Abstract
Replication of hepatitis A virus (HAV) in cultured cells is inefficient and difficult to study due to its protracted and generally noncytopathic cycle. To gain a better understanding of the mechanisms involved, we constructed a subgenomic HAV replicon by replacing most of the P1 capsid-coding sequence from an infectious cDNA copy of the cell culture-adapted HM175/18f virus genome with sequence encoding firefly luciferase. Replication of this RNA in transfected Huh-7 cells (derived from a human hepatocellular carcinoma) led to increased expression of luciferase relative to that in cells transfected with similar RNA transcripts containing a lethal premature termination mutation in 3Dpol (RNA polymerase). However, replication could not be confirmed in either FrhK4 cells or BSC-1 cells, cells that are typically used for propagation of HAV. Replication was substantially slower than that observed with replicons derived from other picornaviruses, as the basal luciferase activity produced by translation of input RNA did not begin to increase until 24 to 48 h after transfection. Replication of the RNA was reversibly inhibited by guanidine. The inclusion of VP4 sequence downstream of the viral internal ribosomal entry site had no effect on the basal level of luciferase or subsequent increases in luciferase related to its amplification. Thus, in this system this sequence does not contribute to viral translation or replication, as suggested previously. Amplification of the replicon RNA was profoundly enhanced by the inclusion of P2 (but not 5′ noncoding sequence or P3) segment mutations associated with adaptation of wild-type virus to growth in cell culture. These results provide a simple reporter system for monitoring the translation and replication of HAV RNA and show that critical mutations that enhance the growth of virus in cultured cells do so by promoting replication of viral RNA in the absence of encapsidation, packaging, and cellular export of the viral genome.
Hepatitis A virus (HAV) is a small, nonenveloped, positive-strand RNA virus belonging to the family Picornaviridae (genus Hepatovirus). It is transmitted via the fecal-oral route and causes acute viral hepatitis. Like other picornaviruses, HAV has a genome consisting of a lengthy 5′ nontranslated region (5′ NTR), a single large open reading frame, and a short 3′ NTR followed by a poly(A) tail (28). The 5′ NTR forms complex secondary and tertiary structures and is likely to contain signals required for viral RNA replication in addition to directing the initiation of translation of the viral polyprotein via an internal ribosome entry site (IRES) within its sequence (5). The P1 segment of the open reading frame encodes the capsid proteins, VP4 (which is small and has not yet been detected in infectious virus particles), VP2, VP3, and VP1. The P2 and P3 regions encode nonstructural proteins that are believed to be involved in replication of the viral RNA. P2 encodes three proteins: 2A, 2B, and 2C. Although the exact function of each is not defined, 2C has an nucleoside triphosphatase motif, while 2B, acting alone or together with 2C, may be involved in directing membrane rearrangements essential for replication of the RNA (19). The P3 segment encodes four proteins: 3A, 3B, 3C, and 3D. 3B (VPg) becomes covalently linked to the 5′ end of genomic RNA (45), while 3Cpro is the only proteinase encoded by the virus and is responsible for most cleavages in the proteolytic processing of the polyprotein (3, 22, 31). Finally, 3Dpol contains sequence motifs suggesting that it is the RNA-dependent RNA polymerase.
Wild-type HAV replicates very poorly in cultured cells but adapts as it is passaged in vitro. The critical mutations responsible for this cell culture adaptation phenotype are located in the P2 region and the 5′ NTR, but mutations elsewhere in the genome may contribute to the replication properties of highly adapted, rapidly replicating, cytopathic virus variants, such as HM175/18f (11, 13, 15, 27, 48). Many of these mutations do not work independently in increasing replication but rather act cooperatively with mutations elsewhere in the genome (48). Different mutations have been identified in HAV strains that have been adapted independently to growth in different cells. However, certain mutations in the 2B and 2C proteins appear to be most important for replication of the virus in cultured cells. In particular, a C-to-T substitution at nucleotide (nt) 3889 that encodes an Ala-to-Val substitution at residue 1052 of the polyprotein (2B) has been identified in several different cell culture-adapted HAV variants (9, 12, 21, 24).
Despite the identification of mutations that enhance the growth of HAV in cultured cells, there is little understanding of the steps in the viral life cycle at which replication of the wild-type virus is restricted. The fact that mutations in the 2B and 2C proteins enhance the growth of virus suggests indirectly that the synthesis of new copies of viral RNA may be restricted in cultured cells (9, 12, 21, 24). However, other than the location of these mutations, there is no direct evidence that supports this hypothesis over a restriction at some other step in the viral life cycle. In contrast, there is direct evidence that sequence changes within the 5′ NTR enhance viral replication by increasing the translational activity of the viral IRES in a cell type-specific fashion (11, 16, 42). Consistent with the notion that RNA replication may be restricted, Anderson et al. have hypothesized further that RNA synthesis is hindered by an unusually high frequency of encapsidation of plus-strand RNA, effectively sequestering potential template molecules and removing them from the pool of replicating RNAs (1).
In an effort to test these two hypotheses, we constructed autonomously replicating, subgenomic HAV RNAs that lack sequence encoding the viral capsid proteins. These replicons are reminiscent of putative defective genomes reported in early descriptions of HAV-infected cell cultures as well as in clinical samples from patients with hepatitis A (35, 36). These RNAs undergo replication in Huh-7 cells, which are derived from human hepatocellular carcinoma cells. Using this novel system, we show that P2 (but not 5′ NTR) mutations associated with adaptation of the virus to growth in cell culture contribute directly to greater efficiency of viral RNA replication in these cells. However, replication of the viral RNA remains very slow and inefficient compared to that of other picornaviruses, even in the absence of protein expression leading to RNA encapsidation.
MATERIALS AND METHODS
Cells.
Huh-7 cells and Frhk-4 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. BSC-1 cells were grown in 1× minimum essential medium supplemented with 10% FBS and antibiotics. BT7-H (46) and Huh-T7 (42) cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, antibiotics, and 400 μg of Geneticin (G418) per ml.
