Abstract
The transcription regulatory sequences (TRSs) of the coronavirus transmissible gastroenteritis virus (TGEV) have been characterized by using a helper virus-dependent expression system based on coronavirus-derived minigenomes to study the synthesis of subgenomic mRNAs. The TRSs are located at the 5′ end of TGEV genes and include a highly conserved core sequence (CS), 5′-CUAAAC-3′, that is essential for mediating a 100- to 1,000-fold increase in mRNA synthesis when it is located in the appropriate context. The relevant sequences contributing to TRS activity have been studied by extending the CS 5′ upstream and 3′ downstream. Sequences from virus genes flanking the CS influenced transcription levels from moderate (10- to 20-fold variation) to complete mRNA synthesis silencing, as shown for a canonical CS at nucleotide (nt) 120 from the initiation codon of the S gene that did not lead to the production of the corresponding mRNA. An optimized TRS has been designed comprising 88 nt from the N gene TRS, the CS, and 3 nt 3′ to the M gene CS. Further extension of the 5′-flanking nucleotides (i.e., by 176 nt) decreased subgenomic RNA levels. The expression of a reporter gene (β-glucuronidase) by using the selected TRS led to the production of 2 to 8 μg of protein per 106 cells. The presence of an appropriate Kozak context led to a higher level of protein expression. Virus protein levels were shown to be dependent on transcription and translation regulation.
Transmissible gastroenteritis virus (TGEV) is a member of the family Coronaviridae, which, together with the family Arteriviridae, forms the order Nidovirales (6). The coronavirus RNA genome has a length ranging from 27.6 to 31.5 kb. About two-thirds of the entire RNA comprises open reading frames (ORFs) 1a and 1ab, encoding the replicase gene. The 3′ one-third of the genome comprises the genes encoding the structural and nonstructural proteins. The organization of the non-ORF 1 nonstructural protein genes, which are interspersed between the known structural protein genes, varies significantly among different coronavirus strains (5).
Coronavirus transcription is based on RNA-dependent RNA synthesis. Coronavirus mRNAs consist of six to eight types of various sizes, depending on the coronavirus strain. The largest mRNA is the genomic RNA, which also serves as the mRNA for ORFs 1a and 1ab; the others are subgenomic mRNAs (sgmRNAs), composed of noncontiguous sequences of the parental genome. The mRNAs and the genomic RNA form a nested set of RNAs of different lengths with common 5′ and 3′ ends. Except for the smallest mRNA, all of the mRNAs are structurally polycistronic; in general, however, only the 5′-most ORF (not present in the next smallest mRNA) is translated. However, there are exceptions: some mRNAs, e.g., mRNA 5 of mouse hepatitis virus (MHV), mRNA 3 of infectious bronchitis virus, and the bovine coronavirus (BCoV) nucleocapsid mRNA, are translated by internal initiation into two or three proteins (21, 25). All the mRNAs possess a 5′ sequence of 70 to 90 nucleotides (nt) that is identical to the leader sequence located at the 5′ end of the viral genome. These special features imply a discontinuous transcription mechanism that is under debate (23, 41).
Sequences at the 5′ end of each gene represent signals for the discontinuous transcription of sgmRNAs (23, 41). These are the transcription regulatory sequences (TRSs), which include a stretch of a highly conserved core sequence (CS), 5′-CUAAAC-3′, for TGEV or a highly related sequence for other coronaviruses. This sequence was previously named the intergenic sequence. Nevertheless, since many coronavirus and arterivirus genes frequently overlap and an intergenic space does not always exist, the acronym CS is used here.
Two major models have been proposed to explain the transcription strategy for coronaviruses and arteriviruses (23, 41). The discovery of transcriptionally active, subgenome-size minus strands that contain the antileader sequence and of transcription intermediates active in the synthesis of mRNAs favors the model of discontinuous transcription during the synthesis of the minus strand (39, 40, 42, 44). According to this model, the TRS acts as a “slow-down” or “stop” signal for the replication complex; the discontinuous mRNA synthesis is governed, at least in part, by a direct base-pairing interaction between the nascent-body (−) TRS and the 3′ end of the leader exposed at the 5′ end of the viral genome (39, 50).
The abundance of the mRNA may be primarily controlled by the extent of the base pairing between the 3′ end of the leader and the TRSs. A second factor, proximity to the 3′ end of the genome, could influence mRNA abundance. Since the TRSs act as slow-down or stop signals for the replicase complex, the smaller mRNAs should be the most abundant. Although this has been shown to be the case for the Mononegavirales (11, 51) and, in coronaviruses, shorter mRNAs are in general more abundant, the relative abundance of coronavirus mRNAs is not strictly related to their proximity to the 3′ end (33, 49). A third factor, the interaction of RNA with viral and cellular proteins, has also been implicated in mRNA transcription (23). The three factors implicated in the control of mRNA abundance assume a key role for the TRSs and the need to define their composition and structure.
The initiation of mRNA transcription originating from sequences that do not conform to the canonical conserved motif has been reported for MHV, BCoV, and arteriviruses (references 18, 22, 23, and 31 and references therein). Also, transcription originating at genomic sites not possessing a canonical CS has been identified for recombinant MHV expressing the foreign green fluorescent protein gene (8). These data reinforce the need for studies to clarify the role of TRSs in the control of transcription.
The role of a TRS has been defined for MHV, infectious bronchitis virus, and BCoV (9, 21, 24, 31, 46, 50). We are interested in studying the extent of the TRSs regulating the expression of the TGEV mRNAs, i.e., to define the role in transcription of the sequences flanking the 5′ and 3′ ends of a CS of 6 nt (5′-CUAAAC-3′). To this end, an expression system based on TGEV-derived minigenomes (12, 30) was used to study TRS length for optimum transcription efficiency. cDNA clones encoding minigenome RNAs (mgRNAs) and including an expression cassette in which the TRS could easily be replaced were constructed. Different combinations of TRSs were inserted preceding a reporter β-glucuronidase (GUS) gene, and the molar ratio of sgmRNA to mgRNA or the total amount of mRNA was determined. The effect of TRSs on the translation of the reporter GUS gene in relation to sgmRNA abundance and the influence of the expression cassette insertion site were also determined.
MATERIALS AND METHODS
Cells and viruses.
TGEV strain PUR46-MAD (38) was grown and titrated as previously described (16). Viruses were grown in swine testis (ST) cells (29a).
Construction of cDNAs encoding RNA minigenomes.
The construction of DI-C-derived cDNAs encoding RNA minigenomes (e.g., M39) was previously described (12). To increase mgRNA expression levels, the cDNAs were preceded by the cytomegalovirus (CMV) promoter (4, 34). The minigenomes were flanked at the 3′ end by the hepatitis delta virus (HDV) ribozyme and the bovine growth hormone (BGH) polyadenylation and termination sequences (34).
To evaluate expression levels by using the minigenomes, the Escherichia coli K-12 GUS gene was used as a reporter gene (13, 43). The GUS gene was amplified by PCR from plasmid pGUS1 (Plant Genetic Systems) by using a forward 40-mer oligonucleotide (5′-GCGGCCGCAGGCCTGTCGACGACCATGGTCCGTCCTGTAG-3′) that included NotI, SgrAI, StuI, and SalI restriction endonuclease sites (bold nucleotides); the GUS initiation codon is underlined, and nucleotides shown in italics were included to fit the consensus motif of the ribosome scanning model (19, 20). The reverse primer was 41 nt long (5′-GGTACCGCGCGCCTGGGCTAGCGCGATCATAGGCGTCTCGC-3′) and included NheI, BssHII, and KpnI restriction sites (bold nucleotides). The GUS gene was cloned into minigenome M39 within the most 3′ deletion introduced during the generation of the minigenome (12). To this end, the GUS gene was cloned into intermediate plasmids flanked at the 5′ end by polylinker L2 (GCGGCCGCGCCGGCGAGGCCTGTCGACGACC) or L3 (GTCGACGACC), including four (NotI, SgrAI, StuI, and SalI) or one (SalI) restriction endonuclease site, respectively (in bold), and an optimized Kozak sequence (in italics). The 3′ end of the GUS gene was flanked by polylinker L4 (GCTAGCCCAGGCGCGCGGTACC), including three restriction endonuclease sites (NheI, BssHII, and KpnI) (in bold).
The 5′ upstream nucleotides flanking the CS (5′-CUAAAC-3′) of the M, N, and S genes of strain PUR46-MAD of TGEV, designated the 5′ TRSs, were synthesized by PCR. Forward oligonucleotides contained the restriction sites BlnI, SwaI, and Bsu36I (in bold) integrated in polylinker L1 (CCTAGGATTTAAATCCTAAGG), as shown in Table 1. Two reverse oligonucleotides were used for PCR amplification; they included either the sequence defined for L2 or the L3 sequence (Table 1). The 5′ TRSs derived from the N and M genes were amplified from TGEV sequences comprising nt 26078 to 26941 and nt 24525 to 26254, respectively (GenBank accession number AJ271965) (33). The 5′ TRS derived from the S gene was amplified from the cDNA encoding minigenome DI-C (pDI-C) (12). The resulting minigenomes were designated mg-N-44, mg-S-69, mg-N-88, mg-M-91, and mg-N-176, followed by L2 or L3, depending on the polylinker introduced 3′ downstream of the CS (Table 1).
