Abstract
Kinetic analyses of infectivity loss during thermal inactivation of reovirus particles revealed substantial differences between virions and infectious subvirion particles (ISVPs), as well as between the ISVPs of reoviruses type 1 Lang (T1L) and type 3 Dearing (T3D). The difference in thermal inactivation of T1L and T3D ISVPs was attributed to the major surface protein μ1 by genetic analyses with reassortant viruses and recoated cores. Irreversible conformational changes in ISVP-bound μ1 were shown to accompany thermal inactivation. The thermal inactivation of ISVPs approximated first-order kinetics over a range of temperatures, permitting the use of Arrhenius plots to estimate activation enthalpies and entropies that account for the different behaviors of T1L and T3D. An effect similar to enthalpy-entropy compensation was additionally noted for the ISVPs of these two isolates. Kinetic analyses with other ISVP-like particles, including ISVPs of a previously reported thermostable mutant, provided further insights into the role of μ1 as a determinant of thermostability. Intact virions, which contain ς3 bound to μ1 as their major surface proteins, exhibited greater thermostability than ISVPs and underwent thermal inactivation with kinetics that deviated from first order, suggesting a role for ς3 in both these properties. The distinct inactivation behaviors of ISVPs are consistent with their role as an essential intermediate in reovirus entry.
The virions of mammalian orthoreoviruses (reoviruses) contain viral proteins arranged in two concentric icosahedral layers, commonly called the outer and inner capsids. During treatments with exogenous proteases in vitro, three proteins from the outer capsid can be sequentially removed to yield two well-characterized disassembly intermediates: the infectious subvirion particle (ISVP) and the core. ISVPs differ from virions in having lost the major outer-capsid protein ς3. In addition, the other major outer-capsid protein, μ1, which appears to have been cleaved near its N terminus in virions to yield particle-bound fragments μ1N and μ1C (29), has been cleaved again near its C terminus in ISVPs to yield the additional particle-bound fragments μ1δ/δ and φ (26). Cores differ from virions in having lost not only ς3 but also μ1 and its fragments as well as the receptor-binding outer-capsid protein ς1. Studies of these subvirion particles have been crucial for localizing proteins within the outer capsid as viewed by cryoelectron microscopy and three-dimensional image reconstruction (14).
In addition to their uses for studies of reovirus structure, ISVPs and cores are thought to represent disassembly intermediates that play essential roles during productive infection. Cores are active at transcription in vitro and according to one hypothesis represent the primary transcriptase particles that gain access to the cytoplasm during entry into cells and first synthesize the viral plus-strand RNAs for translation and packaging (6). Cores are poorly infectious through binding and uptake from the cell surface, since they lack the outer-capsid proteins ς1 and μ1, which have evolved to mediate entry (receptor binding by ς1 and membrane permeabilization by μ1) (reviewed in reference 28). ISVPs, in contrast, remain competent for infection after binding and uptake from the cell surface, since they retain these proteins. For infections in the small intestine of the mouse, ISVPs appear to be specifically required for their unique attachment properties that permit binding and uptake by M cells in the intestinal epithelium (1). In addition, the partial disassembly of virions to yield ISVP-like particles, with μ1 freed from its interactions with ς3, is thought to be required for penetration of the cellular membrane barrier following attachment and uptake, allowing the primary transcriptase particles to enter the cytoplasm (33). While the identification of these two distinct disassembly intermediates has provided a useful model for reovirus entry, a better understanding of the molecular basis of entry will require more complete descriptions of the structural transitions in reovirus particles and their associations with cellular components.
Structural transitions in particle-associated viral proteins can often be induced by chemical or biophysical perturbations, such as changes in temperature. As indicated by the following examples, such induced changes in viral proteins can be related to similar changes that occur during viral entry into cells and can provide new insights into the molecular basis of entry. Irreversible structural changes in the poliovirus capsid resulting in the generation of A-like particles occur with first-order kinetics over a range of temperatures in vitro and appear to be similar to changes that accompany poliovirus entry (34). The rate of these thermally induced changes is decreased by the binding of antiviral drugs that stabilize the capsid primarily through entropic effects and, as a result, block entry into the cell (34). Similarly, irreversible structural changes in the influenza virus hemagglutinin that accompany membrane fusion at acidic pH and physiological temperatures also occur at neutral pH when the temperature is elevated (7). Similarities in the structural changes in hemagglutinin under these two different sets of conditions provide evidence for the inherent metastability of the cleaved form of hemagglutinin that is found in infectious virions (7). Given the insights into viral entry provided by such analyses of thermally induced structural changes in viral proteins, we began an investigation of the thermal inactivation of reovirus particles.
Previous studies of the thermal inactivation of reovirus particles identified important protein determinants within the outer capsid. Drayna and Fields (12, 13) studied the inactivation of virions at 55°C and found differences between reoviruses type 1 Lang (T1L) and either type 2 Jones (T2J) or type 3 Dearing (T3D) that were genetically mapped to the S4 genome segment encoding the outer-capsid protein ς3. Loss of the receptor-binding protein ς1 was seen to accompany the inactivation of T2J virions in one of those studies (12). ISVPs were not analyzed by Drayna and Fields (12, 13), but a similar 52°C-induced elution of ς1 from T2J ISVPs was reported in another study (15). Jané-Valbuena et al. (19) extended analysis of the thermal inactivation of reovirus particles by showing that T1L ISVPs are less thermostable than T1L virions at 52°C but that T1L ISVPs which have been “recoated” with the ς3 protein in vitro are as thermostable as virions at that temperature. These findings indicate that ς3 is an important determinant of the thermostability of virions. Another insight into the thermal inactivation of reovirus particles was obtained from the finding that mutants of reovirus T3D selected for resistance to ethanol and attributed to mutations in the μ1 outer-capsid protein (37) showed an increase in the thermostability of their ISVPs, suggesting that μ1 is an important determinant of the thermostability of particles that lack ς3 (17).
Despite the insights they provided, the preceding studies did not analyze the thermal inactivation of reovirus particles in such a manner as to identify the kinetic or thermodynamic features of this phenomenon. Moreover, the relative roles of the different outer-capsid proteins in thermal inactivation remain unclear. The present experiments were designed to extend our understanding of the thermal inactivation of reovirus particles through studies of its kinetics. Reaction rates were determined over a range of temperatures and used to generate Arrhenius plots for determining the enthalpies and entropies of inactivation according to transition-state theory (30). Genetic and biochemical analyses were also performed to identify important viral determinants of the thermal inactivation behaviors. Although most of the experiments concerned the thermal inactivation of ISVPs, comparisons with virions were also performed. The results indicate that reovirus particles, particularly ISVPs, undergo thermal inactivation in a predictable manner that provides new insights into the determinants of particle stability as well as the roles of these particles and their component proteins in reovirus entry.
MATERIALS AND METHODS
Cells.
Spinner-adapted murine L929 cells were grown in Joklik’s modified minimal essential medium (Irvine Scientific Co., Irvine, Calif.) supplemented to contain 2% fetal bovine serum and 2% bovine calf serum (HyClone Laboratories, Logan, Utah) in addition to 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Irvine). Spodoptera frugiperda clone 21 and Trichoplusia ni TN-BTI-564 (High Five) insect cells (Invitrogen, Carlsbad, Calif.) were grown in TC-100 medium (Gibco BRL, Gaithersburg, Md.) supplemented to contain 10% heat-inactivated fetal bovine serum.
Virions, ISVPs, and cores.