Plasmids.
pT7-18f was the parent recombinant plasmid used to construct HAV replicons. It contains a cDNA copy of the sequence of a rapidly replicating, cytopathic variant of HAV, HM175/18f, under the control of the T7 promoter (2). A silent nucleotide substitution (A to T) was made at nt 843 by site-directed mutagenesis (QuikChange; Stratagene), resulting in creation of a unique StuI site. The firefly luciferase (F-Luc) sequence was amplified by PCR using primers designed to contain StuI and SacI sites at the 5′ and 3′ ends of the fragment. The resulting product was ligated into the StuI and SacI sites of the modified pT7-18f to generate pHAV-Luc, which contains the F-Luc sequence as an in-frame replacement for most of the VP2-VP1-coding sequence. pHAV-LucΔ3D was constructed from this plasmid by further introducing a one-nucleotide change at nt 6084 (G to T), generating a termination codon within the 3Dpol sequence. pHAV-LucΔVP4 is an additional replicon in which the P1 deletion includes all of the VP2 and most of the VP4-coding sequence. For its construction, the F-Luc sequence was amplified by PCR with primers designed to contain NheI and SacI sites at the 5′ and 3′ termini of the product. The resulting fragment was ligated to pT7-18f following its digestion with XbaI and SacI.
pHAV-Luc-wP2P3 was generated from pHAV-Luc by substituting the SacI-XhoI fragment with the related sequence from an infectious wild-type cDNA clone of HM175 virus (14). pHAV-Luc-wP2 and pHAV-Luc-wP2-3889 were constructed in a similar fashion, by substitution of the SacI-EcoRI segments of pHM175-wt and pHM175-8y (12). For construction of pw5′NTR-HAV-Luc, the NcoI-XbaI segment of pHAV-Luc was replaced with the comparable segment from pHM175-wt. All constructs were confirmed by restriction enzyme digestion and limited DNA sequencing.
Plasmids containing bicistronic T7 transcriptional IRES reporter units, pFLuc-WT-RLuc and pFLuc-p16-RLuc, were derived from pLUC-WT-CAT and pLUC-p16-CAT (42), respectively, by digestion with XbaI-NotI and substitution of the chloramphenicol acetyltransferase-coding sequence with sequence encoding Renilla luciferase (R-Luc). To generate an HAV-specific hybridization probe, the small PstI-ApaI fragment of pT7-18f (nt 5134 to 5687) was ligated to the ApaI-PstI sites of pBluescript SK (Stratagene), placing the antisense HAV sequence under the control of the T7 promoter.
In vitro transcription and transfection of replicon RNA.
Replicon plasmids were linearized at a unique XmaI site located at the 3′ end of the HAV sequence. RNA transcription was carried out with a T7 MEGAscript in vitro transcription kit (Ambion) according to the manufacturer’s suggested protocol. Approximately 0.5 μg of RNA was used to transfect cells in each well of a 12-well plate (∼5 × 104 cells) using a liposome-mediated transfection procedure (Lipofectin; Life Technologies). Briefly, 30 μl of Lipofectin was diluted in 400 μl of Optimem (Gibco BRL). Following a 1-h incubation, 3 μg of RNA was added to the Lipofectin mixture and the mixture was incubated for an additional 15 min, after which 70 μl of the RNA-Lipofectin mixture was added to each well of a 12-well culture plate. Following 12 h of incubation, the transfection mixture was removed from the cells, and the cells were washed twice with phosphate-buffered saline and fed with normal growth medium.
DNA transfections.
DNA transfections were carried out with FuGENE 6 transfection reagent (Boehringer Mannheim). About 1 × 105 to 2 × 105 cells/2 ml medium were plated into each well of a six-well plate 1 day prior to transfection. For each transfection, 2 μg of DNA was mixed with 6 μl of FuGENE reagent diluted with 94 μl of Optimem (Gibco BRL) and incubated for 15 min at room temperature. The DNA-FuGENE complex was then added directly to the cells. Cells were harvested after 24 or 48 h for determination of F-Luc and R-Luc activities.
Luciferase assays.
F-Luc activity was assayed with the luciferase reporter assay system (Promega), while the dual-luciferase reporter assay system (Promega) was used to assay both F-Luc and R-Luc activities in the same cell lysate. Briefly, cells were washed twice with phosphate-buffered saline, and 200 μl of passive lysis buffer (Promega) was added to each well of a 12-well plate (500 μl for each well of a six-well plate). The culture plates were placed at room temperature for 30 min prior to collection of the lysate. Aliquots (20 μl) of each lysate were monitored for a luminescent signal in a TD-20/20 luminometer (Turner Design) equipped with a dual injector according to the protocols supplied by the manufacturer.
Guanidine inhibition of replicon amplification.
Guanidine stock solution (50 mM) was diluted to various concentrations in growth medium and stored at 4°C. Twelve hours following transfection of replicon RNAs, the transfection mixture was removed from the cells and replaced with medium containing guanidine. To ensure the continued presence of guanidine in the medium, the guanidine-containing medium was replaced every 48 h.
Dot blot RNA-RNA hybridization.
Total cellular RNA was extracted with TRIzol (Gibco BRL) as recommended by the manufacturer, mixed with formaldehyde loading dye, incubated at 65°C for 15 min, and blotted onto a BrightStar-Plus nylon membrane (NorthernMax; Ambion), using a 96-well filtration manifold (Bio-Dot; Bio-Rad). The membrane was subsequently hybridized in ultrahybridization solution (NorthernMax) against an HAV-specific [α32-P]CTP RNA probe (see above). After a wash in low-stringency buffer at room temperature followed by two washes in high-stringency buffer at 68°C, the membrane was analyzed with a PhosphorImager (Molecular Dynamics) to determine the intensity of the hybridization signal.
RESULTS
Self-amplification of a subgenomic HAV replicon.
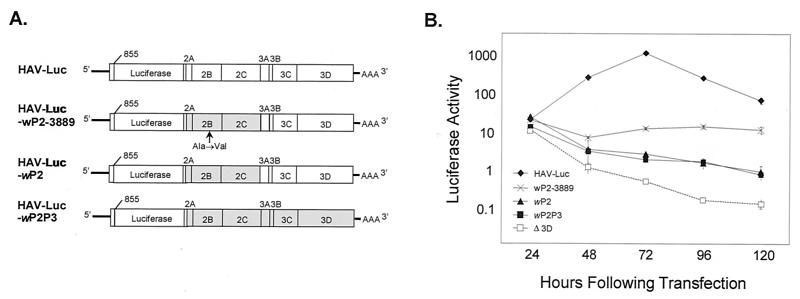
Self-amplifying, subgenomic RNA replicons have been constructed from the genomes of several picornaviruses, including poliovirus, human rhinovirus, and foot-and-mouth disease virus, by the deletion of some or all of the P1 capsid-coding sequence (25, 32, 33). As with their parent viruses, the replication of these subgenomic RNAs occurred rapidly, with maximum amplification of RNA taking place within 24 h of transfection into permissive cells. To determine whether analogous, P1-deletion-containing, subgenomic HAV RNAs are capable of self-amplification, we constructed pHAV-Luc (Fig. 1) by deleting most of the P1 segment from pT7-18f, a genomic-length, infectious cDNA copy of a rapidly replicating, cytopathic variant of HAV HM175. To facilitate detection of RNA amplification, the deleted P1 segment was replaced with an in-frame insertion of sequence encoding the reporter F-Luc. We included in this construct a sequence encoding the small, putative VP4 (1A) protein of HAV as well as the amino-terminal 13 amino acids of VP2 (1B), since a recent report suggests that HAV IRES activity is significantly enhanced when the 114 nt downstream of the initiator AUG codon are present in their natural context (20). In addition, because the precise mechanism of the VP1 (1D)/2A cleavage is not known (22, 31), we included 39 nt of the VP1 sequence upstream of the putative VP1/2A junction. As a negative control, we constructed a mutated form of pHAV-Luc (pHAV-LucΔ3D) (Fig. 1) in which a single nucleotide change results in premature termination of translation within the RNA polymerase (3Dpol)-coding region. All other features of this mutant were identical to those of pHAV-Luc.