TABLE 1.
PCR elements required to obtain the 5′ TRSs
| Minigenomea | Oligonucleotidesb | Oligonucleotide sequencesc | Plasmid or template | Source or referenced |
|---|---|---|---|---|
| mg-N-88-L2 | N vs | AACGCGTCCTAGGATTTAAATCCTAAGGCTATGTAAAATCTAAAGCTGG | TGEV sequence from nt 26078 to 26941 | GenBank AJ271965 |
| N rs | GACGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTTATACCATATG | |||
| mg-M-91-L2 | M vs | TGACGCGTCCTAGGATTTAAATCCTAAGGATTATTGTTCCAGCG | TGEV sequence from nt 24525 to 26254 | GenBank AJ271965 |
| M rs | CAGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTTCAAGC | |||
| mg-S-69-L2 | S vs | CGACGCGTCCTAGGATTTAAATCCTAAGGAATTGAATGAAATGGTC | pDI-C | 12 |
| S rs | CAGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTAACTTACCATTATTTC | |||
| mg-N-88-L3 | N vs | AACGCGTCCTAGGATTTAAATCCTAAGGCTATGTAAAATCTAAAGCTGG | pmg-N-88-L2TGEV sequence from nt 26078 to 26941 | GenBank AJ271965 |
| N-L3 rs | TCGTCGACGTTTAGTTATACCATATGTAATAATTTTTCTTGC | |||
| mg-M-91-L3 | M vs | TGACGCGTCCTAGGATTTAAATCCTAAGGATTATTGTTCCAGCG | TGEV sequence from nt 24525 to 26254 | GenBank AJ271965 |
| M-L3 rs | GCGTCGACGTTTAGTTCAAGCAAGGAGTGCTCC | |||
| mg-S-69-L3 | S vs | CGACGCGTCCTAGGATTTAAATCCTAAGGAATTGAATGAAATGGTC | pDI-C | 12 |
| S-L3 rs | AAGTCGACGTTTAGTAACTTACCATTATTTCTAATG | |||
| mg-N-3-L3 | 6022 M39-GUS vs | CACTGAAGAATTCACATATTAAAACC | pmg-N-88-L2 | GenBank AJ271965 |
| TGEV sequence from nt 26078 to 26941 | ||||
| N-3 rs | GAGTCGACGTTTAGTTACCTTAGGATTTAAATCCTAGGACG | |||
| mg-N-8-L3 | 6022 M39-GUS vs | CACTGAAGAATTCACATATTAAAACC | pmg-N-88-L2 | GenBank AJ271965 |
| TGEV sequence from nt 26078 to 26941 | ||||
| N-8 rs | GAGTCGACGTTTAGTTATACCACCTTAGGATTTAAATCCTAGGACG | |||
| mg-N-44-L3 | 44-N vs | TGCCTAGGATTTAAATCCTAAGGCTGATAATTTGAGTGAGCAAGAAAAATTATTAC | TGEV sequence from nt 26078 to 26941 | GenBank AJ271965 |
| N-L3 rs | TCGTCGACGTTTAGTTATACCATATGTAATAATTTTTCTTGC | |||
| mg-N-176-L3 | 176-N vs | GACCTAGGATTTAAATCCTAAGGACGTAATGGTTGCATTACCTAGCAGG | TGEV sequence from nt 26078 to 26941 | GenBank AJ271965 |
| N-L3 rs | TCGTCGACGTTTAGTTATACCATATGTAATAATTTTTCTTGC | |||
| mg-CS | M vs | TGACGCGTCCTAGGATTTAAATCCTAAGGATTATTGTTCCAGCG | TGEV sequence from nt 24525 to 26254 | GenBank AJ271965 |
| CS-M Bsu36I L3 rs | TGTCGACGTTTAGCCTTAGGAAGGAGTGCTCCATCGGGG |
The first digits indicate the number of nucleotides included in the 5′ TRSs. N, M, and S represent the 5′ TRSs derived from the corresponding viral genes.
vs, virus sense; rs, reverse sense.
Bold nucleotides indicate unique restriction endonuclease sites (MluI, ACGCGT; BlnI, CCTAGG; SwaI, ATTTAAAT; Bsu36I, CCTAAGG; NotI, GCGGCCGC; SgrAI, CGCCGGCG; StuI, AGGCCT; and SalI, GTCGAC). The CS is underlined.
Designations for GenBank are accession numbers.
The effect on transcription of different lengths of 5′ TRSs from the viral N gene was also analyzed. The plasmid coding for minigenome mg-N-88-L2 (pmg-N-88-L2) was used as a template for PCR amplification of the 5′ TRSs from the N gene containing 3 and 8 nt, leading to minigenomes mg-N-3-L3 and mg-N-8-L3, respectively (Table 1). The modular approach used in this study facilitated the exchange of TRSs and genes.
Minigenome mg-CS, containing the GUS gene downstream of the CS but no 5′ TRS, was generated by introducing a 91-nt deletion between two Bsu36I restriction sites in minigenome mg-M-91-L3. One Bsu36I restriction site was present in minigenome mg-M-91-L3. The second Bsu36I restriction site, flanking the CS at the 5′ end, was inserted by PCR mutagenesis with TGEV sequences comprising nt 24525 to 26254, the forward oligonucleotide M vs, and the reverse oligonucleotide CS-M Bsu36I L3 rs (Table 1). Digestion with Bsu36I and subsequent religation generated minigenome mg-CS. Minigenome mg-CS−, in which the CS was deleted, was obtained from minigenome mg-CS after digestion with SwaI and StuI and subsequent religation.
To analyze the effect on transcription of sequences flanking the 3′ end of the CS, a collection of minigenomes was generated (Table 2). All these minigenomes contained 88 nt of the 5′ TRS preceding the CS of the N gene. The 3′ TRSs derived from viral genes S, 3a, 3b, E, M, N, and 7 were introduced by using specific oligonucleotides and standard recombinant DNA techniques (36) (Table 2). The minigenome mg-N-88-L3 (Table 1) is referred to as mg-L3 (Table 2). The plasmid (pmg-N-88-L3) encoding minigenome mg-L3 was used as a template for the synthesis of all these minigenomes, except for mg-L2. Minigenome mg-3b′ contained the CS and 3′ TRS consisting of 24 nt extending from the noncanonical sequence located at position 25344 of the viral genome to the potential initiation codon (AUG) of ORF 3b. Minigenome mg-Ld contained as the 3′ TRS a 12-nt sequence complementary to the 3′ end of the leader sequence present at the 5′ end of the viral genome (Table 2).
TABLE 2.
PCR elements required to obtain the 3′ TRSs
| Minigenomea | Oligonucleotidesb | Oligonucleotide sequencec | Plasmid used as a template | Source |
|---|---|---|---|---|
| mg-S | S-3′ vs | TACATATGGTATAACTAAACTTTGGTAACACTTCGTTAACACACCATGGTCCGTCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-3a | 3a-3′ vs | TACATATGGTATAACTAAACTTACGAGTCATTACAGGTCCTGTATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-3b | 3b-3′ vs | TTACATATGGTATAACTAAACTCCATGCGAAAATGGTCCGTCCTGTAGAAACCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-3b′ | 3b′-3′ vs | TACATATGGTATAACTAAACTTTAAGACGTGTGTCGGCATCTTAATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-E | E-3′ vs | TACATATGGTATAACTAAACGAAATTGACTTAAAAGAAGAAGAAGAAGACCATACCTATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-M | M-3′ vs | TACATATGGTATAACTAAACAAAATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-N | N-3′ vs | TACATATGGTATAACTAAACTTCTAAATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA | |||
| mg-7 | 7-3′ vs | TACATATGGTATAACTAAACGAGATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTGAAACCAATGCCTAAAGAGA | |||
| mg-L2 | N vs | AACGCGTCCTAGGATTTAAATCCTAAGGCTATGTAAAATCTAAAGCTGG | TGEV sequence from nt 26078 to 26941 | GenBank AJ271965d |
| N-L2 rs | GACGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTTATACCATATG | |||
| mg-L3 | N vs | AACGCGTCCTAGGATTTAAATCCTAAGGCTATGTAAAATCTAAAGCTGG | pmg-N-88-L2 | Table 1 |
| N-L3 rs | TCGTCGACGTTTAGTTATACCATATGTAATAATTTTTCTTGC | |||
| mg-Ld | Ld vs | ACATATGGTATAACTAAACGAAATATTTGTCATGGTCCGTCCTGTAGAAACCCC | pmg-N-88-L3 | Table 1 |
| GUS 1076 rs | GCTTCGAAACCAATGCCTAAAGAGA |
Minigenome names were derived from the 3′ TRSs of viral genes (S, 3a, 3b, E, M, N, and 7), from a sequence extending from the noncanonical sequence CAAAC to the potential initiation codon located in ORF 3b (3b′), from a sequence complementary to the 3′ end of the leader (Ld), or from polylinkers L2 and L3, described in the text. mg-L2 and mg-L3 are named mg-N-88-L2 and mg-N-88-L3, respectively, in Table 1.
vs, virus sense; rs, reverse sense.