Virions of reoviruses T1L, T3D, ethanol-resistant mutant 3a9, and the T1L × T3D reassortants in Table 1 were obtained by the standard protocol (15) and stored in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris [pH 7.5]). Purified ISVPs were obtained by the same protocol except that after the second extraction with trichlorotrifluoroethane (Fisher Scientific, Fair Lawn, N.J.), virions were diluted in virion buffer and pelleted by centrifugation at 5°C in an SW28 rotor (Beckman Instruments, Palo Alto, Calif.) at 25,000 rpm for 2 h. The pelleted virions were resuspended in virion buffer at a concentration lower than 1013 particles/ml and treated with 200 μg of N-p-tosyl-l-lysine chloromethyl ketone (TLCK)-treated chymotrypsin (Sigma Chemical Co., St. Louis, Mo.) per ml for 50 min at 37°C. ISVPs were then purified on a preformed CsCl gradient (15) and stored in virion buffer. Nonpurified ISVPs were obtained by digesting virions at a concentration of 1012 particles/ml in 200 μg of TLCK-treated chymotrypsin/ml for 20 min at 37°C. Digestion was stopped by addition of phenylmethylsulfonyl fluoride to 1 mM. Production of ISVPs was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). T1L detergent- and protease-treated particles were prepared by protease digestion of virions in the presence of sodium tetradecyl sulfate (Aldrich Chemical Co., Milwaukee, Wis.) as previously described (8). Cores were prepared by digestion of virions at a concentration in excess of 1013 particles/ml with TLCK-treated chymotrypsin as described elsewhere (9). Particle concentrations were estimated by A260 (11).
TABLE 1.
Genetic analysis of the difference in thermal inactivation between T1L and T3D ISVPs
| Virus isolatea | Allelic origin of genome segment (structural protein in ISVPs)b
|
Infectivity change (log10 PFU/ml)c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 (λ3) | L2 (λ2) | L3 (λ1) | M1 (μ2) | M2 (μ1) | M3 | S1 (ς1) | S2 (ς2) | S3 | S4 | ||
| G2 | L | D | L | L | L | L | D | L | L | L | 0.2 ± 0.2 |
| E3 | D | D | D | D | L | D | D | D | D | D | −0.3 ± 0.4 |
| EB15 | D | D | L | L | L | D | L | D | L | D | −0.3 ± 0.1 |
| T1L | L | L | L | L | L | L | L | L | L | L | −0.4 ± 0.1 |
| EB31 | L | L | L | D | L | L | L | D | D | L | −0.4 ± 0.2 |
| EB143 | Dd | L | L | L | L | L | D | L | L | L | −0.5 ± 0.1 |
| H14 | L | L | D | L | L | L | L | D | D | L | −0.5 ± 0.1 |
| EB13 | D | D | Dd | D | D | D | D | D | D | L | −3.3 ± 0.2 |
| EB144 | L | L | L | L | D | D | L | L | D | L | −3.3 ± 0.3 |
| KC19 | L | L | L | L | D | L | D | L | D | L | −3.4 ± 0.3 |
| EB136 | D | D | D | L | D | L | D | D | D | D | −3.4 ± 0.3 |
| T3D | D | D | D | D | D | D | D | D | D | D | −3.8 ± 0.3 |
| EB87 | L | D | L | L | D | L | L | D | L | L | −4.0 ± 0.1 |
| H27 | L | D | L | L | D | L | L | L | L | L | −4.2 ± 0.1 |
The reassortants chosen for use in this study include all eight possible combinations of T1L and T3D alleles for genome segments M2, L2, and S1, which encode the three proteins in the outer capsids of ISVPs (μ1, λ2, and ς1, respectively). Virus isolates are listed in order of increasing amount of infectivity change.
L and D, T1L and T3D alleles, respectively. The proteins found in ISVPs are indicated beside their encoding genome segments. The μ1 protein is found in ISVPs primarily in the form of cleavage fragments μ1N, δ, and φ (26, 29). The ς1 protein is found in a partially cleaved form in T3D ISVPs but remains uncleaved in T1L ISVPs (25).
Chymotrypsin-generated ISVPs of each isolate were treated at 47°C or room temperature for 30 min, and their infectious titers were then determined by plaque assay. Infectivity change following the treatment at 47°C was calculated as log10(PFU/ml) at 47°C −log10(PFU/ml) at room temperature. The values are means ± the standard errors of the mean from three or more independent experiments with each strain.
The λ3 protein of EB143 and the λ1 protein of EB13 are mobility variants of those proteins derived from T3D.
ISVP-like particles derived from recoated cores.
Recoated cores containing ς1 (r-cores+ς1) were prepared as described previously (10). Briefly, insect cells were separately infected with recombinant baculoviruses expressing μ1/ς3 and ς1 and then harvested at 65 h postinfection. Cells were pooled and lysed to generate a cytoplasmic extract containing μ1, ς3, and ς1. Purified cores were incubated with this lysate, and the resulting r-cores+ς1 were purified by banding on CsCl gradients. Nonpurified ISVP-like particles were prepared from r-cores+ς1 in the same manner as nonpurified ISVPs (see above).
SDS-PAGE and densitometry.
Samples were subjected to SDS-PAGE (10% polyacrylamide) as described elsewhere (8). Proteins were visualized by staining with Coomassie brilliant blue R-250 (Sigma). Estimation of particle concentration for recoated particles was done by using Coomassie staining and laser densitometry as previously described (9).
Plaque assays.
Plaque assays to determine particle/PFU ratios and infectivity after heat treatment were done as described previously (15). For some experiments this protocol was altered by washing the monolayers with 2 ml of phosphate-buffered saline (PBS) containing 2 mM MgCl2 prior to the 1-h viral attachment incubation, after which the monolayers were covered with 2 ml of 1% Bacto Agar and serum-free medium 199 containing 10 μg of trypsin (for strains with T1L ς1) (Sigma) or chymotrypsin (for strains with T3D ς1) per ml. Plaques were counted 2 to 4 days later, depending on the reovirus strain.
Thermal inactivations.
Virion buffer was preheated at the experimental temperature in a water bath for 30 min. Virus particles in virion buffer were then added to a final concentration of 2 × 109 particles/ml and mixed with a vortex mixer. The total volume of each inactivation mixture was 2 ml. Prior to an aliquot of the inactivation mixture being removed at each time point, the entire mixture was again mixed with a vortex mixer. Aliquots were harvested over the length of time required to reduce the titer to 0.1% of starting levels. Upon harvesting, aliquots were immediately diluted in PBS with 2 mM MgCl2 that had been precooled to 4°C. In most cases, a 111-μl aliquot was added to 1 ml of cold PBS at this step; however, for samples expected to have low titers, a 200-μl aliquot was added to 600 μl of cold PBS at this step. Each of these samples was then further diluted and subjected to plaque assay as described above. Samples intended for analysis in the proteolysis assay (see below) were processed as described above, except that virus particles were added to 1 ml of preheated virion buffer at a final concentration of 1011 particles/ml, and the entire inactivation mixture was removed to ice after 30 min at the experimental temperature.
Proteolysis assay for conformational changes in viral proteins.
After incubation at the experimental temperature, samples were chilled on ice for 15 min, and ice-cold TLCK-treated chymotrypsin was added to a final concentration of 200 μg/ml. Reaction mixtures were incubated at 4°C for 40 min, and digestion was stopped by addition of phenylmethylsulfonyl fluoride to 1 mM. Virus particles in the reaction mixtures were pelleted by centrifugation at 5°C in an SW60 rotor (Beckman) at 55,000 rpm for 1 h. Supernatants were discarded, and pellets were resuspended in equal volumes of virion buffer (10 or 20 μl in different experiments). Following addition of 2× Laemmli sample buffer in an equal volume as virion buffer, samples were disrupted by boiling for 5 min and subjected to SDS-PAGE.
RESULTS
Thermal inactivation of reovirus T1L particles.
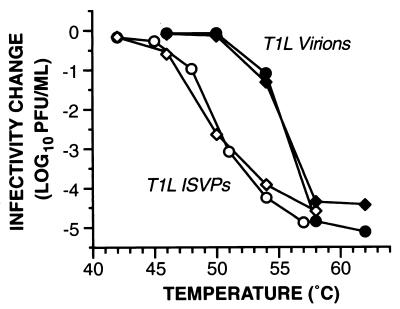
Gradient-purified, chymotrypsin-cleaved ISVPs of reovirus T1L demonstrated an increasingly large drop in infectivity as the temperature of incubation for 30 min in storage buffer was raised past 46°C (Fig. 1). The reproducibility of this behavior was demonstrated with different plaque clones of T1L (Fig. 1 and data not shown). Essentially identical results were obtained with gradient-purified, trypsin-cleaved T1L ISVPs (data not shown) and with nonpurified (see Materials and Methods), chymotrypsin-cleaved T1L ISVPs (Fig. 2), indicating that neither the protease used to generate the ISVPs nor the purification state of the ISVPs substantially affected their response to elevated temperatures. Results of identical experiments with T1L virions indicated that they became subject to a loss of infectivity similar to that of ISVPs but not until the incubation temperature was raised past 50°C (Fig. 1). By comparing the conditions that reduced infectivity to 1% of starting levels, we noted that the inactivation curve of T1L virions was shifted toward higher temperatures by 5 to 6°C from that of T1L ISVPs.