FIG. 1.
Genomic organization of HAV and various replicon constructs. The organization of the HAV genome is at the top, with the P1 sequence highlighted (solid region). Below is the organization of the replicons. In HAV-Luc, the sequence encoding F-Luc (shaded region) was inserted in frame, replacing all but the 5′ 120 nt and 3′ 39 nt of the P1 segment, in an infectious cDNA clone derived from the rapidly replicating, cytopathic HM175/18f virus. HAV-LucΔVP4 is identical to HAV-Luc except that further sequence has been removed from the 5′ end of the P1 segment, leaving only 12 nt of the VP4-coding sequence. Similarly, HAV-LucΔ3D is identical to HAV-Luc except for a single nucleotide change resulting in premature termination of translation within the 3Dpol-coding sequence.
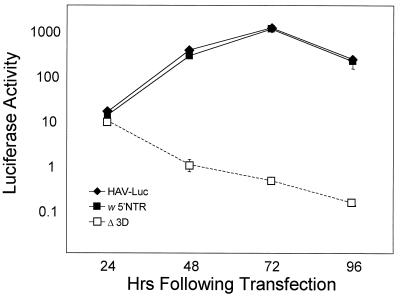
Amplification of the subgenomic RNAs transcribed from these plasmids was monitored by measuring intracellular luciferase activities following transfection of the RNA into cultured Huh-7 cells. As shown in Fig. 2A, transfection of HAV-Luc RNA resulted in an increase in luciferase activity between 24 and 48 h following transfection. Luciferase activity was maximal 72 h following transfection and then declined. In contrast, transfection of HAV-LucΔ3D RNA resulted in no increase in luciferase activity from the baseline value 6 h after transfection, with a steady decline in luciferase activity observed after 24 h. Since there was no difference in the luciferase activities expressed by HAV-Luc and HAV-LucΔ3D up to 24 h after transfection, this early, basal luciferase activity must be derived from translation of the input RNA. The subsequent increases in luciferase expressed from HAV-Luc, but not HAV-LucΔ3D, suggest strongly that HAV-Luc RNA undergoes replication in Huh-7 cells. Compared with similar replicons constructed from other picornaviruses, however, the increase in luciferase activity associated with RNA replication took place after a significantly greater initial delay period (3 h in the case of rhinovirus replicons [33] versus 24 h with HAV-Luc) and with a lower rate of subsequent accumulation.
FIG. 2.
Luciferase activity (in light units) in lysates of Huh-7 cells transfected with the replicon RNAs shown in Fig. 1. Luciferase activity 24 h following transfection reflects translation of input T7 transcript RNA, while subsequent increases in luciferase activity are produced from newly replicated subgenomic RNAs. Note the logarithmic scale for luciferase activity. (A) Replication of HAV-Luc RNA is reflected in increases in luciferase activity following transfection of HAV-Luc (□k) but not HAV-LucΔ3D (□() transcript RNA in Huh-7 cells. (B) VP4-coding sequence is not required for self-amplification of replicon RNAs. □k, HAV-LucΔVP4; □(, HAV-LucΔ3D.
To confirm this difference in kinetics, we transfected Huh-7 cells with ΔP1Luc/cre, a subgenomic human rhinovirus type 14 replicon in which the luciferase-coding sequence replaces most of the P1 capsid-coding RNA (33). Although Huh-7 cells have not been reported previously to be permissive for rhinovirus replication, cells transfected with ΔP1Luc/cre RNA expressed abundant luciferase (data not shown). This luciferase activity peaked at 12 h posttransfection, approximately 60 h prior to the peak luciferase activity detected in Huh-7 cells transfected with HAV-Luc (Fig. 2), and was not observed in cells transfected with a replication-defective rhinovirus RNA, ΔP1Luc, lacking the viral cis-acting replication element (CRE) (33) (data not shown). Thus, the kinetics of HAV-Luc replication reflect the generally slower replication characteristics of HAV, which are evident even with a “rapidly replicating,” cell culture-adapted variant such as HM175/18f (27).
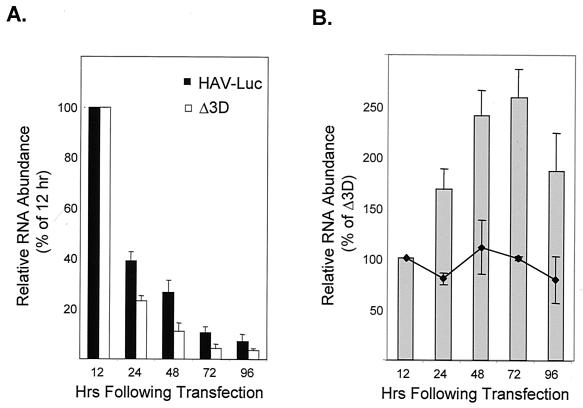
To directly assess the replication of the transfected RNA, we ascertained the abundance of intracellular HAV RNA by dot blot hybridization following transfection of Huh-7 cells. These studies showed that a substantial amount of RNA was associated with the cells 12 h after the start of the transfection procedure but documented only decreasing amounts of HAV-Luc RNA associated with the cells over the ensuing 84 h (Fig. 3A). Nonetheless, there was a significant difference in the rates at which HAV RNA declined in abundance in cells transfected with HAV-Luc and in those transfected with the replication-deficient mutant, HAV-LucΔ3D. This was reflected in an increase in the ratio of RNA abundance in HAV-Luc-transfected cells to that in cells transfected with the mutant RNA at 24 h (Fig. 3B), even though differences in luciferase activity were not yet apparent (Fig. 2). This difference in the relative abundances of RNA in cells transfected with the replicon and its related 3Dpol mutant peaked at 48 to 72 h and declined subsequently, suggesting a pattern of RNA synthesis that preceded increases in luciferase activity by about 12 h but that was consistent with the kinetics of the luciferase response. The difference in RNA abundance observed following transfection with HAV-Luc and HAV-LucΔ3D was not evident when cells were transfected in the presence of 2 mM guanidine (Fig. 3B), suggesting that guanidine inhibits HAV RNA replication at this molar concentration and that the difference in RNA abundance is not due to subtle variation in the stability of these RNAs following transfection. Although these results provide strong support for the replication of the HAV-Luc RNA, they suggest that much of the transfected RNA failed to undergo replication.