Bold nucleotides indicate unique restriction endonuclease sites (MluI, ACGCGT; BlnI, CCTAGG; SwaI, ATTTAAAT; Bsu36I, CCTAAGG; NotI, GCGGCCGC; SgrAI, CGCCGGCG; StuI, AGGCCT; SalI, GTCGAC; SfuI, TTCGAA; and NdeI, CATATG). The CS is underlined. The translation initiation codon is shown in italics.
Accession number.
To evaluate the effect of position on transcription, an expression cassette consisting of 88 nt of the 5′ TRS from the N gene, the CS, the L3 sequence, and the GUS gene was inserted at four different locations along minigenome M39 by using unique endonuclease restriction sites (EcoRV, BglII, BstZ17I, and MluI), leading to minigenomes mg-947, mg-1655, mg-2881, and mg-3337, named according to the nucleotide position of the insertion site. To ensure that the expected plasmids were generated, the constructs were sequenced at the cloning junctions by using an Applied Biosystems 373 DNA sequencer.
Rescue of RNA polymerase II-driven transcripts.
ST cells grown to 50% confluence in 35-mm dishes were transfected with 10 μg of plasmid DNA encoding CMV-driven minigenomes and 15 μl of Lipofectin reagent in Optimem medium (Gibco-BRL) according to the manufacturer’s instructions. The transfected cells were infected with TGEV strain PUR46-MAD (multiplicity of infection, 5) at 4 h posttransfection. Supernatants obtained from these cultures at 22 to 24 h postinfection (h.p.i.) were used to infect fresh ST cell monolayers. Various numbers of passages were performed to amplify the helper virus RNAs and mgRNAs.
RNA analysis by Northern blotting.
Total intracellular RNA was extracted at 16 h.p.i. from DNA-transfected and helper virus-infected ST cells at different passages by using an Ultraspec RNA isolation system (Biotecx) according to the manufacturer’s instructions. RNAs were separated in denaturing 1% agarose-2.2 M formaldehyde gels. Following electrophoresis, RNAs were irradiated for 0.2 min by using a UVP cross-linker (CL-1000) and were blotted onto nylon membranes (Duralon-UV; Stratagene) by using a Vacugene pump (Pharmacia). The nylon membranes were irradiated with two pulses (70 mJ/cm2) and then soaked in hybridization buffer (ultrasensitive hybridization solution [ULTRAhyb]; Ambion). Northern hybridizations were performed with hybridization buffer containing [α-32P]dATP-labeled probes synthesized by using a random-priming procedure (Strip-EZpec DNA; Ambion) according to the manufacturer’s instructions. The 3′ UTR-specific single-stranded DNA probe was complementary to nt 28300 to 28544 of the TGEV strain PUR46-MAD genome (33). The GUS-specific probe was complementary to the first 1,076 nt of the gene. After hybridization, RNA was quantified by using a Personal FX Molecular Imager and Quantity One software (both from Bio-Rad).
The efficiency of transcription from minigenomes was quantified by determining the molar ratio of sgmRNA transcribed to the amount of the corresponding mgRNA. The amount of sgmRNA reflects both minigenome transcription and minigenome replication efficiencies. In order to normalize the effect of minigenome replication efficiency, the abundance of each mgRNA (mgRNAi, where i could be any of the minigenomes including the different TRSs) was determined in relation to the amount of a reference mgRNA (mgRNAR). Also, in order to correct for the different amounts of total RNA that were loaded in the lanes, a correction factor was defined as the inverse of the ratio of the amounts of mRNAs from the N and M viral genes [(N + M)i] in a specific lane to the amounts of these mRNAs in the reference lane [(N + M)R]. The total sgmRNA transcribed from minigenomes was estimated with the formula (sgmRNAi/mgRNAi) × (mgRNAi/mgRNAR) × [(N + M)R/(N + M)i), which includes a fraction representing the transcription efficiency of the specific sgmRNAi (i.e., the subgenomic mRNA encoded by minigenome mgRNAi) and correction factors for the minigenome replication efficiency and the amount of loaded RNA, respectively.
Western blot analysis.
GUS expression in cells transfected with cDNA coding for a minigenome and infected with helper virus was analyzed at passage 4 by Western blotting as described previously (7). Purified GUS protein (Sigma) was used as a positive control. A GUS-specific polyclonal rabbit antibody (5 Prime-3 Prime, Inc.) diluted 1:200 in TBS buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl) was used as the primary antibody to detect GUS protein. A rabbit-specific goat antibody conjugated to peroxidase and diluted 1:8,000 in TTBS buffer (TBS with 0.1% Tween 20) was used as the secondary antibody.
GUS chemiluminescence detection in cell extracts.
The expression of GUS in cell extracts was detected by a chemiluminescence assay (GUS-Light kit; Tropix) according to the manufacturer’s instructions (3). GUS-expressing minigenome-transfected or mock-transfected cells were infected with helper virus (multiplicity of infection, 5). The amount of protein expressed at 22 to 24 h.p.i. was estimated by using standard calibration curves generated with purified GUS (Sigma) and the bicinchoninic acid protein assay (Pierce, Rockford, Ill.), resulting in 106 relative luminometric units per 0.35 ng of GUS.
RT-PCR.
Detection of potential sgmRNA generated from a 5′-CUAAAC-3′ sequence located 120 nt downstream of the S gene initiation codon was performed by reverse transcription (RT)-PCR. Total intracellular RNA was extracted (see above) from TGEV strain PUR46-MAD-infected ST cells at 16 h.p.i. cDNA was synthesized at 42°C for 1 h by using Moloney murine leukemia virus reverse transcriptase (Ambion) and antisense primer S-546rs (5′-GTAAATAAGCAACAACCTCAT-3′), complementary to nt 525 to 546 of the S gene. The cDNA generated was used as the template for an sgmRNA-specific PCR (leader-body PCR). A virus sense primer, leader 15+ (5′-GTGAGTGTAGCGTGGCTATATCTCTTC-3′), complementary to nt 15 to 41 of the TGEV leader sequence and a reverse sense primer, S-449rs (5′-TAACCTGCACTCACTACCCC-3′), complementary to nt 430 to 449 of the S gene were used for the PCR. PCR was performed with a GeneAmp PCR system 9600 thermocycler (Perkin-Elmer) for 35 cycles. Each cycle comprised 30 s of denaturation at 94°C, 45 s of annealing at 57°C, and 1.5 min of extension at 72°C. The 35 cycles were followed by a 10-min incubation at 72°C. RT-PCR products were separated by electrophoresis in 0.8% agarose gels, purified, and used for direct sequencing with the oligonucleotide leader 15+ and a reverse sense primer, S-154 (5′-TAATCACCTAACACCACATC-3′), complementary to nt 154 to 173 of the S gene.
RESULTS
Transcription efficiency with CS 5′ upstream sequences (5′ TRSs) from genes encoding the major structural proteins.
To study the effect of the 5′ TRSs on sgmRNA transcription, a TGEV minigenome of 3.9 kb (M39) was cloned under the control of the CMV promoter; this procedure led to mgRNA amplification in two steps—amplification in the nucleus mediated by CMV promoter recognition by cellular polymerase II and amplification in the cytoplasm by the viral polymerase. The cDNAs encoding the minigenomes were flanked at the 3′ end by a poly(A) tail, the HDV ribozyme, and the BGH termination and polyadenylation sequences (Fig. 1A). These minigenomes included expression cassettes encoding GUS under the control of different TRSs to study transcription and translation efficiencies.
