix. Thus, the key features of the arginine-faced amphipathic α-helix required for E1B 55-kDa protein nuclear localization are conserved among these viruses. The conservation of a positively charged amino acid at the position corresponding to arginine-243 may reflect a more general requirement for a charged amino acid at position 243 for a productive Ad infection.
FIG. 9.
Key features of the amphipathic α-helix are conserved among different adenoviruses. The predicted amino acid sequence of five human Ad E4orf6 proteins and three nonhuman Ad E4 proteins that are similar to the human Ad E4orf6 protein were aligned at the region corresponding to the amphipathic α-helix. The arginine residues in the Ad2 and Ad5 proteins are identified at the top of the alignment. Arginines that occur at the same position in the other proteins are shaded black, basic amino acids in these positions are shaded gray, and divergent amino acids are not shaded.
DISCUSSION
We previously demonstrated that an arginine-faced amphipathic α-helix centered at amino acid 245 is essential for the function of the E4orf6 protein during a productive Ad5 infection (45). The integrity of this amphipathic α-helix is also required for the E4orf6 protein to retain the predominantly cytoplasmic E1B 55-kDa protein in the nucleus of primate cells when both proteins are expressed by transfection (23, 45). We interpret the changes in cellular localization of the E1B 55-kDa protein attributed to the E4orf6 protein to reflect a functional interaction between these proteins in the cell. We and others have suggested that this functional interaction between the E4orf6 and E1B 55-kDa proteins may be correlated with the ability of E4orf6 protein to correct the growth defect of an E4 deletion virus (9, 45, 54). However, the properties of E4orf6 variants created in this study lead us to conclude that the ability to retain the E1B 55-kDa protein in the nucleus is neither necessary nor sufficient to correct the growth defect of an E4 deletion virus.
In this work, we found that arginine-241 and arginine-243 of the E4orf6 protein were critical for the E4orf6 protein to direct nuclear localization of the E1B 55-kDa protein in transfected cells (Fig. 4 and 5). The remaining four arginine residues of the α-helix contribute a net positive charge that is needed to preserve the functional interaction of the E4orf6 and E1B 55-kDa proteins in cells. Notably, homologous proteins of other human and nonhuman Ad serotypes also contain arginine or lysine residues at positions corresponding to 241 and 243 and also maintain a net positive charge among the residues corresponding to the other four arginines (Fig. 9).
Surprisingly, these determinants may not be the same as those required for E4orf6 protein function during productive Ad infection. Four E4orf6 variants that failed to relocalize the E1B 55-kDa protein to the nucleus in transfected cells, R241E, R243E, R240,251E, and, to a limited extent, R240,244,248,251A, corrected the growth defect of an E4 deletion virus that expressed only the orf4 protein of E4 (Fig. 8). If redistribution of the E1B 55-kDa protein to the nucleus reflects a functional interaction with the E4orf6 protein, then these mutant E4orf6 proteins appear to be less dependent on an interaction with the E1B 55-kDa protein to support a productive Ad infection. Additionally, another E4orf6 variant, R243A, retained the E1B 55-kDa protein in the nucleus in transfected cells and during an Ad infection (data not shown) but failed to correct the growth defect of the E4 deletion virus. Thus, the ability to direct nuclear localization of the E1B 55-kDa protein may not be sufficient for E4orf6 protein function during a productive Ad infection.
The E4orf6 variants described here demonstrate that, for the E4orf6 protein, the ability to direct nuclear localization of the E1B 55-kDa protein can be separated from the ability to support a productive infection. Why might some of the mutant E4orf6 proteins support virus growth in the apparent absence of a functional interaction with the E1B 55-kDa protein?
One possible explanation for the behavior reported here is that the E4orf6 protein variants were overexpressed under control of the cytomegalovirus immediate-early promoter for the virus complementation experiments. Under these conditions, the amount of E4orf6 protein in each cell was 20- to 50-fold greater than that present during a normal Ad infection. Perhaps a partial defect in the protein was masked at such nonphysiologically high concentrations. Indeed, when the E1B 55-kDa protein was expressed from the viral chromosome, some of the E4orf6 variants that failed to direct nuclear localization of the E1B 55-kDa protein after transfection did so in a fraction of the Ad-infected cells (Fig. 7 and data not shown).