FIG. 3.
Quantitative analysis of dot blot hybridization assays for cell-associated HAV RNA following transfection of Huh-7 cells with replicon RNA. (A) Relative abundance of HAV-Luc and HAV-LucΔ3D RNA in transfected cells. PhosphorImager values were normalized with respect to the hybridization signal present 12 h after transfection of each transcript. Results are means calculated from triplicate hybridizations against different dilutions of the extracted total cellular RNA. Although there are sequential declines in the abundance of each RNA over time following transfection, the rate of decline in the abundance of HAV-Luc RNA differed from that of HAV-LucΔ3D as shown in panel B. (B) Shaded bars represent the abundance of HAV-Luc RNA relative to the abundance of HAV-LucΔ3D at various times following transfection. Values are normalized to the relative RNA abundance present 12 h after transfection of the transcripts, prior to any evidence of RNA replication. The line represents the relative abundance of the two RNAs in cells treated with 2 mM guanidine. Values are means calculated from duplicate or triplicate hybridizations against different dilutions of extracted total cellular RNA.
Host cell range of subgenomic HAV replicons.
Primary African green monkey kidney (AGMK) cells and the continuous cell lines BSC-1 (derived from AGMK cells) and FRhK-4 (fetal rhesus kidney cells) are typically used for the isolation and propagation of HAV in cell culture. In addition, a number of laboratories prefer the use of FRhK-4 cells for the rescue of virus from infectious, synthetic RNA transcripts. Thus, it might be expected that all of these cell types would be permissive for the replication of HAV-Luc RNA. However, we were unable to demonstrate evidence for the amplification of this replicon except in Huh-7 cells. In multiple experiments with BSC-1 and FRhK-4 cells, there was no difference in the levels of luciferase expressed following transfection of HAV-Luc and HAV-LucΔ3D (data not shown). Importantly, however, the luciferase activity present 24 h after transfection of these cells was less than 0.1% that in Huh-7 cells and either at or very close to background (Fig. 4A). This suggests either much less efficient transfection or very reduced translation of the transfected RNA in BSC-1 and FRhK-4 cells. To distinguish between these possibilities, we determined the abundance of viral RNA in each of these cell types 12 and 48 h after RNA transfection by dot blot hybridization. The cells were washed thoroughly prior to extraction of the RNA in an effort to remove any liposome-enveloped RNA that had not been internalized. These results demonstrated equivalent abundances of RNA associated with all cell types at 12 and 48 h following transfection (Fig. 4B), despite the much lower levels of luciferase present in the BSC-1 and FRhK-4 cells at 24 and 72 h.
FIG. 4.
Host cell range of replicon amplification. (A) Luciferase activity (in light units) in cell extracts 24 h (open bars) and 72 h (solid bars) following transfection of Huh-7, FRhK-4, and BSC-1 cells with HAV-Luc RNA. Note the logarithmic scale for luciferase activity. (B) Dot blot hybridization detection of cell-associated RNA 12 and 48 h following transfection of the indicated cell type with HAV-Luc or HAV-LucΔ3D transcripts showing equivalent abundances of transfected RNA in all cell types.
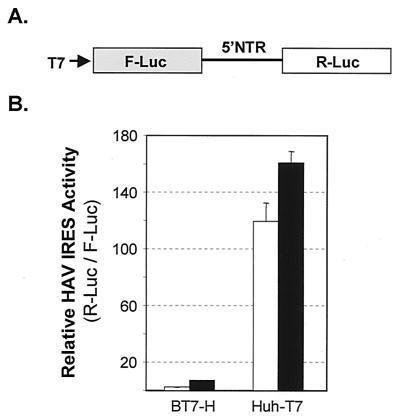
Since translation has been shown previously to be a rate-limiting step for HAV replication in BSC-1 cells, we next sought to determine whether cell-specific differences in the efficiency of translation initiation by the HAV IRES could explain our ability to detect replication of HAV-Luc in Huh-7 but not the other cell types. For this purpose, we transfected BT7-H and Huh-T7 cells (which are derived, respectively, from BSC-1 and Huh-7 cells) with plasmid DNAs containing T7 transcriptional units encoding a dicistronic reporter transcript with F-Luc encoded by the first cistron and R-Luc expressed from the second cistron under translational control of the HAV IRES (Fig. 5A). These two cell lines constitutively express T7 RNA polymerase, leading to the cytoplasmic production of uncapped T7 transcripts from transfected DNA (46). In such experiments, the abundance of F-Luc expressed from the first cistron of these transcripts reflects the efficiency of transfection and T7 transcription in the cells, while the ratio of R-Luc to F-Luc activity represents a normalized measure of HAV IRES activity. We found that the efficiency of HAV IRES-mediated translation initiation, as reflected in the ratio of R-Luc to F-Luc activity, was 24-fold (cell culture-adapted IRES) to 53-fold (wild-type IRES) greater in Huh-T7 cells than in BT7-H cells (Fig. 5B). This suggests that lack of translational activity contributes significantly to the low basal levels of luciferase expressed from the replicon in the BSC-1 cells and probably also the failure of the replicons to undergo demonstrable amplification.
FIG. 5.
HAV IRES-dependent translation in Huh-T7 and BT7-H cells, which constitutively express T7 RNA polymerase and are derived from Huh-7 and BSC-1 cells, respectively. (A) Organization of the dicistronic plasmids, pLuc-wt-RLuc and pLuc-p16-RLuc, that express F-Luc from the 5′ reading frame, which serves as a transfection and transcription control, and R-Luc from the 3′ reading frame under the translational control of the HAV IRES placed within the intercistronic space. This transcription unit is under the control of the T7 promoter. (B) Relative HAV IRES activity in BT7-H and Huh-T7 cells. Values are ratios between F-Luc and R-Luc activities, or the relative activity of HAV IRES in each cell line. Open bars, wild-type IRES; solid bars, cell culture-adapted HM175/p16 IRES (42). Absolute values of F-Luc activity ranged from 3.2 to 11 light units.
VP4 is not required for translation and amplification of replicon RNA.