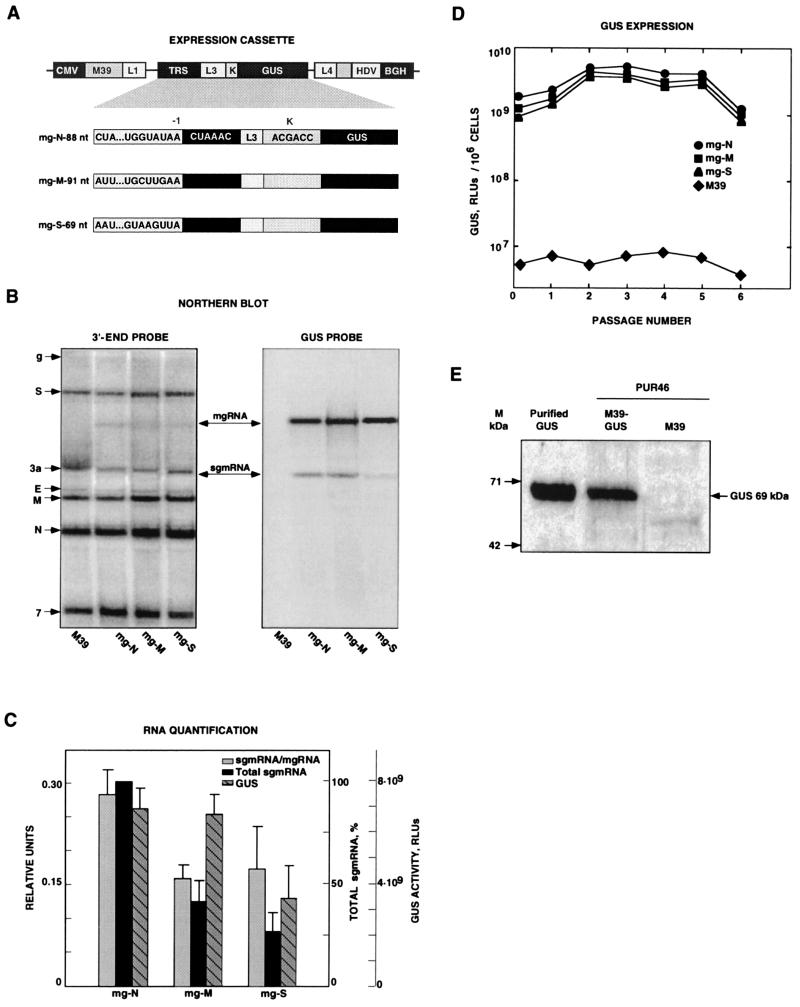
FIG. 1.
Analysis of GUS gene transcription and expression by using minigenomes that contain different virus-derived 5′ TRSs. (A) Schematic structure of expression modules based on TGEV-derived minigenome M39 cloned under the control of the CMV immediate-early promoter (CMV). TRSs include 5′-flanking sequences derived from the TRSs of the major structural genes (N, nucleocapsid gene; M, membrane gene; S, spike gene) and the CS (5′-CUAAAC-3′). The expression cassettes located below the shaded area are flanked by L1 and L4 polylinkers. This cassette includes the TRS, an insertion site (L3), an optimized Kozak sequence (K), and the GUS gene. HDV, HDV ribozyme; BGH, BGH termination and polyadenylation signals. The designations to the left of the expression cassettes indicate the origin of each TRS and the number of nucleotides inserted. (B) Northern blot analysis of intracellular RNAs extracted at passage 2 from minigenome-transfected and TGEV-infected cells. The left and right panels show hybridizations with 3′ UTR- and GUS-specific probes, respectively. The positions of the genomic RNA (g) and mRNAs (S, 3a, E, M, N, and 7) from the helper virus are indicated to the left by their acronyms. M39 RNA overlaps mRNA 3a (first lane). mgRNAs encoding the expression cassettes with different TRSs and the mRNA for the GUS gene (sgmRNA) are indicated with arrows. (C) Quantification of RNAs detected by Northern blotting. RLUs, relative luminometric units. The data shown are the means and standard errors for three independent experiments. (D) GUS activity (per 106 cells) expressed by minigenomes encoding the GUS gene under the control of TRSs derived from the N, M, and S viral genes through six passages after transfection. Background levels are those corresponding to minigenome M39, without an insert. The data shown are averages for at least three experiments with similar results. (E) Western blot analysis of GUS expression by using TGEV-derived minigenomes. Detection of the heterologous GUS protein (69 kDa) expressed by TGEV-derived minigenome M39 (M39-GUS) was performed by Western blot analysis under reducing conditions with a GUS-specific polyclonal rabbit antiserum. Molecular masses (M) are indicated on the left. Purified GUS was used as the positive control. M39-GUS, extracts from ST cells transfected with DNA coding for minigenome mg-N-88-L2 and infected with the helper virus (PUR46-MAD), obtained at passage 4. M39, extracts from ST cells transfected with minigenome M39 and infected with the helper virus.
To rescue the synthetic cDNA minigenome, TGEV strain PUR46-MAD-infected cells were transfected with the corresponding cDNA. Culture supernatants were passed three to six times on fresh ST cell monolayers in order to amplify the viral RNA. Intracellular RNA was extracted, and the levels of mgRNA and sgmRNA were evaluated at passage 2 by Northern blot analysis with two probes (Fig. 1B)—one that was complementary to the 3′ end of the genome and that detected the genomic RNA and the viral mRNAs (S, 3a, E, M, N, and 7) and another that was complementary to the GUS gene and that exclusively hybridized with mgRNA and sgmRNA carrying the GUS gene.
RNA expression levels under the control of TRSs derived from the three virus genes encoding the major structural proteins (S, M, and N) were studied. The TRSs were inserted at nt 3337 of the minigenome and contained the highly conserved CS flanked at the 3′ end by a 10-nt sequence of nonviral origin, 5′-GUCGACGACC-3′ (L3), including a unique restriction endonuclease site and an optimized Kozak sequence (19, 20) (Fig. 1A). These expression cassettes differ at the 5′ end of the CS, where 5′ TRSs with similar lengths (88, 91, and 69 nt), derived from the three major viral genes (N, M, and S genes), were inserted (Fig. 1A). The three minigenomes were efficiently rescued, as shown by Northern blot analysis. Hybridization was stronger with the probe complementary to the GUS gene than with the probe specific for the 3′ end of the genome (Fig. 1B). A fivefold increase in the amount of the probe complementary to the 3′ end of the genome did not significantly improve the visualization of the mgRNA or the sgmRNA.
Transcription efficiency under the control of each TRS was determined by studying the molar ratio of sgmRNA to the corresponding mgRNA; this ratio was about twofold higher for the minigenome including the N gene 5′ TRS than for that including the M gene or S gene 5′ TRS (Fig. 1C). In order to compare the total quantities of mRNA produced, mgRNA amounts were normalized, since the insertion of TRSs into minigenomes could affect their replication level. To this end, an average value of M and N mRNAs was taken as an internal standard. The total sgmRNA level was maximum when the 5′ TRS derived from the N gene was used. This level was about two- and fourfold higher than those produced when the 5′ TRSs were derived from the M and S genes, respectively (Fig. 1C). Apparently, the amounts of sgmRNA are the same for the N and M genes (Fig. 1B), but due to the normalization of the amounts of RNA loaded in each lane, it was estimated that the amount of the N gene sgmRNA was higher. The ratios of sgmRNA to mgRNA were maintained essentially identical between passages 2 and 5. Then, smaller minigenomes were generated, and these ratios were not further estimated because of the instability of the minigenomes.
The insertion of TRSs including sequences from the N, M, and S genes in expression cassettes in minigenome M39 had little effect on helper virus growth, with titers ranging from 3 × 108 to 6 × 108 PFU/ml. These titers were maintained through six passages. The expression of GUS activity through these passages was between 500- and 1,000-fold over the background level (Fig. 1D) and was about 2- to 4-fold higher for minigenomes containing the N gene 5′ TRS than for those containing the M or S gene 5′ TRS. The GUS activity detected corresponds to protein expression levels of about 2 to 8 μg/106 cells. These GUS expression levels were confirmed by Western blot analysis with a GUS-specific rabbit antiserum (Fig. 1E).
Influence of the length of the N gene 5′ TRS on sgmRNA transcription.
To study the effect of the 5′ TRS on sgmRNA transcription, seven expression cassettes were constructed by using minigenome M39 with different fragments from the N gene TRS (Fig. 2A). A cassette lacking the CS (5′-CUAAAC-3′) (mg-CS−) was constructed as a negative control. The other six minigenomes included the CS alone (mg-CS) or flanked at the 5′ end by an increasing number (3, 8, 44, 88, and 176) of nucleotides derived from the N gene TRS (mg-N-3, mg-N-8, mg-N-44, mg-N-88, and mg-N-176, respectively). In all these expression cassettes, the sequences (L3) 3′ to the CS were kept constant and consisted of 10 nt including a unique endonuclease restriction site and an optimized Kozak sequence (Fig. 2A).
FIG. 2.
Influence of TRS length on transcription. (A) Schematic structures of expression modules (shown below the shaded area) based on TGEV-derived minigenome M39. The expression cassettes are flanked by L1 and L4 polylinkers. The TRS includes N gene 5′-flanking sequences of different lengths, the CS, the L3 polylinker, an optimized Kozak sequence (K), and the GUS gene. HDV, HDV ribozyme; BGH, BGH termination and polyadenylation signals. Designations to the left of the expression cassettes indicate the composition of the minigenome (mg), including the gene providing the TRS and the number of nucleotides of this TRS. (B) Northern blot analysis of intracellular RNAs extracted at passage 2 from minigenome-transfected and TGEV-infected cells. The left and right panels show hybridizations with 3′ UTR- and GUS-specific probes, respectively. The positions of the genomic RNA (g) and mRNAs (S, 3a, E, M, N, and 7) from the helper virus are indicated to the left by their acronyms. M39 RNA overlaps mRNA 3a (first lane). mgRNAs including expression cassettes with TRSs of 3, 8, 44, 88, and 176 nt from the N gene (mg-N-3, mg-N-8, mg-N-44, mg-N-88, and mg-N-176, respectively) and the mRNA for the GUS gene (sgmRNA) are indicated with arrows. (C) GUS activity (per 106 cells) expressed by the minigenomes under the control of the CS alone (mg-CS) or preceded by TRSs of different lengths from the N gene through six passages after transfection. Background levels are represented by mg-CS−. RLUs, relative luminometric units. The data shown are averages for at least three experiments with similar results. (D and E) Quantification of RNAs detected by Northern blotting in passage 2, shown as the means and standard errors for three independent experiments. (D) Ratio of sgmRNA to mgRNA. (E) Total amount of mRNA. This amount was corrected by taking into account the different amounts loaded in the lanes. (F) Quantification of the expression of GUS at virus passage 2 for the experiment shown in panel C. Data are means and standard errors of the means.