Another possible explanation for the apparent separation of E4orf6-E1B 55-kDa protein interaction and virus complementation among the mutant E4orf6 proteins is that the mutant E4orf6 proteins may bind different cellular factors to perform various functions. It seems plausible that the R243A variant could bind cellular factors needed for E4orf6-E1B 55-kDa protein interaction in cells, but not those factors required for a productive Ad infection. Conversely, it would have to be argued that the R241E, R243E, and R240,251E variants bind cellular factors required for virus growth but not those required for interaction with the E1B 55-kDa protein. Note that this hypothesis requires that the glutamic acid substitution E4orf6 variants have the ability to bind the cellular factors independently of the E1B 55-kDa protein. Perhaps, for example, the negative charge on the hydrophilic face of the glutamic acid replacement E4orf6 protein somehow mimicked a change brought about by association with the E1B 55-kDa protein.
A third possible explanation for the behavior of the R241E, R243E, and R240,251E variants is that their function during an Ad infection is less dependent on the E1B 55-kDa protein than the wild-type E4orf6 protein. Perhaps, for example, one role for the E4orf6-E1B 55-kDa protein complex is to promote proper folding and presentation of the C-terminal amphipathic α-helix in the E4orf6 protein. Therefore, changes to the E4orf6 protein that stabilize this amphipathic α-helix without destroying key contacts for other interactions may diminish the need for a physical interaction with the E1B 55-kDa protein. We suggest that this admittedly helix-centric view is not entirely without merit. Indeed, the charge substitutions in R241E, R243E, and R240,251E are predicted to enhance the intrinsic stability of the α-helix through electrostatic interactions between oppositely charged residues separated by three or four positions on the α-helix (33, 39). Thus, if the ability of the E4orf6 amphipathic α-helix to form properly is a limiting step for E4orf6 protein function during Ad infection, the role of the E1B 55-kDa protein in the E4orf6-E1B 55-kDa complex may be to establish or stabilize the C-terminal amphipathic α-helix in the E4orf6 protein.
This hypothesis may also provide an explanation for the cold-sensitive phenotype of E1B mutant viruses (see, for example, reference 22). Proper formation of the amphipathic α-helix in the E4orf6 protein probably depends on hydrophobic interactions within the E4orf6 protein. If the proper configuration of the E4orf6 protein is promoted by an interaction with the E1B 55-kDa protein, then this configuration might be cold-labile in the absence of the E1B 55-kDa protein. At low temperatures, at which hydrophobic interactions are weaker, the E4orf6 protein would fold ineffectively, and the yield of an E1B mutant virus would be reduced compared to the yield at 37°C. Conversely, at higher temperatures, at which hydrophobic interactions are stronger, the E4orf6 protein may fold independently of the E1B 55-kDa protein, allowing the E1B mutant virus to replicate to nearly wild-type levels.
Recently, Shen and associates described variant E1B 55-kDa proteins that failed to bind the E4orf6 protein in a coimmunoprecipitation assay but still supported late viral gene expression and host cell shutoff in Ad-infected A549 cells (51). This finding suggests that mutant E1B 55-kDa proteins exist that may support virus growth in the absence of a functional interaction with the E4orf6 protein. Additionally, these investigators described one E1B 55-kDa protein variant that bound the E4orf6 protein less effectively than the wild-type E1B 55-kDa protein. Intriguingly, the corresponding mutant virus displayed a cold-sensitive phenotype intermediate between that of the E1B 55-kDa protein-null virus and wild-type virus (51).
Although clearly speculative, these hypotheses provide readily tested implications, the pursuit of which should increase our understanding of the relationship between the E1B 55-kDa and E4orf6 proteins. One immediate goal is to determine the function of these mutant E4orf6 proteins when they are expressed from the viral chromosome. Some of these mutant genes are being introduced into both the wild-type Ad background and viruses bearing additional mutations for this purpose.