FIG. 1.
Temperature dependence of thermal inactivation of T1L particles. Gradient-purified T1L virions and ISVPs were diluted in storage buffer and then equally divided into separate microtubes for treatment for 30 min at different temperatures. At 30 min, an aliquot was removed from each sample into PBS with 2 mM MgCl2, and viral infectivity in the sample was determined by plaque assay. Infectivity after incubation for 30 min at each temperature (T) was expressed as log10(PFU/ml)T − log10(PFU/ml)RT, where RT (room temperature) was the lowest temperature tested. Each data point is the average of duplicate infectivity measurements on the same sample. Results for particles from two different plaque clones of reovirus T1L are shown (circles and diamonds).
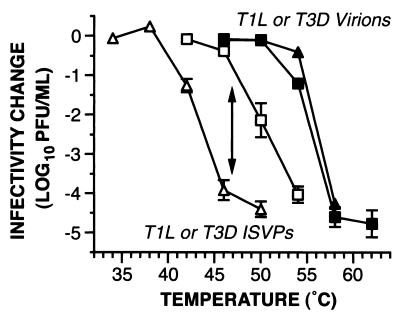
FIG. 2.
Temperature dependence of thermal inactivation of T1L and T3D particles. Gradient-purified virions and nonpurified ISVPs of T1L (squares) and T3D (triangles) were analyzed as described for Fig. 1. Each data point represents the mean ± standard error of the mean from three independent experiments. Each pair of curves is labeled according to particle type. The double-headed arrow indicates the difference between T1L and T3D ISVPs in the amount of infectivity remaining after incubation at 47°C for 30 min. This difference was subjected to genetic analysis, as shown in Table 1.
Thermal inactivation of reovirus T3D particles.
Nonpurified, chymotrypsin-cleaved ISVPs of reovirus T3D demonstrated an increasingly large drop in infectivity as the temperature of incubation for 30 min in storage buffer was raised past 40°C (Fig. 2), i.e., at substantially lower temperatures than observed for T1L ISVPs. By comparing the conditions that reduced ISVP infectivity to 1% of starting levels, we determined that the inactivation curve of T3D ISVPs was shifted toward lower temperatures by 6 to 7°C from that of T1L ISVPs. Thus, the ISVPs of reovirus T3D were less thermostable than those of T1L under these conditions. Results of identical experiments with T3D virions indicated that they became subject to approximately the same loss of infectivity, over approximately the same range of temperatures, as T1L virions (Fig. 2). This differs from findings in a previous study (13), in which T3D virions exhibited somewhat greater inactivation at 55°C than did those of T1L. The discrepancy may reflect a difference in the specific conditions (e.g., different buffers) or the specific plaque clones of reoviruses T1L and T3D that were used in the two studies. In either case, by comparing the conditions in this study that reduced infectivity to 1% of starting levels, we determined that the inactivation curve of T3D virions was shifted toward higher temperatures by 12 to 13°C from that of T3D ISVPs. The findings with both T1L and T3D particles thus showed that virions were more thermostable than ISVPs of the same isolate.
Genetic analysis of the difference in thermal inactivation of T1L and T3D ISVPs.
A previously generated panel of reassortant viruses containing defined mixtures of genome segments from reoviruses T1L and T3D (27) permitted a genetic analysis of the different temperatures at which their ISVPs were inactivated. For these experiments, purified virions of 12 different reassortants were converted to ISVPs by chymotrypsin cleavage. The nonpurified ISVPs were then tested for infectivity after incubation for 30 min at 47°C in storage buffer, conditions at which the behaviors of T1L and T3D ISVPs were clearly distinct (Fig. 2 and Table 1). Based on their behaviors in this assay, the reassortants were readily segregated into two groups, each approximating the behavior of one of the parents (Table 1). Upon analysis of the genome segment origins of these reassortants, the parental origin of a single segment, M2, was found to segregate with the sensitivity of the ISVPs to inactivation at 47°C. M2 encodes protein μ1, the primary constituent of the outer capsid of ISVPs. No evidence for involvement of any other genome segment was obtained in this experiment (Table 1). To confirm the latter point, we tested the ISVPs of reassortant E3 (which has the T1L M2 segment on a background of all other segments from T3D) and reassortant EB144 (which has the T3D M2 segment on a background of all other segments that encode structural proteins from T1L) for inactivation over a range of temperatures. Each behaved very similarly to the ISVPs of the parent from which its M2 genome segment is derived (data not shown, but see Fig. 6), consistent with the role of μ1 as the primary determinant of the different thermostabilities of T1L and T3D ISVPs under these conditions.
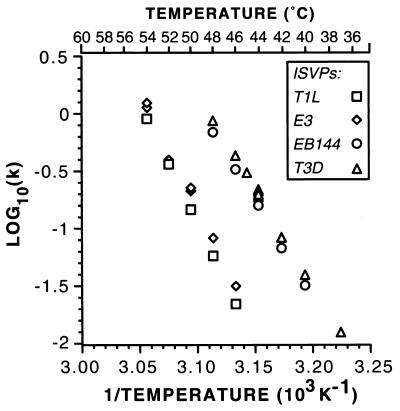
FIG. 6.
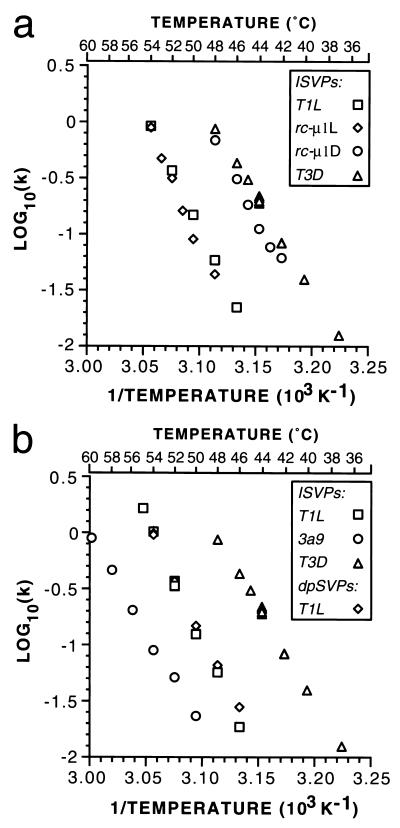
Arrhenius plots for thermal inactivation of T1L, T3D, and reassortant ISVPs. Thermal inactivation time courses for nonpurified ISVPs of reoviruses T1L, T3D, E3, and EB144 were fit by linear regression analysis to the equation for decay with first-order kinetics, and the rate constant, k, was determined for each temperature from these fits. The rate constants were plotted as log10(k) versus the reciprocal of the temperature in degrees Kelvin, according to the equation describing the Arrhenius relationship. In some cases, such as for T3D ISVPs at 44°C, data from more than one independent time course were collected and used to generate separate k values, which are plotted as discrete data points. A scale showing temperature in degrees Celsius is included for reference.
Biochemical changes in the μ1 protein of ISVPs at elevated temperatures.