All the minigenomes were efficiently replicated and packaged when transfected in ST cells infected with the helper virus, as determined by Northern blot analysis (Fig. 2B). In addition, the synthesis of helper virus RNAs was not significantly altered by the presence of the mgRNAs (Fig. 2B). mgRNA and sgmRNA were clearly detected by using the probe complementary to the GUS gene.
The absence of transcription after CS removal was initially evaluated by testing GUS activity (Fig. 2C). Restoration of the CS led to an increase in GUS expression of up to 1,000-fold over the background level. GUS expression further increased about fourfold when the CS was extended 88 nt on its 5′ flank (Fig. 2C and F), in agreement with the increase in the total sgmRNA amount (Fig. 2E). Interestingly, GUS activity levels decreased when the 5′ TRS was extended by 176 nt. Helper virus titers (about 9 × 108 PFU/ml) remained constant upon passage (data not shown); however, GUS expression levels remained constant until passage 5, and then a reduction in GUS expression levels was observed (Fig. 2C). This reduction in GUS expression levels with minigenome passage most likely was a consequence of the instability of the minigenome with GUS, since after passage 5 the appearance of minigenomes of a smaller size was observed (2).
In accordance with GUS expression levels, the absence of the CS led to a complete abrogation of GUS sgmRNA transcription, as expected. Insertion of the CS restored the sgmRNA levels, up to 1,000-fold over the background level. The extension of the CS with 3 and 8 nt led to an initial twofold reduction of the molar ratio of sgmRNA to mgRNA. There was a two- to threefold increase in sgmRNA synthesis (in comparison to the results obtained with constructs containing the CS alone) when 44 or 88 nt derived from the 5′ TRS of the TGEV N gene were added to the CS (Fig. 2D). Interestingly, when the 5′ TRS was further extended by 176 nt derived from the N gene, the ratio of sgmRNA to mgRNA was fourfold lower than that produced by the 5′ TRS flanking sequence of 88 nt. Total sgmRNA levels in the presence of the TRS containing 88 nt plus the CS were fourfold higher than those in the presence of the CS alone (Fig. 2C). A comparison of the amount of total mRNA accumulated (Fig. 2E) and the amount of GUS expressed (Fig. 2F) indicates that once a minimum amount of mRNA is produced, there is a relatively smaller increase in the amount of GUS expressed.
These results indicate that the CS (5′-CUAAAC-3′) is essential for the transcription of TGEV-derived sgmRNAs, that the addition of nucleotides derived from the 5′ TRS of the N gene 5′ upstream of the CS influences transcription efficiency, and that maximum sgmRNA levels are provided by CS 5′-flanking sequences that are about 88 nt long.
Effect of sequences 3′ downstream of the CS (3′ TRSs) on sgmRNA transcription.
To study whether the 3′ TRSs influence sgmRNA levels, we engineered GUS expression cassettes in which the CS 5′-flanking sequences were maintained constant (88 nt from the N gene 5′ TRS). In seven constructs, the CS was flanked at the 3′ end by each of the sequences flanking the CS in viral genes S, 3a, 3b, E, M, N, and 7. In addition, two additional expression cassettes (L2 and L3) were constructed with sequences of nonviral origins inserted at the 3′ end of the CS. These sequences comprised 31 nt (L2) and 10 nt (L3) including four and one unique restriction endonuclease sites, respectively, and an optimized Kozak sequence (Fig. 3A). A third construct (mg-Ld) was designed to extend with an additional 12 nt the potential base pairing between the 3′ end of the leader and the sequence complementary to the TRS (Fig. 3A). The added nucleotides were identical to those present in the genomic RNA behind the CS at the 3′ end of the leader.
FIG. 3.
Influence of 3′ TRSs on transcription, as determined by using 3′ TRSs of viral and nonviral origins. (A) Schematic structures of expression cassettes cloned into TGEV-derived minigenome M39. The TRS includes 88 nt of the 5′ TRS from the N gene preceding the CS. The sequences flanking the 3′ end of the CS are derived from viral genes (S, 3a, 3b, E, M, N, and 7) or from nonviral sequences (L2 and L3). L3 consists of 10 nt including one restriction site and an optimized Kozak sequence (K). L2 consists of 31 nt including four restriction endonuclease sites and an optimized Kozak sequence. In addition, a sequence (Ld) comprising 12 nt complementary to the 3′ end of the leader was analyzed. (B) Northern blot analysis of intracellular RNAs extracted at passage 2 from cells transfected with the indicated minigenomes and infected with TGEV. The left and right panels show Northern blot analysis with 3′ UTR- and GUS-specific probes, respectively. The positions of the genomic RNA (g) and mRNAs (S, 3a, E, M, N, and 7) from the helper virus are indicated to the left by their acronyms. M39 RNA overlaps mRNA 3a (first lane). mgRNAs encoding expression cassettes with different TRSs and the mRNA for the GUS gene (sgmRNA) are indicated with arrows. (C) Quantification of RNAs detected by Northern blotting and of GUS activity at passage 2. The data are the means and standard errors for three independent experiments. (D) GUS activity (per 106 cells) expressed by minigenomes encoding the GUS gene under the control of 3′ TRSs of viral and nonviral origins through three passages after transfection. Background levels are those observed with minigenome M39. RLUs, relative luminometric units. The data are averages for at least three experiments with similar results.
Amplification of the helper virus RNAs remained unaltered in coinfections with all the minigenomes studied (virus titers, about 4 × 108 PFU/ml) and remained stable upon passage (data not shown). Viral mRNAs, minigenomes, and sgmRNA were evaluated by Northern blotting by using probes for the 3′ UTR and GUS (Fig. 3B). The minigenomes were rescued, and similar molar ratios of sgmRNA to mgRNA were produced for all of them, except for those encoding the M and L2 3′ TRSs, which gave molar ratios two- to threefold higher and fivefold lower than those for the other minigenomes, respectively. Similar amounts of total sgmRNA were produced, except for minigenomes encoding the 3a 3′ TRS (two- to threefold increase) and the L2 3′ TRS (fivefold decrease) (Fig. 3C), probably because the different inserts affected the efficacy of minigenome replication in different ways.
GUS expression by minigenomes with the CS 3′-flanking sequences derived from different viral genes (S, 3a, E, M, N, and 7) differed by 10-fold, being maximum for minigenomes mg-Ld and mg-M. GUS activity synthesized by minigenome mg-L2 throughout virus passage was between 10- and 100-fold lower than that for the other constructs (Fig. 3D). The reduction in mRNA and GUS levels was also observed when the L3 3′ TRS was replaced by that from L2 in the minigenomes mg-M-91-L2 and mg-S-69-L2, containing 5′ TRSs derived from the M and S genes, respectively (data not shown). These data indicate that CS 3′-flanking sequences have a great deal of influence on sgmRNA accumulation and protein synthesis.
Reconstruction of the CS 5′ upstream of the TGEV ORF 3b CS and of a noncanonical CS within this ORF led to mRNA synthesis.
In strain PUR46-MAD of TGEV, the CS of gene 3b has a mutation in the sixth nucleotide (U instead of C), and the corresponding mRNA, named 3-1, is not synthesized. To study expression with minigenome M39, containing a TRS derived from ORF 3b, this CS was repaired to reconstitute the canonical CS in the minigenome (including the CS 3′-flanking sequences of this gene). In fact, the expression of the corresponding sgmRNA was restored to levels similar to those driven by ORF 3a (data not shown). This result is in accordance with observations made by using strain MIL65-AME of TGEV (37). This strain has a canonical CS upstream of gene 3b, and synthesis of the expected mRNA takes place (52).
To determine whether sgmRNA could be driven by modified CSs generated in the viral genome, a noncanonical CS (C_AAAC) located inside gene 3b was repaired by inserting the missing U. By using minigenome M39, an expression cassette was constructed with a 5′ TRS from the N gene, a standard CS, and the flanking sequences at the 3′ end of the imperfect CS. Interestingly, sgmRNA levels obtained with the helper virus-dependent expression system were as high as those obtained with the 3′ TRS present in the native viral ORFs, even though the repaired TRS contains CS 3′-flanking sequences from a variant TRS in the TGEV genome (data not shown).
