Abstract
This report compares gene transfer efficiencies as well as durations and levels of gene expression for human immunodeficiency virus (HIV) and equine infectious anemia virus (EIAV) lentiviral vectors in a variety of human cell types in vitro. EIAV and HIV vectors transduced equivalent numbers of proliferating and G1/S- and G2/M-arrested cells, and both had very low efficiencies of transduction into G0-arrested cells. Analysis of the levels of both the enhanced green fluorescent protein (EGFP) and mRNA demonstrated that the HIV-transduced cells expressed greater levels of EGFP protein and RNA than the EIAV-transduced cells. Measurements of vector-derived EGFP RNA half-lives were fourfold higher with the HIV vector than with the EIAV vector. Long-term culture of EIAV-transduced human cells showed a significant decrease in the number of cells expressing the transgene; however, no corresponding loss was found in EIAV-transduced equine cells. In contrast, only a moderate decrease in the number of transgene-expressing cells was seen with the HIV vectors. Taken together, these results demonstrate that the EIAV vectors transduced human cells with efficiencies similar to those of the HIV vectors. However, our data indicate that transgene expression from EIAV vectors is limited by the instability of vector-derived RNA transcripts and silencing of the EIAV vectors over time.
Lentiviruses are efficient gene transfer vehicles due to their ability to transduce both dividing and nondividing cells. The most thoroughly investigated of the lentiviral gene therapy vectors are derived from human immunodeficiency virus (HIV) type 1. While significant effort has gone into HIV vector development to maximize the biosafety of HIV vectors and minimize the possibility of generating replication-competent lentiviruses, significant questions remain about the biosafety of HIV-based vectors for use in human clinical trials.
Due to these biosafety concerns, a series of gene therapy vectors based on other primate and nonprimate lentiviruses have been developed (1, 2, 5, 6, 10). Gene transfer vectors based on a lentivirus, equine infectious anemia virus (EIAV), were developed and shown to be effective in transducing proliferating and G1-arrested cells in vitro and mouse neurons in vivo (11, 14). However, the ability of EIAV vectors to transduce and express in human-derived cells has not been extensively investigated. The goal of this study was to directly compare gene transfer efficiencies as well as levels of transgene expression and persistence in a variety of human cell types by using vectors derived from recombinant EIAV and HIV.
EIAV transduces human cells in vitro with an efficiency similar to that of HIV vectors.
To directly compare the transduction efficiencies of HIV- and EIAV-based vectors, we transduced the cell lines SupT1 (T cell; American Type Culture Collection [ATCC] strain no. CRL1942), HepG2 (hepatocellular carcinoma; ATCC no. HB8065), NB8 (neuroblastoma; a generous gift of Vincent Kidd, St. Jude Children’s Research Hospital, Memphis, Tenn.), 293 (fibroblast; ATCC no. CRL1573), and NBL-6 (horse fibroblast; ATCC no. CCL57). EIAV, HIV, and murine leukemia virus (MLV) vectors with the internal cytomegalovirus (CMV) promoter driving the enhanced green fluorescent protein (EGFP) were produced by triple transient transfection of 293 T cells, pseudotyped with the vesicular stomatitis virus envelope glycoprotein, and concentrated by ultracentrifugation as described previously (14, 15). HIV and MLV vectors, non-self-inactivating vectors, were chosen to more accurately compare gene expression levels with that of the non-self-inactivating EIAV vector. Proliferating and G1/S (treated with the DNA polymerase inhibitor aphidicolin at 10 μg/ml)- and G2/M (treated with the microtubulin inhibitor nocodazole at 5 μg/ml)-arrested cells were transduced at a multiplicity of infection (MOI) of 1 and analyzed for EGFP expression at 48 h posttransduction. As shown in Table 1, there was little difference in the levels of transduction of proliferating human cells with EIAV, HIV, and MLV vectors (40 to 97%). Experimental data demonstrated that the numbers of EGFP-positive cells did not significantly differ when analyzed at 14 days posttransduction, suggesting that the vector systems were integrated (data not shown). The levels of gene transfer for both the EIAV and HIV vectors were also comparable in cells arrested in G1/S (63 to 87%) and G2/M (23 to 80%) (Table 1). As expected, the MLV-based vectors were ineffective at transducing nondividing cells. To compare the gene transfer levels in G0-arrested cells, the growth of cells of the rat fibroblast cell line 208F was arrested by density-dependent growth arrest for either 7 or 14 days and then the cells were transduced at an MOI of 5 and assayed for EGFP 48 h posttransduction. Both lentiviral vector systems were significantly compromised in their ability to transduce G0-arrested cells (the transduction efficiency of EIAV was 18%, and that of HIV was 14%) compared to the ability of proliferating cells (Table 2). Also, with both vectors, a 40% decrease in the level of cells transduced was observed in cells arrested for 14 days compared to that of cells arrested for 7 days, suggesting a negative correlation between the duration of quiescence and gene transfer efficiency. The reason for the decreased EIAV transduction of G0 fibroblasts may be defective reverse transcription or inefficient nuclear translocation of the preintegration complex, both of which have been observed with HIV vectors (7, 13). G0-arrested cells were resistant to transduction by the MLV vectors.