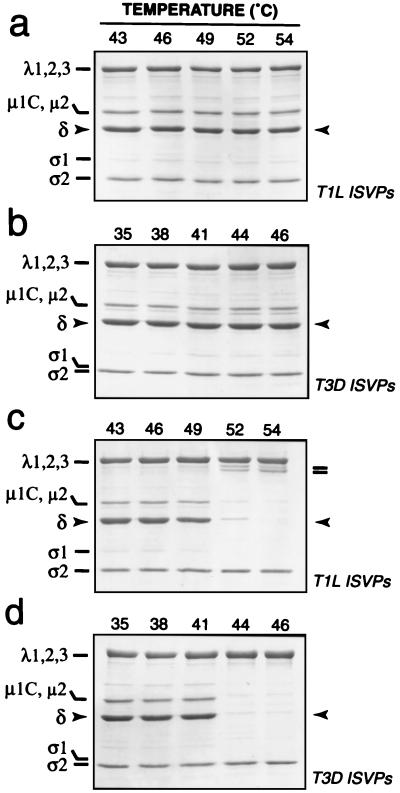
To gain further insight into the biochemical basis of ISVP inactivation at elevated temperatures, we investigated the effect of different temperatures on the state of μ1 in these particles. After incubation in storage buffer for 30 min, T1L and T3D ISVPs were pelleted by centrifugation, and the protein composition of the pelleted material was analyzed by SDS-PAGE and Coomassie staining (Fig. 3a and b). We observed that δ, the predominant fragment of μ1 in T1L and T3D ISVPs, was present at similar levels in the viral pellet at all temperatures tested, including those at which less than 1% of the original infectivity remained after 30 min (Fig. 1 and 2). Thus, the central δ fragment of μ1 was not substantially eluted from ISVPs in association with thermal inactivation.
FIG. 3.
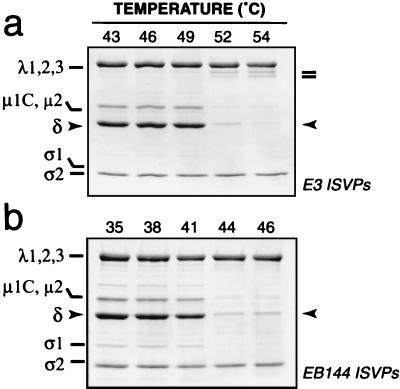
Status of μ1 protein after incubation of T1L and T3D ISVPs at elevated temperatures. ISVPs of reoviruses T1L and T3D were incubated in storage buffer at various temperatures for 30 min. Each sample was then removed to ice and allowed to chill for 15 min. Chilled samples containing T1L or T3D ISVPs were either immediately subjected to centrifugation (a and b) or treated with TLCK-treated chymotrypsin at 4°C and then subjected to centrifugation (c and d). Pelleted material was resuspended in Laemmli sample buffer, and viral proteins were resolved by SDS-PAGE and visualized by Coomassie staining. The position of μ1 fragment δ is highlighted with arrowheads. Fragments derived from the λ2 protein are highlighted by bars (c).
We next considered the possibility that μ1 is irreversibly altered at increased temperatures in a manner that does not result in the elution of δ from ISVPs. After incubation in storage buffer for 30 min at elevated temperatures, T1L and T3D ISVPs were removed onto ice, and a 4°C proteolysis assay was used to determine whether μ1 had changed conformation during the course of the incubation. In this assay, ISVPs were treated with chymotrypsin at 4°C prior to pelleting by centrifugation to concentrate the particles. The pelleted material was then analyzed by SDS-PAGE and Coomassie staining. We found that the δ fragment of μ1 in T1L ISVPs became increasingly sensitive to digestion by chymotrypsin at 4°C as the initial incubation temperature was raised past 46°C (Fig. 3c). In addition, the core protein λ2 became susceptible to cleavages near its C terminus that generated stable fragments, with Mrs of approximately 120,000, as the incubation temperature was raised past 49°C (Fig. 3c; also data not shown). Thus, both μ1 and λ2 in T1L ISVPs underwent irreversible conformational changes over the range of temperatures at which loss of infectivity occurred. When we tested T3D ISVPs in this assay, we found that the δ fragment of μ1 in these particles became increasingly sensitive to digestion by chymotrypsin as the incubation temperature was raised past 38°C (Fig. 3d). However, λ2 in T3D ISVPs did not become susceptible to cleavage between 38°C and the highest temperature tested in these experiments, 46°C (Fig. 3d). Thus, μ1 but not λ2 in T3D ISVPs underwent irreversible conformational changes over the range of temperatures at which loss of infectivity occurred. The results with T1L and T3D ISVPs thus revealed a strong, temperature-dependent correlation between the loss of infectivity and the increased sensitivity of μ1 to chymotrypsin digestion. These findings suggest that the difference in thermostability of T1L and T3D ISVPs was determined by the different propensities of the T1L and T3D μ1 proteins to undergo irreversible conformational changes in a temperature-dependent manner.
A study with the T1L × T3D reassortant viruses E3 and EB144 was next performed to examine the genetic determinants of the different behaviors of the μ1 and λ2 proteins in thermally inactivated T1L and T3D ISVPs. ISVPs of the reassortants were incubated over a range of temperatures and then analyzed with the 4°C proteolysis assay. The T1L δ fragment and T3D λ2 protein in E3 ISVPs became subject to chymotrypsin cleavage after the temperature was raised past 46 and 49°C, respectively (Fig. 4a), very similarly to the T1L δ and λ2 proteins in T1L ISVPs (Fig. 3c). In contrast, the T3D δ fragment in EB144 ISVPs became subject to chymotrypsin cleavage after the temperature was raised past 38°C, but little or no proteolysis of the T1L λ2 protein in EB144 ISVPs was observed up to the highest temperature tested in these experiments, 46°C (Fig. 4b), similarly to the T3D δ and λ2 proteins in T3D ISVPs (Fig. 3d). The results thus indicate that the different temperatures at which the T1L and T3D μ1 proteins in ISVPs became subject to proteolysis were determined by genetic differences in their M2 genome segments. In addition, with each type of ISVP, the protease sensitivity of μ1 was correlated with the loss of infectivity. Proteolysis of λ2 following incubation of ISVPs at temperatures above 49°C was common to both T1L (Fig. 3c) and T3D λ2 (Fig. 4a) and did not correlate with the infectivity losses of T3D and EB144 ISVPs (see Fig. 6). Moreover, both T1L and T3D λ2 became sensitive to chymotrypsin digestion following incubation of core particles, which lack the μ1 protein, at 48 to 50°C (23; T. F. Severson, unpublished data). On the basis of those findings, we concluded that proteolysis of λ2 is a separate phenomenon relating to the higher temperatures required for inactivation of ISVPs containing the T1L μ1 protein.
FIG. 4.
Status of protein μ1 after incubation of E3 (a) and EB144 (b) ISVPs at elevated temperatures. ISVPs of T1L × T3D reassortants E3 and EB144 were analyzed as described for T1L and T3D ISVPs for Fig. 3. The position of μ1 fragment δ is highlighted with arrowheads. Fragments derived from the λ2 protein are highlighted by bars (a).
First-order kinetics for thermal inactivation of ISVPs.
To learn more about the difference in thermal inactivation of T1L and T3D ISVPs, we performed kinetic analyses. We first defined the time courses with which T1L and T3D ISVPs were inactivated at different temperatures. For each temperature at which inactivation occurred at a practically measurable rate, we found that log10(infectious titer) decreased linearly with time, with only a small or no apparent lag at the outset of each incubation (representative time courses are shown in Fig. 5). A reaction for which log10(concentration of substrate) decreases linearly with time exhibits first-order kinetics. Thus, the data indicate that the inactivation of both T1L and T3D ISVPs approximated a first-order process. This first-order thermal inactivation of ISVPs is described by
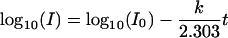 |
(1) |
where I is the infectious titer of ISVPs at time t, I0 is the infectious titer of ISVPs at time zero, and k is the overall rate constant for the reaction.
FIG. 5.
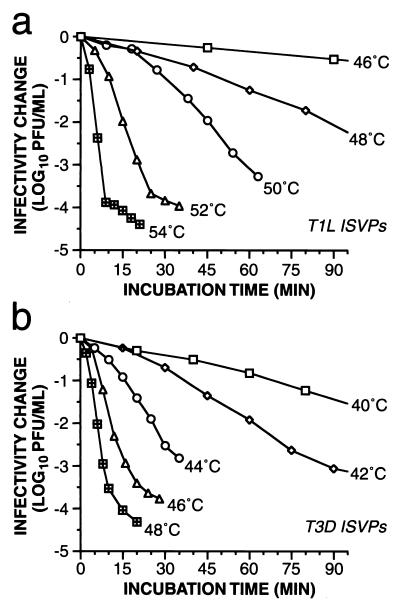
Kinetics of thermal inactivation of T1L (a) and T3D (b) ISVPs. Purified ISVPs of reovirus T1L and nonpurified ISVPs of reovirus T3D were incubated in storage buffer at various temperatures. An aliquot was removed from each sample into ice-cold PBS with 2 mM MgCl2 at each time point, and viral infectivity in the sample was determined by plaque assay. Infectivity after incubation for time interval t was expressed as log10(PFU/ml)t − log10(PFU/ml)t0, where (PFU/ml)t0 is the infectivity of a sample incubated at 4°C throughout the time course. Each data point represents the average of two determinations for that sampled aliquot. Data from later time points with T1L at 46 and 48°C and T3D at 40 and 42°C were omitted for simplicity.