These results on the reconstruction of noncanonical sequences located in positions where viruses of the same group have a canonical CS 5′ upstream of a gene or inside a gene, in locations that are not expected to lead to the synthesis of viral mRNA, indicate that the insertion of a CS in the appropriate context of a minigenome promotes transcription. The same observation was made when a CS was inserted in a full-length genome by engineering an infectious cDNA clone (results not shown).
Expression of TGEV sgmRNAs from TRSs along the virus genome.
TGEV has nine CSs along the genome, one at the 3′ end of the leader, one at the 5′ end of each of the seven viral genes, and one additional CS located inside the coding sequence of the S gene (at nt 120 of this gene). The CS present at the 5′ end of each gene seems to be used to express the corresponding mRNA (Fig. 1B, 2B, 3B, 4B, and 5B), unless mutated, as happens with ORF 3b of TGEV strain PUR46-MAD. We were interested in knowing whether the second canonical CS present within the coding sequence of the S gene was used to generate a small S gene mRNA. RT-PCR analysis to identify the standard and potential second S gene mRNAs was performed (Fig. 4) by using a primer complementary to the leader sequence and another one complementary to positions of the S gene 3′ downstream of the internal CS. Interestingly, only one band was amplified, corresponding to the full-length mRNA (mRNA S1) (Fig. 4). The mRNA controlled by the CS located 5′ upstream of the S gene was at least 200-fold more abundant than the mRNA starting at the internal CS of the S gene, as determined by RT-PCR of twofold dilutions of the expressed mRNAs (data not shown). This observation indicated that the presence of the CS may not be sufficient to induce transcription because it can be silenced by flanking sequences or because of the absence of sequences required to activate transcription in this context.
FIG. 4.
RT-PCR analysis of S gene mRNA expression. (A) Schematic diagram depicting the structure of the viral genomic RNA (gRNA) providing the position of the S gene CSs. RT-PCR was performed for the detection of sgmRNAs (mRNA S1 and mRNA S2) potentially generated from two CSs located 27 nt 5′ upstream (CS-S1) and 120 nt 3′ downstream (CS-S2) of the initiation codon of the S gene. Arrows indicate the approximate positions of the primers used for RT (gray arrow) and PCR (black arrows). The black box represents the leader sequence. (B) Analysis by electrophoresis in an agarose gel of RT-PCR products of the mRNAs produced. Lane M, molecular markers; lanes RT-PCR and PCR, bands observed after PCR amplification alone (PCR) or preceded by RT (RT-PCR).
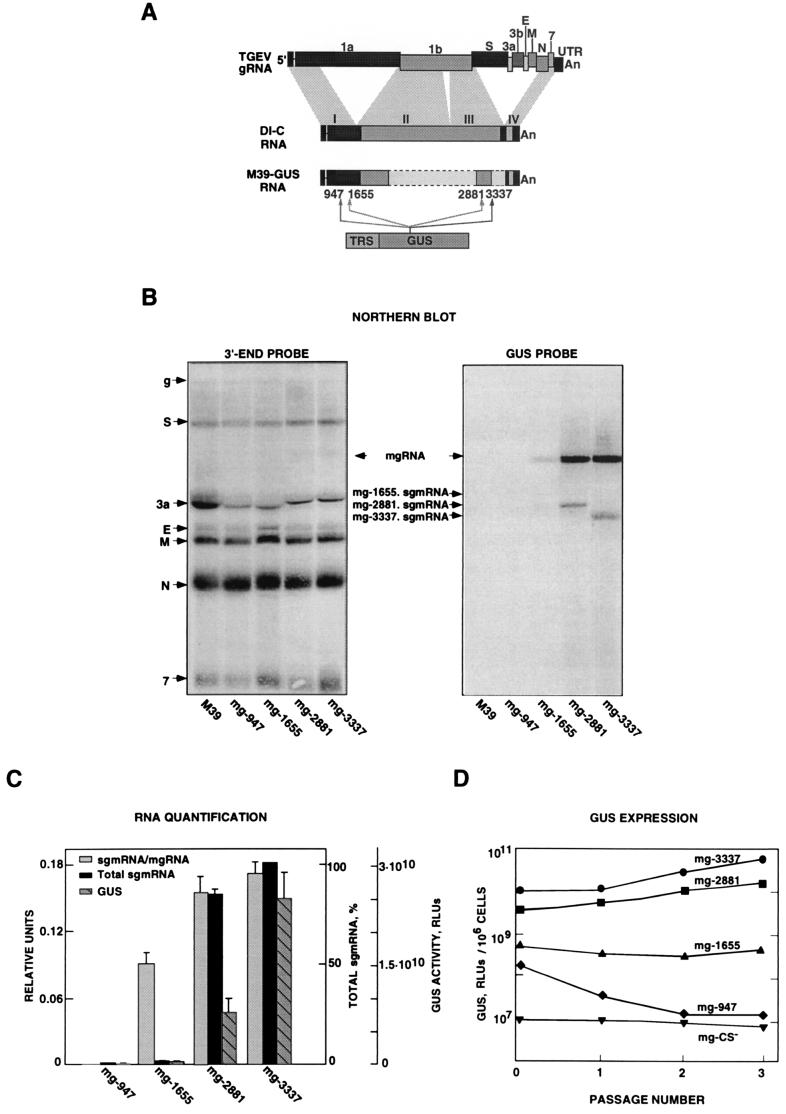
FIG. 5.
Influence of expression cassette insertion site on transcription. (A) Diagram showing the origins in the virus genome of fragments I, II, III, and IV (shaded areas), cloned in order to assemble the cDNA encoding DI-C RNA. The bars defined by broken lines indicate the areas deleted in minigenome DI-C to obtain M39. The expression cassette comprising the TRS and the GUS gene was cloned in four different positions along minigenome M39. The TRS includes 88 nt from the 5′ TRS of the N gene, the CS, and polylinker L3 (Fig. 2). The numbers below the bars indicate the four positions (947, 1655, 2881, and 3337) where the expression cassette was inserted in relation to the first nucleotide of the minigenome. S, 3a, 3b, E, M, N, and 7 indicate the positions of the corresponding genes. An, poly(A). (B) Northern blot analysis of intracellular RNAs extracted at passage 2 from minigenome-transfected and TGEV-infected cells. The left and right panels show Northern blot analysis with 3′ UTR- and GUS-specific probes, respectively. The positions of the genomic RNA (g) and mRNAs (S, 3a, E, M, N, and 7) from the helper virus are indicated to the left by their acronyms. M39 RNA overlaps mRNA 3a (first lane). The data shown are the means and standard errors for three independent experiments. The positions of mgRNAs inserted at different positions of M39 and GUS sgmRNA are indicated by arrows. (C) Quantification of RNAs detected by Northern blotting and of GUS activity at passage 2. (D) GUS activity (per 106 cells) expressed by minigenomes including the GUS gene at different positions through three passages after transfection. Background levels are represented by minigenome mg-CS−. RLUs, relative luminometric units. The data shown are averages for three experiments with similar results.
Influence of the insertion site on transcription.
To examine whether the location of an expression cassette within a minigenome affects transcription efficiency and expression of a heterologous gene, four minigenomes were constructed. In these constructs, an expression cassette encoding the GUS gene under the control of a TRS containing 88 nt of the N gene, the CS, and 10 nt including an optimized Kozak sequence (L3) was inserted at different distances (947, 1,655, 2,881, and 3,337 nt) from the minigenome M39’s 5′ end (Fig. 5A). The presence of the minigenome did not significantly affect the amplification of the helper virus RNAs during virus passage; helper virus titers ranged from 2.6 × 108 to 4.2 × 108 PFU/ml (data not shown). The presence of mgRNA or sgmRNA was weakly or strongly detected with the probes complementary to the 3′ end of the genome and the GUS gene, respectively (Fig. 5B). Insertion of the expression cassette in different positions of the minigenome had a strong effect on its rescue (replication and packaging) by the helper virus. Minigenome replication was completely or partially abolished when the expression cassette was inserted 947 nt or 1,655 nt from the 5′ end, respectively. In contrast, insertion at nt 2881 or 3337 led to a high level of minigenome amplification (Fig. 5B).
Insertion of the expression cassette at position 947 abrogated minigenome amplification and consequently reduced the sgmRNA level to the background level (i.e., that obtained with a minigenome without a TRS upstream of the GUS gene). In contrast, insertions at the other two positions (2881 and 3337) led to the synthesis of large amounts of the minigenome and to the production of high molar ratios of sgmRNA to mgRNA (Fig. 5C). Although the sgmRNA/mgRNA ratio was relatively high when the expression cassette was inserted at nt 1655, the absolute amount of sgmRNA produced was very low, since the mgRNA level was also very low (Fig. 5C).