Graff and Ehrenfeld (20) suggested recently that a 114-nt RNA segment comprised mostly of the VP4-coding sequence located at the 5′ end of the long open reading frame and immediately downstream of the IRES may serve to promote viral translation. For this reason, we retained the entire VP4-coding sequence in designing the first candidate replicon, HAV-Luc (Fig. 1). To determine whether this sequence was necessary for replicon translation and replication, we deleted most of the VP4-coding sequence from pHAV-Luc, creating pHAV-LucΔVP4 (Fig. 1). RNA transcribed from this construct contains only the 5′-most 12 nt of the VP4 sequence upstream of the luciferase sequence. However, the luciferase activity expressed from this RNA in Huh-7 cells was indistinguishable from that expressed by HAV-Luc RNA or HAV-LucΔ3Dpol 12 h after transfection (compare Fig. 2B and 2A). This suggests the absence of a requirement for more than 12 nt of the VP4 sequence for optimal HAV IRES-directed translation in these cells. Moreover, the kinetics of the increase in luciferase activity expressed by the VP4 deletion replicon was similar to that of HAV-Luc over the ensuing 84 h, indicating that the VP4-coding sequence does not contribute materially to RNA replication. The results obtained with the constructs shown in Fig. 2 indicate that HAV resembles poliovirus in that the P1 capsid protein-coding sequences are not required for RNA replication. It is distinct from human rhinovirus 14 and the cardioviruses, other picornaviruses in which the P1 sequences contain an essential cis-acting RNA replication element (30, 33, 34).
Effect of guanidine on replicon amplification.
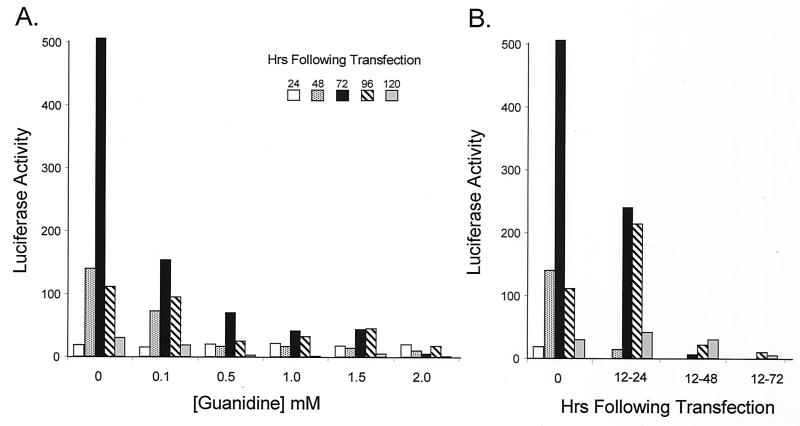
Although the replication of many picornaviruses is inhibited by guanidine, it is not clear what effects guanidine exerts on the replication of HAV. Previous reports are conflicting and suggest that the replication of some strains of HAV may be resistant to guanidine while others may be sensitive to high concentrations of the drug (8, 43). The results shown in Fig. 3B, however, suggest that the replication of HAV RNA is indeed sensitive to the drug at a concentration of 2 mM. Further studies demonstrated that the replication of HAV-Luc was inhibited by guanidine in a dose-dependent manner (Fig. 6A). Replication was inhibited by as little as 0.1 mM guanidine when it was added to the medium 12 h following transfection of the RNA, resulting in a substantial reduction in the subsequent levels of luciferase activity expressed by these cells (Fig. 6A). This result differs from that of Siegl and Eggers (43), who reported that the replication of HAV was not inhibited by guanidine, and also contrasts with that of Cho and Ehrenfeld (8), who found that replication of the HM175 strain of HAV in BSC-1 cells was inhibited by guanidine only at concentrations greater than 1 mM.
FIG. 6.
Guanidine inhibits replication of the HAV-Luc replicon in Huh-7 cells. (A) Effect of 0.1, 0.5, 1.0, 1.5, and 2.0 mM guanidine on the expression of luciferase at various time points following transfection. The time of transfection was considered the point at which the liposome-RNA mixture was initially applied to the cells. Guanidine was added 12 h later (when the transfection mixture was removed from the cells) and maintained at the indicated concentration subsequently. Each bar represents a specific time point. (B) Transient suppression of RNA replication by 2.0 mM guanidine added to the cell culture medium for 12 (12 to 24 h), 36 (12 to 48 h), or 60 (12 to 72 h) h following the removal of the liposome-RNA mixture from the cells.
The inhibitory effect of guanidine was partially reversible, as there was amplification of the replicon (evidenced by increases in luciferase activity) following a transient 12-h period of treatment with 2.0 mM guanidine (from 12 to 24 h following transfection) (Fig. 6B). Interestingly, reversal of the guanidine inhibitory effect was possible only after a relatively brief period of exposure. When treatment was continued for 36 h (12 to 48 h posttransfection), there was little evidence of replicon amplification as reflected in expression of luciferase. This result differs from the report by Cho and Ehrenfeld (8), in which virus replication was suspended for up to 10 days by addition of guanidine and was followed by robust replication with a shortened apparent replication cycle after removal of the drug. As shown in Fig. 6B, there was no apparent change in the kinetics of luciferase accumulation following release from guanidine suppression at 24 h posttransfection. These observations were reproduced in separate experiments.
P2 mutations that enhance viral growth in cultured cells promote replicon amplification.
Either primary or continuous (BSC-1 cell) cultures of AGMK cells have typically been used for the isolation of HAV in cell culture. Wild-type HAV replicates very slowly and inefficiently in these cells. However, the virus adapts to this new cellular environment over successive passages with accompanying changes in its nucleotide sequence. The most important nucleotide substitutions associated with adaptation of the virus to growth in cell culture have been mapped to the P2 and 5′ NTR segments of the genome (11, 13, 15). Because we used cDNA derived from a highly cell culture-adapted strain of HAV to construct the replicons shown in Fig. 2, it was of interest to determine whether replicons constructed from cDNA of the parental wild-type virus would have similar or different replication properties in Huh-7 cells. As shown in Fig. 7, a replicon containing the wild-type P2P3 sequence (HAV-Luc-wP2P3) was severely impaired in its ability to replicate in Huh-7 cells. The luciferase activity present 24 h after transfection of this replicon into Huh-7 cells was similar to that expressed by HAV-Luc. This indicates similar levels of translation of the input RNA, as would be expected since these replicons (as well as the HAV-Luc3Dpol mutant) share identical 5′ NTR sequences. However, in contrast to the subsequent increase in luciferase activity observed in cells transfected with HAV-Luc, the luciferase activity declined gradually over the ensuing 96 h in cells transfected with HAV-Luc-wP2P3. Nonetheless, despite this decline, the luciferase activity expressed by HAV-Luc-wP2P3 was significantly greater than that expressed from HAV-LucΔ3D between 48 and 120 h following transfection, providing evidence for low-level replication of the HAV-Luc-wP2P3 RNA.