For temperatures at which T1L and T3D ISVPs were incubated for enough time, another phenomenon was observed; namely, after infectivity had fallen to about 0.01% of the starting level, the rate of inactivation decreased dramatically so that an approximate plateau of residual infectivity was reached (Fig. 5). These findings suggest that each original population of T1L or T3D ISVPs included small subpopulations of virus particles that underwent inactivation at a substantially lower rate than the predominant population. Possible identities for the inactivation-resistant particles include undigested virions, ISVPs in distinct microenvironments (e.g., aggregated or trapped at the air-liquid interface), and ISVPs containing a mutant form of one or more capsid proteins. The presence of thermostable mutants among the residually infectious subpopulations has been confirmed, and these mutants are being characterized (J. K. Middleton, unpublished data).
Arrhenius plots for temperature-dependent inactivation of ISVPs.
As seen in equation 1, the rate constant of ISVP inactivation at a specific temperature is defined as −2.303 × the slope of the line in a plot of log10(infectious titer of ISVPs) versus time. The values for log10(k) measured at a number of temperatures could then be plotted against 1/T to determine whether the inactivation reaction obeyed the Arrhenius relationship over that range of temperatures, as shown by
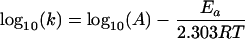 |
(2) |
where R is the ideal gas constant, T is the temperature in degrees Kelvin, k is the rate constant, A is the pre-exponential factor, and Ea is the activation energy. In fact, the Arrhenius relationship was found to hold well (r2 > 0.997) for the inactivation of both T1L and T3D ISVPs incubated in storage buffer at temperatures between 37 and 54°C (Fig. 5).
As equation 2 indicates, the Arrhenius relationship was useful for providing two thermodynamic values for comparing the inactivation reactions with different preparations of reovirus ISVPs: Ea, which was obtained as the negative slope of the linear fit of the data to the Arrhenius equation, and A, where log10 A is obtained as the y intercept of the linear fit of the data to the Arrhenius equation. These data could also be analyzed with the Eyring absolute rate equation from transition-state theory (30), as shown by
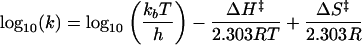 |
(3) |
where k is the rate constant, kb is Boltzmann’s constant, h is Planck’s constant, R is the ideal gas constant, T is the temperature in degrees Kelvin, ΔH‡ is the enthalpy of activation, and ΔS‡ is the entropy of activation. By comparing equations 2 and 3, values for Ea and A were thus convertible into values for ΔH‡ and ΔS‡ of each inactivation reaction with the different ISVPs (Table 2). ΔH‡ and −TΔS‡ could then be combined to provide an estimate of the free energy of activation (ΔG‡) of each inactivation reaction at a particular temperature, as shown in the following equation.
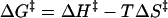 |
(4) |
TABLE 2.
Thermodynamic values for inactivation of reovirus ISVPs and ISVP-like particles
| Virus isolate | Particle type | Calculated value ± SDf
|
ΔG‡a (kcal mol−1) | |
|---|---|---|---|---|
| ΔH‡ (kcal mol−1) | ΔS‡ (kcal mol−1 K−1) | |||
| T1L | ISVPb | 102 ± 2 | 0.251 ± 0.007 | 22 |
| T1L | ISVPc | 94.8 ± 0.3 | 0.231 ± 0.001 | 20.9 |
| E3 | ISVPc | 91 ± 3 | 0.22 ± 0.01 | 21 |
| rc-μ1Ld | ISVP-likec | 107 ± 6 | 0.27 ± 0.02 | 21 |
| T1L | dpSVPb | 92 ± 2 | 0.224 ± 0.006 | 20 |
| T3D | ISVPc | 77 ± 1 | 0.182 ± 0.004 | 19 |
| EB144 | ISVPc | 76 ± 1 | 0.179 ± 0.003 | 19 |
| rc-μ1Dd | ISVP-likec | 84 ± 4 | 0.20 ± 0.01 | 20 |
| 3a9e | ISVPc | 78 ± 2 | 0.174 ± 0.006 | 22 |
At 47°C, the temperature at which the phenotypic difference between T1L and T3D ISVPs was subjected to genetic analysis (Table 1). The value is calculated from ΔH‡ and ΔS‡ (equation 4).
Purified.
Nonpurified.
r-cores + ς1 containing μ1 protein of the indicated origin. All other proteins in these particles were from T1L.
Ethanol-resistant mutant derived from T3D.
In sum, these calculations suggest that the different inactivation behaviors of T1L and T3D ISVPs reflected a difference in the ΔG‡ values of their inactivation reactions, which was determined by differences in their ΔH‡ and ΔS‡ values. Thus, changes in both enthalpy and entropy appear to make important contributions to ISVP inactivation. (See Discussion for further considerations.)
We also performed kinetic studies at different temperatures, calculated the respective rate constants, and generated Arrhenius plots for the T1L × T3D reassortants E3 and EB144 described above (Fig. 6). The values for ΔH‡ and ΔS‡ derived from the Arrhenius plots for each were found to approximate those of the parent virus from which its M2 genome segment and μ1 protein were derived (Table 2), T1L for E3 and T3D for EB144. These data, coupled with the more extensive genetic analysis in Table 1, provided evidence that the different inactivation behaviors of T1L and T3D ISVPs are based in these thermodynamic properties of their M2-encoded μ1 proteins.
Inactivation of ISVP-like particles derived from recoated cores.
Reovirus cores can be recoated in vitro through the addition of recombinant reovirus outer-capsid proteins μ1, ς3, and ς1 (9, 10), thereby reconstituting virion-like particles termed r-cores+ς1. Furthermore, the ς3 and μ1 proteins in r-cores+ς1 can be cleaved by chymotrypsin in vitro to generate ISVP-like particles (9, 10). For the present study we purified T1L cores and recoated them with recombinant T1L or T3D μ1, T1L ς3, and T1L ς1 proteins. We then subjected the r-cores+ς1 to in vitro digestion with chymotrypsin and tested them for inactivation at elevated temperatures. The ISVP-like particles derived from r-cores+ς1 containing all T1L proteins were inactivated over the same temperature range and with approximately the same rate at each temperature as the ISVPs derived from T1L virions (data not shown). In contrast, the ISVP-like particles derived from r-cores+ς1 containing all T1L proteins except for μ1 from T3D were inactivated over the same temperature range and with approximately the same rate at each temperature as the ISVPs derived from T3D virions (data not shown). Arrhenius plots from these data showed that the inactivation behavior of the ISVP-like particles obtained from r-cores+ς1 segregated with the parental origin of their μ1 protein (Fig. 7a). The ΔH‡ and ΔS‡ values reflected by these plots also segregated with the parental origin of μ1 (Table 2). Although these values were somewhat higher for the ISVP-like particles derived from r-cores+ς1 than for the homologous ISVPs derived from virions, the basis or significance of this trend remains unknown. Despite this unexplained feature, the findings with r-cores+ς1 confirmed that the μ1 protein was the primary determinant of the differences in thermostability of T1L and T3D ISVPs.
FIG. 7.