Arginine-rich, amphipathic α-helices have been implicated in protein-protein interaction in at least three other proteins. First, the Nef protein of human immunodeficiency virus contains an amino-terminal arginine-rich amphipathic α-helix that binds to the Lck kinase and a serine kinase that phosphorylates both Nef and Lck (5). The interaction between the Nef amphipathic α-helix and the two kinases appears to be necessary for maximum virion infectivity. Second, the NS-1 protein of influenza A virus is a dimeric, helix-rich protein that binds RNA (36). Helix 3 of NS-1 is an amphipathic α-helix that appears to contribute to dimer formation (53). Third, the 14-3-3 proteins are ubiquitous proteins involved in a number of processes, including mitogenic signal transduction, cell cycle progression, and oncogenesis (reviewed in references 4 and 19).
The 14-3-3 proteins bind a wide variety of protein ligands, including the cellular Raf-1 kinase and a bacterial ADP-ribosyltransferase. The three-dimensional crystal structure of the 14-3-3 ζ-isoform shows that the ligand-binding surface, which is present in all 14-3-3 proteins, is lined by amphipathic α-helices rich in basic amino acids (35). Changes to the conserved basic residues on the hydrophilic face selectively disrupted the interaction with some ligands but not with others (57). Perhaps the E4orf6 protein can also bind multiple and diverse ligands through similar interactions via the C-terminal arginine-faced amphipathic α-helix.
In conclusion, we have elucidated some of the requirements of the arginine-faced amphipathic α-helix of the E4orf6 protein for a functional interaction with the E1B 55-kDa protein and for E4orf6 protein function during Ad infection. We suggest that the requirements for these two properties are not identical. Perhaps one role for the arginine-faced amphipathic α-helix of the E4orf6 protein is to mediate protein-protein interactions with multiple cellular factors through different arginine residues on the hydrophilic face. Clearly, however, additional work is needed to confirm the structure proposed by Flint and associates (13), to identify the cellular factors used by the E4orf6 protein to promote virus growth (6), and to understand the relationship between the E1B 55-kDa and E4orf6 proteins that contributes to the multiple activities of the E4orf6 oncoprotein.
Acknowledgments
This work was supported by Public Health Service grants AI35589 from the National Institute of Allergy and Infectious Diseases and CA77342 from the National Cancer Institute. J.S.O. was supported in part by NIH training grant T32 AI07401 to the Department of Microbiology and Immunology. Tissue culture reagents and services were provided by the Tissue Culture Core Laboratory, and DNA sequencing was performed by the DNA Synthesis Core Laboratory, both of the Comprehensive Cancer Center of Wake Forest University, supported in part by NIH grant CA12197.
We gratefully acknowledge Al Claiborne, Mark Lively, Maria McGee, and Leslie Poole for help with molecular modeling and protein structure analysis and Ken Grant of the Micromed facility for assistance with confocal laser scanning microscopy. We thank Susan Sun and Jeff Childers for technical assistance and Amy Chastain, Felicia Goodrum, Roy Hantgan, Doug Lyles, Griff Parks, and Robin Shepard for providing valuable advice on the work in progress and on manuscript preparation.
REFERENCES
- 1.Alexander, I. E., D. W. Russell, and A. D. Miller. 1994. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 68:8282–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armentano, D., C. C. Sookdeo, K. M. Hehir, R. J. Gregory, J. A. St. George, G. A. Prince, S. C. Wadsworth, and A. E. Smith. 1995. Characterization of an adenovirus gene transfer vector containing an E4 deletion. Hum. Gene Ther. 6:1343–1353. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915. [DOI] [PubMed] [Google Scholar]
- 4.Baldin, V. 2000. 14-3-3 proteins and growth control. Prog. Cell Cycle Res. 4:49–60. [DOI] [PubMed] [Google Scholar]
- 5.Baur, A. S., G. Sass, B. Laffert, D. Willbold, C. Cheng-Mayer, and B. M. Peterlin. 1997. The N terminus of Nef from HIV-1/SIV associates with a protein complex containing Lck and a serine kinase. Immunity 6:283–291. [DOI] [PubMed] [Google Scholar]
- 6.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boles, E., and T. Miosga. 1995. A rapid and highly efficient method for PCR-based site-directed mutagenesis using only one new primer. Curr. Genet. 28:197–198. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307–312. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, J. L., and G. Ketner. 2000. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 275:14969–14978. [DOI] [PubMed] [Google Scholar]