FIG. 7.
Cell culture-adaptive mutations in the P2-coding region are important determinants for HAV RNA replication. (A) Schematic representation of replicon constructs containing P2 or P2P3 sequences from wild-type HAV (shaded segments) or the cell culture-adapted, cytopathic HM175/18f virus. The arrow indicates the site of a cell culture-adaptive Ala-to-Val substitution in the 2B protein encoded by a mutation at nt 3889 of the genome. (B) Luciferase activity expressed by the replicon RNAs shown in panel A, following transfection of the respective RNAs into Huh-7 cells. Δ3D, HAV-LucΔ3D (Fig. 1).
The P2P3 sequences of HAV-Luc and HAV-Luc-wP2P3 differ at 32 nucleotide positions (27). To map the locations of the nucleotide substitutions in HAV-Luc that contribute to its enhanced replication capacity relative to HAV-Luc-wP2P3, we replaced the P3 sequence in HAV-Luc-wP2P3 with the P3 segment of HAV-Luc. As shown in Fig. 7B, the expression of luciferase by this replicon (HAV-Luc-wP2) was identical to that by HAV-Luc-wP2P3, indicating that the nucleotide substitutions in the P3 sequence do not contribute to the greater replication capacity of HAV-Luc. Since previous studies suggest that a single nucleotide change at nt 3889 of the wild-type sequence, resulting an Ala-to-Val amino acid change in the 2B protein, enhances the ability of the virus to replicate in cultured cells (12), we created an additional replicon (HAV-Luc-wP2-3889) containing this mutation within the background of HAV-Luc-wP2. As shown in Fig. 7B, luciferase expression and hence replication of the RNA was clearly improved by the addition of this single nucleotide substitution. In aggregate, these data indicate that the capacity for replicon amplification in cultured Huh-7 cells is dependent upon nucleotide substitutions that were selected during the original adaptation of virus to growth in AGMK cells. These data confirm the importance of the P2 sequence, and in particular this 2B residue, to replication of viral RNA in a broad range of cultured cells. They also serve to validate the use of these subgenomic replicons to further dissect the molecular mechanisms underlying the process of cell culture adaptation.
Adaptive mutations in the 5′ NTR do not enhance replicon amplification in Huh-7 cells.
Unlike mutations associated with cell culture adaptation in the P2 segment of the HAV genome, mutations in the 5′ NTR that are selected during passage in cultured cells typically promote cap-independent translation of the viral RNA, and thus viral replication, in a cell-type-specific fashion. Different clusters of such mutations that enhance the ability of the virus to replicate specifically in BSC-1 cells (42) or in MRC-5 cells (16) have been identified. To determine the influence of such 5′ NTR mutations on the amplification of the HAV-Luc replicon in Huh-7 cells, we replaced the 5′ NTR sequence (nt 45 to 744) in HAV-Luc with the corresponding sequence of the wild-type virus (HAV-Luc-w5′NTR). This sequence substitution had no effect on the basal level of luciferase expressed 24 h after replicon transfection of Huh-7 cells, when the luciferase activity present was derived entirely from translation of the input RNA (Fig. 8). There was also no difference in the subsequent levels of luciferase expressed in cells transfected with HAV-Luc-w5′NTR and HAV-Luc. Thus, in contrast to their effect in BSC-1 cells (42), the cell culture-adaptive mutations within the 5′ NTR of the HM175/18f virus (HAV-Luc replicon) do not enhance translation driven by the HAV IRES in Huh-7 cells and thus have no effect on replication efficiency in these cells.
FIG. 8.
Cell culture-adaptive mutations within the 5′ NTR have no effect on replication of the HAV-Luc replicon in Huh-7 cells. The RNA transcript HAV-Luc-w5′NTR is identical to HAV-Luc except for the 5′ NTR sequence, which is derived from wild-type virus. Values are means from replicate cultures.
DISCUSSION
The hepatoviruses are distinct from most other well-studied picornaviruses in several important respects. The IRES structure of HAV, while related to that of the aphthoviruses and cardioviruses, is unique, as is its requirement for intact eIF-4G for internal initiation of translation (4, 5, 46). The capsid structure, while incompletely understood, appears to differ from other picornavirus capsids with respect to the very small size and probable absence of myristylation of VP4 (44). The 2A protein lacks homology with any other picornaviral 2A protein, and it appears that a cellular proteinase directs cleavage at the VP1/2A (22, 31). Perhaps most striking, however, is the fact that the HAV replication cycle is unusually protracted, with one-cycle growth studies indicating a replication cycle on the order of 24 h even for isolates that are well adapted to growth in cell culture (11, 27). This slow and generally noncytopathic growth of the virus has made it difficult to study HAV replication, and for that reason the mechanisms underlying the differences between HAV and other picornaviruses remain poorly defined. Assays for replication are generally dependent upon the detection of viral antigen or viral RNA, as plaque assays are technically difficult and typically require a week or more for detection of infection in cells (26). In contrast, the subgenomic HAV replicons described in this communication offer a relatively rapid and convenient approach to the assessment of RNA replication and are certain to prove useful in studies of HAV replication.
One hypothesis that has been advanced to explain the slow and inefficient replication of HAV has been the idea that newly replicated positive-strand RNAs are rapidly sequestered by a high affinity interaction with the capsid proteins, effectively removing potential template molecules from the pool of replicating viral RNAs (1). Supporting this hypothesis is evidence that only a very small proportion of intracellular HAV RNA is of the negative sense. If this hypothesis is correct, however, the replication of subgenomic RNAs lacking the P1-coding sequence should be markedly enhanced and approach that of replicons derived from other picornaviruses. This was not the case with the replicons we constructed, providing strong evidence against this hypothesis. The initial delay and subsequent kinetics of the increase in luciferase activity observed in cells transfected with HAV-Luc and HAV-LucΔVP4 RNA mirror closely what has been observed in one-step growth analyses of the parent virus, HM175/18f (27).
Poliovirus replicons that lack the entire P1 region are capable of efficient replication in transfected cells (10, 25), but similar subgenomic rhinovirus 14 and cardioviral RNAs fail to replicate due to the presence of a critical RNA replication element within the capsid-coding sequence (30, 33). This CRE forms a stem-loop structure and is believed to be required for initiation of negative-strand RNA synthesis (34). Poliovirus possesses a similar CRE, but it is located within the P2-coding region (17). The results obtained with our replicons demonstrate that HAV is similar to poliovirus with respect to the absence of a CRE within the P1 segment of the genome. HAV-LucΔVP4 lacks all of the P1 segment except for 12 nt of VP4-coding sequence downstream of the AUG codon and 39 nt encoding the carboxy terminus of VP1, yet it is capable of replication in Huh-7 cells (Fig. 2B). If there is a CRE in HAV, it must reside in the RNA outside the P1 domain.