Arrhenius plots for thermal inactivation of ISVPs and ISVP-like particles derived from r-cores+ς1, dpSVPs, and mutant ISVPs. The plots were generated as in Fig. 6. (a) Arrhenius plots are shown for thermal inactivation of nonpurified ISVP-like particles derived from r-cores+ς1 containing T1L μ1 (rc-μ1L) or T3D μ1 (rc-μ1D) on a background of all other proteins from T1L. Arrhenius plots for nonpurified ISVPs of reoviruses T1L and T3D are repeated from Fig. 6 for comparison. (b) Arrhenius plots are shown for thermal inactivation of purified T1L ISVPs, purified T1L dpSVPs, and nonpurified mutant 3a9 ISVPs. Arrhenius plot for nonpurified ISVPs of reovirus T3D are repeated from Fig. 6 for comparison.
Inactivation of ISVPs from a reovirus mutant selected for ethanol resistance.
Ethanol-resistant mutants were previously selected from reovirus T3D and, in the case of one of these mutants, 3a9, shown by reassortant analysis and nucleotide sequencing to involve a single resistance-conferring mutation in the M2 genome segment and μ1 protein (37). Other mutants from this panel, although not analyzed with reassortants, were shown also to contain distinct mutations in M2 and μ1, which were also concluded to confer ethanol resistance. In a later study, ISVPs derived from 3a9 and other ethanol-resistant mutants were shown to be more resistant to inactivation during a time course of treatment at 50°C as well (17). For the present study we analyzed the ethanol-resistant mutant 3a9, whose μ1 protein differs from that of T3D by a single amino acid substitution at position 459. In our experiments, 3a9 ISVPs were found to be inactivated with approximately first-order kinetics but at higher temperatures than required for either T3D or T1L ISVPs (inactivation of 3a9 ISVPs was shifted toward higher temperatures by 11 to 12°C from that of T3D ISVPs and by 4 to 6°C from that of T1L ISVPs). The distinct behavior of 3a9 ISVPs was evident in the Arrhenius plot derived from these data (Fig. 7b). Interestingly, the ΔH‡ and ΔS‡ values derived from this plot for 3a9 ISVPs remained similar to those for T3D ISVPs (3a9 was originally selected from a T3D parent) (Table 2). The calculated ΔG‡ value was of course higher for 3a9 ISVPs, reflecting their increased thermostability (Table 2). The increased thermostability of 3a9 ISVPs, relative to that of T3D ISVPs, was thus attributable to much smaller changes in ΔH‡ and/or ΔS‡ than those demonstrated between T1L and T3D ISVPs. (See Discussion for further considerations.)
Inactivation of ISVP-like particles that lack the δ:φ cleavage of μ1/μ1C.
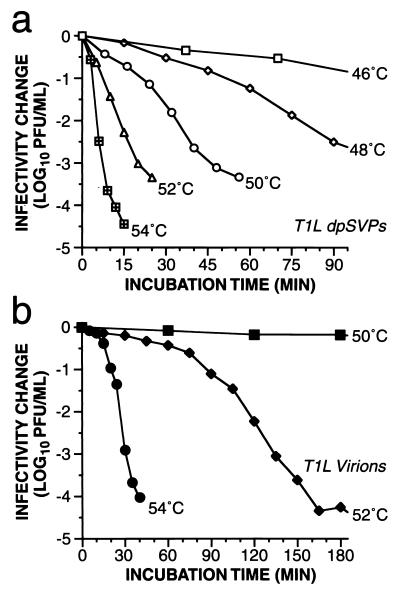
A novel type of reovirus ISVP-like particle, in which the ς3 protein has been removed by proteolysis but the μ1/μ1C protein has not yet been cleaved to yield the μ1δ/δ and φ fragments as in ISVPs (26), was recently described and named the dpSVP (detergent- and protease-treated subvirion particle) (8). By comparing the temperature-dependent inactivation of ISVPs and dpSVPs, we sought to determine whether cleavage of μ1/μ1C at the δ:φ junction contributes to the higher rate of inactivation exhibited by ISVPs than by virions. In particular, if the μ1/μ1C cleavage contributes to this difference, then dpSVPs should have an inactivation profile either very similar to that of virions or intermediate between that of virions and ISVPs. In initial experiments (19), we found that purified T1L dpSVPs were inactivated to approximately the same extent as T1L ISVPs after 30 min at 52°C. In the present study, we extended these analyses to show in time course experiments that T1L and EB144 dpSVPs were inactivated with approximately first-order kinetics and with approximately the same rates over a range of temperatures as T1L and EB144 ISVPs, respectively (Fig. 8a [T1L dpSVPs] and data not shown [EB144 dpSVPs]). T3D dpSVPs could not be analyzed in this experiment due to a large drop in infectivity during preparation, tentatively attributable to ς1 protein degradation (K. Chandran, unpublished data). An Arrhenius plot from these data for T1L dpSVPs (Fig. 7b) furthermore yielded ΔH‡ and ΔS‡ values that were very similar to those for T1L ISVPs (Table 2). These findings therefore indicate that the δ:φ cleavage of μ1/μ1C made little if any contribution to the different thermostabilities of virions and ISVPs and to the different thermostabilities of ISVPs containing a T1L or T3D μ1 protein. In addition, the μ1C protein in T1L dpSVPs became sensitive to chymotrypsin digestion at 4°C following incubation of those particles at elevated temperatures, thereby extending the correlation between conformational changes in μ1 and loss of infectivity (data not shown).
FIG. 8.
Kinetics of thermal inactivation of T1L dpSVPs (a) and virions (b). Gradient-purified T1L dpSVPs (see the text for a description of these particles) and virions were incubated in storage buffer at the indicated temperatures, and time courses of viral infectivity were determined as described for Fig. 5. Each data point represents the average of two determinations for that sampled aliquot. Data from later time points with dpSVPs at 46 and 48°C and virions at 50 and 52°C were omitted for simplicity.
Non-first-order kinetics for thermal inactivation of virions.
For comparison with the results for ISVPs, we also performed kinetic studies of the inactivation of T1L and T3D virions. We first defined the time courses with which T1L virions (representative experiments shown in Fig. 8b) and T3D virions (data not shown) were inactivated during incubations at different temperatures. Interestingly, we found that log10(infectious titer) did not decrease linearly with respect to incubation time. Instead, for temperatures at which inactivation occurred at a practically measurable rate, the inactivation curve for virions exhibited an extended initial slow phase (lag), followed by an accelerated phase, and ended with a second slow phase. Possible explanations for the second slow phase are the same as those suggested for ISVPs above, including the presence of thermostable mutants. The initial slow and following accelerated phases, however, are distinct from the pattern exhibited by ISVPs and indicate that the thermal inactivation of virions did not exhibit first-order kinetics. In addition, the range of temperatures over which the inactivation of virions changed from very slow to very fast was not only higher but also narrower than that observed for ISVPs and dpSVPs (compare Fig. 8b with Figs. 5a and 8a). Future studies will aim at better defining the nature and molecular basis of the distinct kinetic features of virion inactivation.
DISCUSSION
Why seek a better understanding of virus particle stability and inactivation?
In addition to providing new insights into the process of virus entry (see below), studies of inactivation such as those performed with reovirus particles here may have practical benefits. Agents for inactivating infectious virus particles on surfaces and in liquids are in widespread use, but the mechanisms and important determinants of inactivation by many or most of these agents remain poorly described at the molecular level. The potential for selection of viral mutants that are resistant to these agents, are fit enough for survival in nature, and may be altered in their disease-causing properties also remains for the most part unexplored. Other important arguments for seeking a better understanding of virus stability concern virus particle vaccines, such as how to design these vaccines for greater stability during production and in the field. Lastly, the increasing use of viral therapeutic agents, in the treatment of genetic deficiencies or cancer, for example, also calls for improvements in our understanding of virus stability and the ways in which we might aim to increase or decrease particle stability as the therapeutic situation dictates.
First-order thermal inactivation of reovirus ISVPs, role of surface protein μ1, and basis of infectivity loss.