GUS expression levels at passage 2 were very low for the two expression cassettes inserted closer to the 5′ end of the minigenome (at nt 947 and 1655) (Fig. 5D) and increased 1,000- and 3,000-fold (in relation to the background level) for the insertions closer to the 3′ end of the minigenome. GUS expression levels were also analyzed at different passages. When minigenome mg947 (encoding the GUS gene at position 947) was used, GUS activity sharply decreased to the background level at passage 2 (Fig. 5D), suggesting that the replication or packaging of the minigenome was inhibited. Expression from a cassette inserted at position 1655 remained at levels between 10- and 100-fold lower than the levels of expression from cassettes inserted at positions 2881 and 3337.
With changing of the insertion site of the expression cassette, a potential second variable was introduced: the sequences flanking the expression module. Nevertheless, since the CS is flanked at the 3′ end by the heterologous GUS gene (1,812 nt) and at the 5′ end by 88 nt, the possibility of modifying the structure of the CS is greatly reduced, although it cannot be completely excluded.
DISCUSSION
In this report, we have partially delimited the extension of the essential TRS required for efficient expression of heterologous genes in TGEV by analyzing sequence requirements at the 5′ and 3′ flanks of the highly conserved CS of TGEV that improved RNA synthesis (accumulation). Overall, the molar ratio of sgmRNA to total mgRNA was increased 12.5-fold by engineering the TRS by using CS 5′-flanking sequences derived from the N gene, the CS, and CS 3′-flanking sequences derived from the M gene. We have shown that the hexanucleotide 5′-CUAAAC-3′ can drive transcription in the context of the minigenome and in the context of the full-length genome (data not shown) and that sequences flanking this CS influenced transcription. This conclusion is in agreement with conclusions drawn from a series of studies with MHV defective interfering (DI) RNAs (10, 14, 17, 21, 27, 34, 49, 51) and with DI RNAs derived from BCoV (31). In addition, it has been shown that when a sequence closely related to the CS of the viral genome was engineered to reconstitute the canonical sequence 5′-CUAAAC-3′, the corresponding mRNA was synthesized. TGEV CS seems very sensitive to changes, since deletion of the U or mutation of the last C abrogated transcription. In addition, it was shown that the sequences flanking the CS or the site of insertion of the expression cassette influenced mgRNA levels, ranging from a discrete (two- to threefold) modulation of relative sgmRNA synthesis to the complete abrogation of transcription (such as for the S gene internal CS); this result was due either to inhibition of this transcription or to the lack of an activating domain. It has also been shown that viral protein expression levels are dependent on both transcriptional and translational regulation. The translational regulation role is deduced from the different amounts of GUS expressed by similar amounts of total mRNA. This result was expected, because the different mRNAs have immediately 5′ upstream of the translation initiation codon Kozak motifs that match to different extents the ones optimized by Kozak (19, 20). Higher expression levels correlated with the presence of canonical Kozak motifs.
TGEV has nine highly conserved CSs with the sequence 5′-CUAAAC-3′, including one at the 5′ end of each ORF (Rep, S, 3a, 3b, E, M, N, and 7) and an internal CS in the S gene. ORF 3b CS is mutated to 5′-CUAAAU-3′ in TGEV of the Purdue cluster, leading to abrogation of the corresponding mRNA. Interestingly, when the mutated 5′-CUAAAU-3′ sequence of the PUR46-MAD clone of TGEV was replaced by the canonical 5′-CUAAAC-3′ sequence, as happens in strain MIL65, the corresponding mRNA was produced (37, 52). Furthermore, within the 3b gene there is a sequence, 5′-C_AAAC-3′, starting at nt 246 from the first AUG of the 3b gene which could be transformed to the canonical 5′-CUAAAC-3′ sequence by the insertion of a U. When this expression cassette was generated, maintaining the CS 3′-flanking sequences downstream of the imperfect CS, mRNA expression levels were comparable to those produced when natural CS 3′-flanking sequences were used. Altogether, these data suggest that the 5′-CUAAAC-3′ sequence (CS) is an essential determinant of mRNA transcription in TGEV and that there is flexibility for the sequences flanking the CS at the 3′ end.
The S gene has two canonical CSs associated with it, one 5′ upstream of the first codon and a second one 120 nt downstream, within the coding region. Interestingly, only the mRNA corresponding to the full-length mRNA was amplified by RT-PCR, strongly suggesting that the mRNA corresponding to the internal CS is produced in minimal amounts or not at all. This result indicates that not all the CSs are used to promote mRNA synthesis and that expression from a canonical CS may be prevented by a nonfavorable context due to its neighbor sequences. Predictions of secondary structures for all the TGEV TRSs were made by using the M-fold program (29, 53) (data not shown), and a favored structure that could differentiate functional from nonfunctional TRSs was not identified. Therefore, the nature of the flanking sequences themselves and not their secondary structures could be responsible for silencing the S gene internal CS. These results are in agreement with recently reported data, obtained with BCoV DI RNAs, suggesting that a secondary structure surrounding a CS motif might not be a primary determinant in the choice of that site for leader fusion (31). Experiments to determine the nature of the suppressor sequences are in progress. The lack of expression of the S gene internal CS indicates that the corresponding mRNA ought to be nonessential for TGEV replication. Consistent with this observation is the fact that the internal CS is present in some but not all TGEV isolates (37).
Like the TGEV CS, the MHV CS contains the conserved sequence 3′-UCUAAAC-5′ or a closely related one (49). Cloning of short oligonucleotides ranging from 10 to 18 nt, comprising the CSs in MHV minigenomes, showed that these sequences alone were sufficient to direct DI sgmRNA synthesis (48). Deletions within the TRS reduced its strength, implying that efficient sgmRNA synthesis requires a TRS larger than the heptamer CS (5′-UCUAAAC-3′) (18, 26, 27, 48). With TRSs of 13 or 17 nt, the synthesis of mRNA in MHV is affected only slightly when a single nucleotide is mutated, and MHV transcription regulation is sufficiently flexible to recognize altered CSs. In contrast, the introduction of the same substitutions in some positions of a 10-nt TRS results in a more than 10-fold reduction of transcription (18, 48).
We showed that the nature and length of the sequences 5′ upstream of the TGEV CS influenced transcription. When the CS length was increased by 88 nt, a two- to threefold enhancement of the molar ratio of sgmRNA to total mgRNA was observed relative to the results obtained with the CS alone. By extending the 5′ TRS with 176 nt derived from the TRS of the N gene, this ratio was decreased, indicating that, at least for TRSs based on the N gene, maximum transcription levels were provided by CS 5′-flanking sequences of about 88 nt.
Sequences 3′ downstream of the CS also influenced mRNA transcription levels in TGEV. Expression modules in which the 5′-flanking sequence of the CS was kept constant and the CS 3′-flanking sequences were taken from each of the TGEV genes (S, 3a, 3b, E, M, N, and 7) led to a fourfold change in the molar ratio of sgmRNA to total mgRNA. This ratio did not correlate with the distances from the CS to the translation initiation codon (AUG) of each gene, ranging from 3 to 37 nt. Interestingly, in these constructs the level of translation (i.e., GUS activity) was related to the presence of a favorable Kozak motif within the 3′-flanking sequences (i.e., maximum expression levels for a given amount of RNA were obtained for the optimum Kozak context), most likely because of the proximity of these sequences to the translation initiation codon. Translation levels only partially correlated with the total amount of sgmRNA produced. Once a certain amount of viral mRNA was produced, GUS levels were influenced by the translation efficiency of each mRNA, suggesting that viral protein production was regulated at both the transcriptional and the translational levels.
The minor changes in the sequences of mgRNAs most likely did not affect translocation of these RNAs from the nucleus to the cytoplasm, since the sequences of these RNAs have been slightly modified inside the RNA molecule and changes in RNA translocation probably are not responsible for variations in expression levels. Although a hypothetical effect of RNA translocation efficiency on the amount of mgRNA available in the cytoplasm cannot be completely ruled out, that alteration would affect very little the ratio of sgmRNA to mgRNA provided in this report, since sgmRNA and the majority of mgRNA are synthesized within the cytoplasm by the RNA-dependent RNA polymerase of the helper virus.
To further test whether the extent of the complementarity between the 3′ end of the leader and the sequences flanking the 3′ end of the CS could influence mRNA levels, an expression cassette (Ld) was constructed to extend by an additional 12 nt the complementarity of the sequence downstream of the CS with the 3′ end of the leader. Although the expression levels were among the highest, neither the molar sgmRNA/mgRNA ratio nor the amount of total sgmRNA was higher than those observed for other TRSs with a lower potential for base pairing (such as the M gene-derived sequences). In addition, it was previously reported (33) that TGEV mRNA abundance is not directly proportional to the potential base pairing between the 3′ end of the viral genomic leader and the sequence complementary to the CS in the TRS. Overall, our results show that transcription in TGEV requires a duplex of minimum stability. Once this condition is met, increasing the potential base pairing between the TRS and the leader 3′ end does not enhance transcription. This conclusion is in agreement with previous results reported for MHV. In this system, TRS activity became optimal when a CS of 18 nt from a region showing full complementarity to the 3′ end of the MHV genomic leader was used, and extension of this complementarity did not increase expression levels (10, 24, 28, 45).