Graff and Ehrenfeld (20) suggested that the 5′-most 114 nt of the P1 sequence acts as a translational enhancer, increasing the efficiency of cap-independent translation initiation when included in reporter transcripts in its natural position immediately downstream of the IRES. This finding is reminiscent of the claim by Reynolds et al. (38) that the IRES of hepatitis C virus (HCV), which is unrelated in structure or sequence to that of HAV but also operates in hepatocytes, extends downstream of the AUG codon. We could not confirm this effect of the VP4-coding sequence on HAV translation, however, since we found no difference in the luciferase activity following transfection of Huh-7 cells with HAV-Luc or HAV-LucΔVP4 (Fig. 2). The luciferase activity at this early time point is derived exclusively from translation of the input RNA, since it is similar in cells transfected with replication-competent RNAs and those transfected with RNAs with a premature termination mutation created within the 3Dpol-coding sequence (Fig. 2). However, consistent with the lack of an effect on translation, the VP4-coding sequence also did not contribute materially to replication of HAV RNA in Huh-7 cells, since subsequent increases in luciferase expression were similar in cells transfected with HAV-Luc or HAV-LucΔVP4 RNA (Fig. 2). We have shown recently that there is no requirement for specific sequence downstream of the HCV IRES, only a requirement for an absence of secondary structure in the RNA (40). The situation is probably similar for the HAV IRES, since the 40S ribosome subunit enters on the RNA at or very close to the initiator codon, as it does in the case of HCV (6, 23, 39).
Previous work has indicated that mutations in the P2-coding region play an important role in determining the ability of cell culture-adapted HAV strains to replicate in cultured cells (12, 13). Our results demonstrated the contributions of these mutations to replication of HAV RNA in Huh-7 cells, as replication was substantially reduced, almost to the level of the nonreplicating HAV-LucΔ3D mutant, by replacing the P2 sequence in HAV-Luc with that from wild-type virus (Fig. 7). The creation of a single mutation within the 2B-coding region partially but not completely restored the replication capacity of the RNA. This mutation at nt 3889 has been identified in several independently isolated cell culture-adapted strains of HAV (9, 21, 24). Because the 2B protein is likely to be part of the HAV replicase complex, it has been assumed to play a role in adapting the replicase to function in a novel cellular environment. However, the data presented in Fig. 7 provide formal proof that this and other cell culture-adaptive mutations in the P2 segment specifically enhance RNA synthesis in the absence of encapsidation, cell-to-cell spread, and unpackaging of the virion RNA. In contrast, sequence changes within the P3 segment of HAV-Luc had little apparent effect on RNA replication in Huh-7 cells, as there was no difference in the luciferase activities expressed by wP2P3HAV-Luc and wP2HAV-Luc (Fig. 7).
There are several mutations within the 5′ NTR of HAV-Luc that were selected during initial adaptation of the parental virus to growth in BSC-1 cells (24). The removal of these mutations from the 5′ NTR of HAV-Luc had no effect on the level of luciferase either early after transfection (24 h, when luciferase activity represents translation of input RNA only) or at subsequent time points (reflecting RNA replication) (Fig. 8). This is not surprising, since these 5′ NTR mutations enhance translation and replication in a cell-type specific fashion in BSC-1 cells and have been shown previously to have no effect on translation of reporter transcripts in Huh-7 cells (11, 42). One of these mutations, a deletion of two uridine residues within domain III of the wild-type HAV IRES, reduces the affinity of the RNA for binding to glyceraldehyde 3′-phosphate dehydrogenase (GAPDH), thereby preserving the structure of the IRES which is required for internal initiation of translation (41, 47). The fact that these 5′ NTR mutations are functionally silent in Huh-7 cells may be due to the high abundance of polypyrimidine tract-binding protein (PTB) in these cells. PTB abundance is much higher than in BSC-1 cells, and PTB binds to similar RNA elements within the IRES in competition with GAPDH (41). Unlike GAPDH, which is the major BSC-1 protein identified in UV cross-linking studies with probes from the HAV IRES (7), PTB stimulates translation directed by the HAV IRES in vivo (18). The cellular PTB abundance also likely contributes to the relatively increased efficiency of the IRES in Huh-7 cells (Fig. 5).
Although an effective vaccine was licensed in the United States almost a decade ago, immunization has had little impact on the incidence of acute hepatitis A or on the proportion of cases of acute hepatitis that are due to this virus (29). There remains a need for less expensive vaccines. Early studies demonstrated that some mutations that are selected during adaptation of the virus attenuate its ability to replicate in the liver and to cause disease in nonhuman primates as well as humans (37). Further studies of the mechanisms of attenuation are warranted. In addition, a better understanding of the restrictions underlying the slow and inefficient replication of the virus may also be helpful in producing vaccine strains that are capable of replicating to higher titers during production of inactivated vaccines. The subgenomic replicons described in this communication should facilitate such studies of HAV proteins and RNA structures involved in replication of the virus.
Acknowledgments
This work was supported in part by grant RO1-AI32599 from the National Institutes of Health.
We are grateful to Terri Chapa for expert technical assistance and to Annette Martin for helpful discussions.