- 10.Bridge, E. 2000. The nuclear export signal within the adenovirus E4orf6 protein contributes to several steps in the viral life cycle. J. Virol. 74:12000–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345–353. [DOI] [PubMed] [Google Scholar]
- 12.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, L. M., R. A. Gonzalez, J. Novotny, and S. J. Flint. 2001. Structure of the adenovirus E4 Orf6 protein predicted by fold recognition and comparative protein modeling. Proteins 44:97–109. [DOI] [PubMed] [Google Scholar]
- 14.Chou, P. Y., and G. D. Fasman. 1978. Empirical predictions of protein conformation. Annu. Rev. Biochem. 47:251–276. [DOI] [PubMed] [Google Scholar]
- 15.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470–1473. [DOI] [PubMed] [Google Scholar]
- 17.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25–54. [DOI] [PubMed] [Google Scholar]
- 18.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617–647. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74. [DOI] [PubMed] [Google Scholar]
- 25.Grifman, M., N. N. Chen, G. P. Gao, T. Cathomen, J. M. Wilson, and M. D. Weitzman. 1999. Overexpression of cyclin A inhibits augmentation of recombinant adeno-associated virus transduction by the adenovirus E4orf6 protein. J. Virol. 73:10010–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higashino, F., J. M. Pipas, and T. Shenk. 1998. Adenovirus E4orf6 oncoprotein modulates the function of the p53-related protein, p73. Proc. Natl. Acad. Sci. USA 95:15683–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683–689. [DOI] [PubMed] [Google Scholar]
- 30.Jones, N., and T. Shenk. 1978. Isolation of deletion and substitution mutants of adenovirus type 5. Cell 13:181–188. [DOI] [PubMed] [Google Scholar]
- 31.Ketner, G., E. Bridge, A. Virtanen, C. Hemstrom, and U. Pettersson. 1989. Complementation of adenovirus E4 mutants by transient expression of E4 cDNA and deletion plasmids. Nucleic Acids Res. 17:3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850–857. [DOI] [PubMed] [Google Scholar]
- 33.Lacroix, E., A. R. Viguera, and L. Serrano. 1998. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 284:173–191. [DOI] [PubMed] [Google Scholar]
- 34.Leppard, K. N. 1997. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 78:2131–2138. [DOI] [PubMed] [Google Scholar]
- 35.Liu, D., J. Bienkowska, C. Petosa, R. J. Collier, H. Fu, and R. Liddington. 1995. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376:191–194. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896–899. [DOI] [PubMed] [Google Scholar]
- 37.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz, V., and L. Serrano. 1995. Elucidating the folding problem of helical peptides using empirical parameters. III. Temperature and pH dependence. J. Mol. Biol. 245:297–308. [DOI] [PubMed] [Google Scholar]
- 40.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 94:1206–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 2000. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J. Virol. 74:5168–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevels, M., T. Spruss, H. Wolf, and T. Dobner. 1999. The adenovirus E4orf6 protein contributes to malignant transformation by antagonizing E1A-induced accumulation of the tumor suppressor protein p53. Oncogene 18:9–17. [DOI] [PubMed] [Google Scholar]
- 43.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 75:3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicolas, A. L., P. L. Munz, E. Falck-Pedersen, and C. S. Young. 2000. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology 266:211–224. [DOI] [PubMed] [Google Scholar]
- 45.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha-helix is required for adenovirus type 5 e4orf6 protein function. J. Virol. 73:4600–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabino, C., A. Aspegren, K. Corbin-Lickfett, and E. Bridge. 2000. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 74:6684–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404–407. [PubMed] [Google Scholar]
- 50.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517. [DOI] [PubMed] [Google Scholar]
- 51.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steegenga, W. T., A. Shvarts, N. Riteco, J. L. Bos, and A. G. Jochemsen. 1999. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol. Cell. Biol. 19:3885–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinberg, D. H., and G. Ketner. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 80:5383–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44–58. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, L., H. Wang, D. Liu, R. Liddington, and H. Fu. 1997. Raf-1 kinase and exoenzyme S interact with 14-3-3ζ through a common site involving lysine 49. J. Biol. Chem. 272:13717–13724. [DOI] [PubMed] [Google Scholar]