To study the effect of TRS position within the minigenome, an expression cassette that included 88 nt derived from the N gene 5′ TRS was inserted at different distances (947, 1,655, 2,881, and 3,337 nt) from the 5′ end of a TGEV minigenome of 3.9 kb. The mRNA levels were high at the two insertion sites closest to the 3′ end of the minigenome. In contrast, in a minigenome derived from human coronavirus (HCoV) 229E, an expression cassette cloned into three different positions (1.1, 1.3, and 1.8 kb from the 5′ end of a minigenome of about 2.1 kb) resulted in decreased mRNA levels for inserts located closer to the 3′ end (47). Experiments performed with TGEV and HCoV 229E led to different results. With TGEV and possibly with HCoC 229E, some insertion sites were too close to the ends of the minigenome and affected essential primary or secondary structures required for their replication. It is possible that in helper virus-dependent expression systems based on coronavirus-derived minigenomes encoding a limited number of sgmRNAs, variation in the expression level with the insertion site is influenced mainly by the sequences flanking the TRS in each position and that the relative position itself plays a less prominent role. In fact, in a systematic study with MHV, a TRS of 12 nt flanked by 0.2-kb upstream and 0.2-kb downstream sequences was inserted at seven different positions of a 2.2-kb minigenome (15). The 12-nt TRS core was flanked by upstream and downstream sequences in order to prevent the influence of variable flanking sequences within the different insertion sites. In all insertion sites, the level of the minigenome and the level of the mRNA produced were similar, suggesting that the position of the insert along the minigenome had little influence on the mRNA expression level. The lack of an insertion site effect on transcription levels probably does not apply to expression levels in full-length genomes with several sgmRNAs.
The helper virus-dependent expression system used has the advantage of high cloning capacity, which theoretically is higher than 22 kb, since the total length of the TGEV genome is 28.5 kb and the length of the minigenome is less than 6 kb. In contrast, it has limited stability, mainly due to the foreign gene, since TGEV minigenomes of 9.7, 5.4, 3.9, and 3.3 kb, in the absence of the heterologous gene, were amplified, packaged, and efficiently propagated for at least 30 passages without the generation of new dominant sgmRNAs (12, 30).
The helper virus-dependent expression system described in this report produced large quantities of heterologous antigen. The expression of the reporter GUS gene (2 to 8 μg/106 cells) and the expression of porcine respiratory and reproductive syndrome virus ORF 5 (1 to 2 μg per 106 cells) have been shown by using a TGEV-derived minigenome (2). The expression of GUS was detected in the lungs of swine immunized with the virus vector. Interestingly, strong humoral immune responses to both GUS and porcine respiratory and reproductive syndrome virus ORF 5 were induced in swine with this virus vector. The large cloning capacity and the tissue specificity of the TGEV-derived minigenomes suggest that these virus vectors are very promising for vaccine development. This helper virus-dependent expression system has also been very useful for the characterization of TGEV TRSs. Fortunately, infectious cDNAs are available for TGEV (1) and HCoV 229E (46), and that availability will permit verification of the conclusions reached with minigenomes by use of full-length genomes. This aspect is particularly important, since most of the information on coronavirus transcription has been generated by using helper virus-dependent expression systems based on minigenomes encoding sgmRNAs (12, 21, 23, 27, 35, 48, 49), and it has been shown that the relative flexibility of the TRS requirement in the DI RNA system significantly differs from that seen in viral genomic RNA (18, 23, 32).
Acknowledgments
We thank F. Almazán, J. Ortego, and S. Zúñiga for critically reading the manuscript.
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects). A.I. and S.A. received fellowships from the Department of Education, University and Research of the Gobierno Vasco. I.S. received a postdoctoral fellowship from the Community of Madrid and the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects: QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874).
REFERENCES
- 1.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., I. Sola, H. Wege, J. Teifke, M. Balach, J. Plana-Durán, and L. Enjuanes. Heterologous gene expression in tissue culture and in vivo using a transmissible gastroenteritis coronavirus helper dependent system. J. Gen. Virol., in press.
- 3.Bronstein, I., J. J. Fortin, J. C. Voyta, R.-R. Juo, B. Edwards, C. E. M. Olenses, N. Lijam, and L. J. Kricka. 1994. Chemiluminescent reporter gene assays: sensitive detection of the GUS and SEAP gene products. BioTechniques 17:172–177. [PubMed] [Google Scholar]
- 4.Dubensky, T. W., D. A. Driver, J. M. Polo, B. A. Belli, E. M. Latham, C. E. Ibanez, S. Chada, D. Brumm, T. A. Banks, S. J. Mento, D. J. Jolly, and S. M. W. Chang. 1996. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J. Virol. 70:508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Coronaviridae, p.835–849. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, Inc., New York, N.Y.
- 6.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p.827–834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, Inc., New York, N.Y.
- 7.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann, M. A., S. D. Senanayake, and D. A. Brian. 1993. A translation-attenuating intraleader open reading frame is selected on coronavirus messenger RNAs during persistent infection. Proc. Natl. Acad. Sci. USA 90:11733–11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477–484. [DOI] [PubMed] [Google Scholar]
- 12.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Méndez, J. Plana-Durán, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong, Y. S., and S. Makino. 1992. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J. Virol. 66:3339–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong, Y. S., J. F. Repass, Y.-N. Kim, S.-M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez, G., I. Correa, M. P. Melgosa, M. J. Bullido, and L. Enjuanes. 1986. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo, M., and S. Makino. 1992. Mutagenic analysis of the coronavirus intergenic consensus sequence. J. Virol. 66:6330–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867–19870. [PubMed] [Google Scholar]
- 21.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, M. M. C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1–12. [DOI] [PubMed] [Google Scholar]
- 23.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Monica, N., K. Yokomori, and M. M. C. Lai. 1992. Coronavirus mRNA synthesis: identification of novel transcription initiation signals which are differentially regulated by different leader sequences. Virology 188:402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapps, W., B. G. Hogue, and D. A. Brian. 1987. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology 157:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus messenger RNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino, S., C.-K. Shieh, L. H. Soe, S. C. Baker, and M. C. Lai. 1988. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology 166:550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940. [DOI] [PubMed] [Google Scholar]
- 29a.McClurkin, A. W., and J. O. Norman. 1966. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Vet. Sci. 30:190–198. [PMC free article] [PubMed] [Google Scholar]
- 30.Méndez, A., C. Smerdou, A. Izeta, F. Gebauer, and L. Enjuanes. 1996. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology 217:495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasternak, A. O., A. P. Gultyaev, W. J. Spaan, and E. J. Snijder. 2000. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 74:11642–11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penzes, Z., J. M. González, A. Izeta, M. Muntion, and L. Enjuanes. 1998. Progress towards the construction of a transmissible gastroenteritis coronavirus self-replicating RNA using a two-layer expression system. Adv. Exp. Med. Biol. 440:319–327. [DOI] [PubMed] [Google Scholar]
- 35.Penzes, Z., C. Wroe, T. D. K. Brown, P. Britton, and D. Cavanagh. 1996. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 70:8660–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sánchez, C. M., F. Gebauer, C. Suñé, A. Méndez, J. Dopazo, and L. Enjuanes. 1992. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 190:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez, C. M., G. Jiménez, M. D. Laviada, I. Correa, C. Suñé, M. J. Bullido, F. Gebauer, C. Smerdou, P. Callebaut, J. M. Escribano, and L. Enjuanes. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174:410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, D. L., T. Wang, and S. G. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:386–396. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215–220. [DOI] [PubMed] [Google Scholar]
- 42.Schaad, M., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlaman, H. R. M., E. Risseeuw, M. E. I. Franke-van Dijk, and P. J. J. Hooykaas. 1994. Nucleotide sequence corrections of the uidA open reading frame encoding β-glucuronidase. Gene 138:259–260. [DOI] [PubMed] [Google Scholar]
- 44.Sethna, P. B., S.-L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shieh, C.-k., L. H. Soe, S. Makino, M.-F. Chang, S. A. Stohlman, and M. M. C. Lai. 1987. The 5′-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology 156:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel, V., J. Herold, B. Schelle, and S. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273–1281. [DOI] [PubMed] [Google Scholar]
- 47.Thiel, V., S. G. Siddell, and J. Herold. 1998. Replication and transcription of HCV 229E replicons. Adv. Exp. Med. Biol. 440:109–114. [DOI] [PubMed] [Google Scholar]
- 48.van der Most, R. G., R. J. De Groot, and W. J. M. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p.11–31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 50.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056–12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesley, R. D., A. K. Cheung, D. M. Michael, and R. D. Woods. 1989. Nucleotide sequence of coronavirus TGEV genomic RNA: evidence of 3 mRNA species between the peplomer and matrix protein genes. Virus Res. 13:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p.11–43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO ASI Series, Kluwer Academic Publishers, New York, N.Y.