REFERENCES
- 1.Anderson, D. A., B. C. Ross, and S. A. Locarnini. 1988. Restricted replication of hepatitis A virus in cell culture: encapsidation of viral RNA depletes the pool of RNA available for replication. J. Virol. 62:4201–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard, M. R., L. Cohen, S. M. Lemon, and A. Martin. 2001. Characterization of recombinant hepatitis A virus genomes containing exogenous sequences at the 2A/2B junction. J. Virol. 75:1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, E. M., S. C. Mosimann, M. M. Chernaia, B. A. Malcolm, and M. N. G. James. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71:2436–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., and K. M. Kean. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129–136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. A., S. P. Day, R. W. Jansen, and S. M. Lemon. 1991. The 5′ nontranslated region of hepatitis A virus: secondary structure and elements required for translation in vitro. J. Virol. 65:5828–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, K. H., E. A. Brown, and S. M. Lemon. 1993. Cell type-specific proteins which interact with the 5′ nontranslated region of hepatitis A virus RNA. J. Virol. 67:6716–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, M. W., and E. Ehrenfeld. 1991. Rapid completion of the replication cycle of hepatitis A virus subsequent to reversal of guanidine inhibition. Virology 180:770–780. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. I., B. Rosenblum, J. R. Ticehurst, R. J. Daemer, S. M. Feinstone, and R. H. Purcell. 1987. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc. Natl. Acad. Sci. USA 84:2497–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collis, P. S., B. J. O’Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day, S. P., P. Murphy, E. A. Brown, and S. M. Lemon. 1992. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J. Virol. 66:6533–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson, S. U., Y. K. Huang, C. McRill, M. Lewis, and R. H. Purcell. 1992. Mutations in both the 2B and 2C genes of hepatitis A virus are involved in adaptation to growth in cell culture. J. Virol. 66:650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson, S. U., Y. K. Huang, and R. H. Purcell. 1993. 2B and 2C mutations are essential but mutations throughout the genome of HAV contribute to adaptation to cell culture. Virology 194:475–480. [DOI] [PubMed] [Google Scholar]
- 14.Emerson, S. U., M. Lewis, S. Govindarajan, M. Shapiro, T. Moskal, and R. H. Purcell. 1992. cDNA clone of hepatitis A virus encoding a virulent virus: induction of viral hepatitis by direct nucleic acid transfection of marmosets. J. Virol. 66:6649–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson, S. U., C. McRill, B. Rosenblum, S. Feinstone, and R. H. Purcell. 1991. Mutations responsible for adaptation of hepatitis A virus to efficient growth in cell culture. J. Virol. 65:4882–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funkhouser, A. W., D. E. Schultz, S. M. Lemon, R. H. Purcell, and S. U. Emerson. 1999. Hepatitis A virus translation is rate-limiting for virus replication in MRC-5 cells. Virology 254:268–278. [DOI] [PubMed] [Google Scholar]
- 17.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosert, R., D. Egger, and K. Bienz. 2000. A cytopathic and a cell culture adapted hepatitis A virus strain differ in cell killing but not in intracellular membrane rearrangements. Virology 266:157–169. [DOI] [PubMed] [Google Scholar]
- 20.Graff, J., and E. Ehrenfeld. 1998. Coding sequences enhance internal initiation of translation by hepatitis A virus RNA in vitro. J. Virol. 72:3571–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graff, J., C. Kasang, A. Normann, M. Pfisterer-Hunt, S. M. Feinstone, and B. Flehmig. 1994. Mutational events in consecutive passages of hepatitis A virus strain GBM during cell culture adaptation. Virology 204:60–68. [DOI] [PubMed] [Google Scholar]
- 22.Graff, J., O. C. Richards, K. M. Swiderek, M. T. Davis, F. Rusnak, S. A. Harmon, X. Y. Jia, D. F. Summers, and E. Ehrenfeld. 1999. Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus. J. Virol. 73:6015–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda, M., E. A. Brown, and S. M. Lemon. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen, R. W., J. E. Newbold, and S. M. Lemon. 1988. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology 163:299–307. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, G., and V. R. Racaniello. 1988. Construction and characterization of poliovirus subgenomic replicons. J. Virol. 62:1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon, S. M., L. N. Binn, and R. H. Marchwicki. 1983. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J. Clin. Microbiol. 17:834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemon, S. M., P. C. Murphy, P. A. Shields, L.-H. Ping, S. M. Feinstone, T. Cromeans, and R. W. Jansen. 1991. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J. Virol. 65:2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemon, S. M., and B. H. Robertson. 1993. Current perspectives in the virology and molecular biology of hepatitis A virus. Semin. Virol. 4:285–295. [Google Scholar]
- 29.Lemon, S. M., and D. L. Thomas. 1997. Vaccines to prevent viral hepatitis. N. Engl. J. Med. 336:196–204. [DOI] [PubMed] [Google Scholar]
- 30.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, A., D. Benichou, S. F. Chao, L. M. Cohen, and S. M. Lemon. 1999. Maturation of the hepatitis A virus capsid protein VP1 is not dependent on processing by the 3Cpro proteinase. J. Virol. 73:6220–6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInerney, G. M., A. M. King, N. Ross-Smith, and G. J. Belsham. 2000. Replication-competent foot-and-mouth disease virus RNAs lacking capsid coding sequences. J. Gen. Virol. 81(Pt. 7):1699–1702. [DOI] [PubMed] [Google Scholar]
- 33.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuesch, J., S. Krech, and G. Siegl. 1988. Detection and characterization of subgenomic RNAs in hepatitis A virus particles. Virology 165:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuesch, J. P. F., J. de Chastonay, and G. Siegl. 1989. Detection of defective genomes in hepatitis A virus particles present in clinical specimens. J. Gen. Virol. 70:3475–3480. [DOI] [PubMed] [Google Scholar]
- 37.Provost, P. J., F. S. Banker, P. A. Giesa, W. J. McAleer, E. B. Buynak, and M. R. Hilleman. 1982. Progress toward a live, attenuated human hepatitis A vaccine. Proc. Soc. Exp. Biol. Med. 170:8–14. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds, J. E., A. Kaminiski, H. J. Kettinen, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds, J. E., A. Kaminski, A. R. Carroll, B. E. Clarke, D. J. Rowlands, and R. J. Jackson. 1996. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 40.Rijnbrand, R. C. A., P. J. Bredenbeek, P. C. J. Haasnoot, J. S. Kieft, W. J. M. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz, D. E., C. C. Hardin, and S. M. Lemon. 1996. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′ nontranslated RNA of hepatitis A virus. J. Biol. Chem. 271:14134–14142. [DOI] [PubMed] [Google Scholar]
- 42.Schultz, D. E., M. Honda, L. E. Whetter, K. L. McKnight, and S. M. Lemon. 1996. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J. Virol. 70:1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegl, G., and H. J. Eggers. 1982. Failure of guanidine and 2-(alpha-hydroxybenzyl)benzimidazole to inhibit replication of hepatitis A virus in vitro. J. Gen. Virol. 61:111–114. [DOI] [PubMed] [Google Scholar]
- 44.Tesar, M., S. A. Harmon, D. F. Summers, and E. Ehrenfeld. 1992. Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology 186:609–618. [DOI] [PubMed] [Google Scholar]
- 45.Weitz, M., B. M. Baroudy, W. L. Maloy, J. R. Ticehurst, and R. H. Purcell. 1986. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J. Virol. 60:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whetter, L. E., S. P. Day, O. Elroy-Stein, E. A. Brown, and S. M. Lemon. 1994. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J. Virol. 68:5253–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi, M., D. E. Schultz, and S. M. Lemon. 2000. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 74:6459–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, H. C., S. F. Chao, L. H. Ping, K. Grace, B. Clarke, and S. M. Lemon. 1995. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effect. Virology 212:686–697. [DOI] [PubMed] [Google Scholar]