All preparations of reovirus ISVPs and ISVP-like particles in this study approximated first-order kinetics with respect to their decrease in infectivity during thermal inactivation. This phenomenon seems somewhat remarkable given the structural complexity of these particles (14); however, first-order inactivation of a variety of different viruses has been known for many years (30). The simplest mechanistic explanation for a process exhibiting first-order kinetics involves a single reactant undergoing a single-step change. In the case of a multicomponent viral particle, this might translate to a single protein subunit undergoing denaturation or elution from the particle, thereby rendering the particle noninfectious. This explanation seems unlikely in the case of reovirus ISVPs, since an irreversible increase in chymotrypsin sensitivity was exhibited by most or all of the 600 copies of μ1 protein in association with thermal inactivation, consistent with an inactivation mechanism involving a larger number of individual protein subunits. It is well established that first-order kinetics can be exhibited by more complex reactions than unimolecular ones (3). In the case of reovirus ISVPs, we hypothesize that rate-limiting conformational changes in as few as one μ1 subunit per particle are translated to other protein subunits around the surface in a concerted manner such that the whole particle behaves as a single reactant exhibiting first-order kinetics. The μ1 protein is identified as the rate-limiting component because of the strong genetic evidence that it determines the different inactivation rates of T1L and T3D ISVPs (Table 1). Conformational changes in μ1 are indicated by the irreversible increase in chymotrypsin sensitivity of its central δ fragment. Loss of infectivity may be a direct consequence of the conformational changes in the δ region, which may render μ1 nonfunctional for its role(s) in viral entry such as membrane penetration (28). However, other changes in the virus particle, such as conformational changes in the μ1N and/or φ regions of μ1, or elution of the receptor-binding protein ς1 (12, 15), may occur in concert with conformational changes in the δ region of μ1 and may contribute to the loss in infectivity. Further analyses of biochemical features of the inactivated particles are in progress in an effort to address these questions.
Thermodynamic parameters of ISVP inactivation.
All preparations of reovirus ISVPs and ISVP-like particles in this study not only approximated first-order thermal inactivation but also obeyed the Arrhenius relationship over the examined temperature range, 37 to 55°C, yielding ΔH‡ and ΔS‡ values for the inactivation reactions from classical transition-state theory (30). ΔH‡ and ΔS‡ for protein conformational changes can reflect contributions from two sources: interactions within the protein and interactions between the protein and solvent (16, 21). Within the protein, interactions among amino acid residues contribute to ΔH‡ through van der Waals, hydrogen-bonding, and electrostatic forces and to ΔS‡ by affecting structural flexibility. Contributions to ΔH‡ and ΔS‡ from the solvent include enthalpy and entropy changes associated with changes in solvation of newly exposed or newly buried amino acids following the conformational change (24, 31). In combination, ΔH‡ and ΔS‡ define ΔG‡ at a particular temperature (equation 4), which reflects the energy needed to cross the activation barrier of a reaction.
In the case of T1L and T3D ISVPs, the higher ΔG‡ value of T1L ISVPs indicates a larger input energy required for their inactivation and thus their greater thermostability. Although the difference in ΔG‡ between T1L and T3D ISVPs appears to be relatively small (2 kcal/mol) (Table 2), it is more than large enough to provide clear differences in the temperatures required for thermal inactivation (e.g., Fig. 2, 6, and 7). Interestingly, the relative differences in the ΔH‡ and ΔS‡ values of T1L and T3D ISVPs are much higher, representing more substantial differences in the respective contributions of ΔH‡ and ΔS‡ to the free energies of activation of the two isolates. The difference in ΔH‡ values for T1L and T3D ISVPs is consistent with the higher ΔG‡ value and thus greater thermostability of T1L ISVPs, with ΔH‡(T1L) being higher than ΔH‡(T3D) (equation 4). In contrast, ΔS‡(T1L) is higher than ΔS‡(T3D), which provides a greater opposition to the enthalpic contribution to ΔG‡. The higher ΔH‡ value for T1L than T3D ISVPs is therefore partially balanced by a higher ΔS‡ value for T1L than T3D ISVPs. This increase in entropy may have been necessary for the T1L ISVP to remain infectious. For example, if an ISVP had a higher ΔG‡ value than seen in these experiments [e.g., as provided by ΔH‡(T1L) and ΔS‡(T3D), or a ΔG‡ of ∼37 kcal/mol at 47°C], it may be too stable to undergo the conformational change in μ1 required for infection. On the contrary, if an ISVP had a lower ΔG‡ value [e.g., as provided by ΔH‡(T3D) and ΔS‡(T1L), or a ΔG‡ of ∼3 kcal/mol at 47°C], it may be too unstable, allowing μ1 to change conformation before properly encountering the cellular membrane. It may thus be that nature selects for a particular range of stability of reovirus particles, reflected by the observed range of ΔG‡ values that constrain the relative values of ΔH‡ and ΔS‡. This balancing of changes in ΔH‡ and ΔS‡ to limit changes in ΔG‡, resulting in an enthalpy-entropy compensation effect, has been observed in a variety of chemical and biological systems and attributed to both structural and solvent effects (2, 20–22). It is uncertain at present whether structural or solvent effects are responsible for the compensation seen with reovirus particles, and determination of the contributing factors requires further study using different solution conditions and mutant μ1 proteins (see below).
From the perspective of intramolecular interactions and solvent effects, the balancing of enthalpy and entropy contributions in T1L and T3D ISVPs might be explained in several ways. Relative to the T3D μ1 protein, the T1L μ1 protein in ISVPs may have stronger van der Waals, hydrogen-bonding, and electrostatic interactions, which would necessitate more input energy to break those bonds during the inactivation transition, i.e., a higher ΔH‡ value. This stronger bonding, however, would be accompanied by a restriction in the flexibility of T1L μ1 in ISVPs so that when μ1 changes conformation during the inactivation transition, there would be a greater gain in entropy, i.e., a higher ΔS‡ value. In addition or alternatively, the T1L μ1 protein may have more ordered water molecules bound within ISVPs than does T3D μ1. The conformational change of T1L μ1 at the inactivation transition would then require more input energy to break the ordered water bonds, resulting in both a higher ΔH‡ and a higher ΔS‡ upon release of the ordered waters.
What is the molecular basis of the difference in thermostability of T1L and T3D ISVPs? Genetic analyses in this study indicate that the differences in ΔG‡, ΔH‡, and ΔS‡ for T1L and T3D ISVPs are determined by their μ1 proteins. The μ1 proteins of the wild-type T1L and T3D isolates in use in our lab differ in sequence at only 15 amino acid positions (9; Chandran, unpublished), one or more of which must determine the differences in thermodynamic properties of these proteins. We will dissect these determinants in a subsequent study, but in the meantime the results with ethanol-resistant mutant 3a9 are instructive. The 3a9 μ1 protein differs from that of T3D by a single amino acid change at position 459, Lys in T3D to Gln in 3a9 (37). Since the ethanol-resistance phenotype of 3a9 was shown by reassortant analysis to be linked to the M2 genome segment, the single substitution in μ1 almost certainly determines the ethanol resistance of this clone. Based on findings with related mutants (17, 37), the single substitution in 3a9 μ1 is likely as well to determine the increase in thermostability of 3a9 ISVPs. Substitutions in the μ1 proteins of 3a9 and other ethanol-resistant mutants are all located within the δ region of μ1, suggesting, together with the conformational changes in δ observed in this study, a role for this central region of μ1 in regulating particle stability and infectivity.
The greater thermostability of 3a9 than T3D ISVPs (ΔG‡ ≈ 22 kcal/mol for 3a9 ISVPs versus 19 kcal/mol for T3D ISVPs at 47°C) makes them relatively more similar to T1L ISVPs (ΔG‡ ≈ 21 kcal/mol for T1L ISVPs at 47°C), yet the ΔH‡ and ΔS‡ values for 3a9 ISVPs remain very similar to those for T3D ISVPs, indicating that only subtle changes in the enthalpy and entropy of activation have given rise to the phenotypically significant increase in the free energy of activation of 3a9. In addition to suggesting that large changes in thermostability can result from single amino changes, the findings suggest that the different inactivation behaviors of T1L and T3D ISVPs reflect evolution of their different enthalpic and entropic properties by accumulation of several relevant mutations within μ1. Analyses of additional reoviruses, both wild-type isolates and thermostable mutants, as well as chimeric and site-directed-mutant μ1 proteins in recoated cores, should provide a better description of this evolutionary process and the range of ΔG‡, ΔH‡, and ΔS‡ values that are consistent with reovirus infectivity.
Distinct inactivation behavior of reovirus virions and role of ς3.
In addition to requiring higher temperatures for inactivation than ISVPs do, virions underwent thermal inactivation with kinetics that deviated from first order. A likely explanation for the greater thermostability of virions is that the ς3 protein in these particles, by binding the underlying μ1 protein (14, 19), prevents or delays the conformational changes in μ1 that were seen to be associated with the thermal inactivation of ISVPs in this study. Through binding to μ1, ς3 may also alter the nature of the inactivation-associated conformational changes that occur at higher temperatures with virions as well as the type of kinetics with which they occur. Since the thermal inactivation of virions is not fit well by equations for either first-order or second-order kinetics (Middleton, unpublished), the mechanism of virion inactivation cannot be modeled by simple bimolecular reaction schemes such as involving ς3 and μ1 as the two reactants. Further studies are thus needed to explain both the inactivation kinetics of virions and the manner in which ς3 provides particle stabilization.
Relevance for understanding reovirus entry into cells and host animals.
The μ1 protein is thought to play a direct role in reovirus penetration of the cellular membrane barrier during the entry phase of infection (28). Moreover, this activity is hypothesized to involve a conformational change in μ1, analogous to the entry-related conformational changes in the fusion proteins of enveloped viruses and the capsid proteins of picornaviruses, for example (3, 7, 34). Indeed, recent evidence from our lab has demonstrated conformational changes in the ISVP-associated μ1 protein that accompany membrane interactions in several different assays both in vitro and in cells (Chandran, unpublished). As a result, we believe that the present demonstration of an approach for defining kinetic and thermodynamic parameters of ISVPs will prove useful for ongoing studies of reovirus entry and mutations that affect that process. We moreover hypothesize that the temperature-dependent conformational changes in μ1 apparent in Fig. 3 and 4 are closely related to those that occur during entry. The low rate of ISVP inactivation and associated conformational changes in μ1 that we observed at physiological temperatures in this study may indicate the involvement of a cellular or environmental cofactor or catalyst (ion, sugar, lipid, protein, etc.) to promote these conformational changes in cells. For example, recent evidence indicates that binding of the poliovirus receptor to poliovirus virions reduces the virions’ ΔH‡ by 50 kcal/mol (from 145 to 95), thereby promoting uncoating (35).
An early study using a small panel of reassortant viruses identified the M2 genome segment encoding μ1 as the primary determinant of the different efficiencies with which T1L and T3D infect newborn mice via the gastrointestinal tract (32). A subsequent study using a different and larger panel of reassortants instead identified the L2 and S1 genome segments encoding proteins λ2 and ς1, respectively, as the determinants of this difference (4). Despite the latter results, evidence that ISVPs are generated from inoculated virions by proteolysis within the lumen of the newborn mouse intestine (5) is consistent with a potential role of μ1 in determining particle stability and survival within the gut lumen. Other genetic studies have shown a role for M2/μ1 in determining differences between reovirus isolates in neurovirulence in newborn mice (18) and induction of apoptosis in cell culture (36), which may also reflect the role of μ1 as an important determinant of particle stability.
Acknowledgments
J.K.M. and T.F.S. made equivalent contributions to this study.
We thank Laura Breun for excellent technical support and the other members of our laboratories for helpful discussions.
This work was supported by a research grant from the Lucille P. Markey Charitable Trust (to the Institute for Molecular Virology), NSF BES-9896067 (to J.Y.), a Shaw Scientist Award from the Milwaukee Foundation, a Steenbock Career Development Award from the Department of Biochemistry, and NIH grants R29 AI39533 and R01 AI46440 (to M.L.N.). J.K.M. received additional support from NIH grant T32 GM08349 to the Biotechnology Training Program. K.C. was additionally supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
REFERENCES
- 1.Amerongen, H. M., G. A. R. Wilson, B. N. Fields, and M. R. Neutra. 1994. Proteolytic processing of reovirus is required for adherence to intestinal M cells. J. Virol. 68:8428–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, R., H. Vogel, and I. Gordon. 1969. Temperature of compensation: significance for virus inactivation. Proc. Natl. Acad. Sci. USA 62:263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentz, J. 1992. Intermediates and kinetics of membrane fusion. Biophys. J. 63:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodkin, D. K., and B. N. Fields. 1989. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J. Virol. 63:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodkin, D. K., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191–200. [DOI] [PubMed] [Google Scholar]
- 7.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306–14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandran, K., and M. L. Nibert. 1998. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J. Virol. 72:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and ς3. J. Virol. 73:3941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, K., X. Zhang, N. H. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 75:5335–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218–228. [DOI] [PubMed] [Google Scholar]
- 12.Drayna, D., and B. N. Fields. 1982. Biochemical studies on the mechanism of chemical and physical inactivation of reovirus. J. Gen. Virol. 63:161–170. [DOI] [PubMed] [Google Scholar]
- 13.Drayna, D., and B. N. Fields. 1982. Genetic studies on the mechanism of chemical and physical inactivation of reovirus. J. Gen. Virol. 63:149–159. [DOI] [PubMed] [Google Scholar]
- 14.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunwald, E. 1997. Thermodynamics of molecular species. John Wiley & Sons, Inc., New York, N.Y.
- 17.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrdy, D. B., D. H. Rubin, and B. N. Fields. 1982. Molecular basis of reovirus neurovirulence: role of the M2 gene in avirulence. Proc. Natl. Acad. Sci. USA 79:1298–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jané-Valbuena, J., M. L. Nibert, S. M. Spencer, S. B. Walker, T. S. Baker, Y. Chen, V. E. Centonze, and L. A. Schiff. 1999. Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed ς3 protein: an approach for analyzing ς3 functions during virus entry. J. Virol. 73:2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leffler, J. E. 1955. The enthalpy-entropy relationship and its implications for organic chemistry. J. Org. Chem. 20:1202–1231. [Google Scholar]
- 21.Liu, L., C. Yang, and Q. X. Guo. 2001. Isokinetic relationship, isoequilibrium relationship, and enthalpy-entropy compensation. Chem. Rev. 101:673–695. [DOI] [PubMed] [Google Scholar]
- 22.Lumry, R., and S. Rajender. 1970. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers 9:1125–1227. [DOI] [PubMed] [Google Scholar]
- 23.Luongo, C. L., K. A. Dryden, D. L. Farsetta, R. L. Margraf, T. F. Severson, N. H. Olson, B. N. Fields, T. S. Baker, and M. L. Nibert. 1997. Localization of a C-terminal region of λ2 protein in reovirus cores. J. Virol. 71:8035–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makhatadze, G. I., and P. L. Privalov. 1993. Contribution of hydration to protein-folding thermodynamics. 1. The enthalpy of hydration. J. Mol. Biol. 232:639–659. [DOI] [PubMed] [Google Scholar]
- 25.Nibert, M. L., J. D. Chappell, and T. S. Dermody. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved ς1 protein. J. Virol. 69:5057–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nibert, M. L., R. L. Margraf, and K. M. Coombs. 1996. Nonrandom segregation of parental alleles in reovirus reassortants. J. Virol. 70:7295–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p.1679–1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard, E. C. 1953. The physics of viruses. Academic Press, New York, N.Y.
- 31.Privalov, P. L., and G. I. Makhatadze. 1993. Contribution of hydration to protein-folding thermodynamics. 2. The entropy and Gibbs energy of hydration. J. Mol. Biol. 232:660–679. [DOI] [PubMed] [Google Scholar]
- 32.Rubin, D. H., and B. N. Fields. 1980. Molecular basis of reovirus virulence. Role of the M2 gene. J. Exp. Med. 152:853–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang, S. K., P. Danthi, M. Chow, and J. M. Hogle. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J. Mol. Biol. 296:335–340. [DOI] [PubMed] [Google Scholar]
- 35.Tsang, S. K., B. M. McDermott, V. R. Racaniello, and J. M. Hogle. 2001. Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol. 75:4984–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyler, K. L., M. K. Squier, A. L. Brown, B. Pike, D. Willis, S. M. Oberhaus, T. S. Dermody, and J. J. Cohen. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J. Virol. 70:7984–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wessner, D. R., and B. N. Fields. 1993. Isolation and genetic characterization of ethanol-resistant reovirus mutants. J. Virol. 67:2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]