TABLE 1.
Average levels of transduction (n = 3) of proliferating or G1 or G2 growth-arrested human cell lines with EIAV, HIV, and MLV vectors
| Cell line | Cell type | Cell statusa | % of cells that were EGFP positive (avg ± SD) after transduction with:
|
||
|---|---|---|---|---|---|
| EIAV vector | HIV vector | MLV vector | |||
| D17 | Canine osteosarcoma | Proliferating | 81.6 ± 0.02 | 85.5 ± 2.0 | 84.2 ± 2.2 |
| G1/S arrested | 80.5 ± 10.6 | 70.5 ± 3.5 | 0.83 ± 0.3 | ||
| G2/M arrested | 32.5 ± 5.8 | 25.5 ± 6.3 | 15.0 ± 0.1 | ||
| NB8 | Neuroblastoma | Proliferating | 79.5 ± 1.4 | 84.2 ± 3.8 | 71.6 ± 7.8 |
| G1/S arrested | 63.8 ± 5.2 | 72.5 ± 5.6 | 2.3 ± 1.5 | ||
| G2/M arrested | 80.6 ± 2.7 | 80.1 ± 15.1 | 9.2 ± 6.8 | ||
| HepG2 | Hepatic carcinoma | Proliferating | 39.7 ± 0.4 | 59.0 ± 12.1 | 57.7 ± 3.8 |
| G1/S arrested | 64.6 ± 1.8 | 71.7 ± 2.6 | 2.0 ± 1.4 | ||
| G2/M arrested | 40.2 ± 1.9 | 82.4 ± 0.8 | 7.8 ± 2.5 | ||
| 293 | Embryonic fibroblast | Proliferating | 58.0 ± 4.5 | 84.5 ± 3.4 | 34.3 ± 6.0 |
| G1/S arrested | 80.2 ± 5.2 | 85.9 ± 1.8 | 0.4 ± 0.3 | ||
| G2/M arrested | 44.3 ± 1.3 | 69.2 ± 0.6 | 5.2 ± 5.4 | ||
| Sup T1 | T cell | Proliferating | 94.2 ± 0.2 | 97.5 ± 0.8 | 81.5 ± 1.2 |
| G1/S arrested | 78.6 ± 0.6 | 87.9 ± 0.6 | 4.9 ± 1.5 | ||
| G2/M arrested | NDb | ND | ND | ||
| NBL-6 | Horse fibroblast | Proliferating | 50.9 ± 4.0 | 70.2 ± 0.2 | 59.0 ± 21.1 |
| G1/S arrested | 80.8 ± 3.9 | 85.7 ± 4.4 | 1.3 ± 1.5 | ||
| G2/M arrested | 58.1 ± 13.5 | 92.7 ± 5.5 | 9.1 ± 4.2 | ||
G1/S-phase cells arrested with aphidocolin, and G2/M-phase cells arrested with nocodazole.
ND, not done due to loss of cell viability.
TABLE 2.
Average levels of transduction (n = 3) of G0-arrested rat 208 fibroblast cells with EIAV, HIV, and MLV vectors
| Cells | Transduction rate of quiescent 208F rat fibroblast EGFP-positive cells relative to that of proliferating cells (avg ± SD) after transduction with:
|
||
|---|---|---|---|
| EIAV vector | HIV vector | MLV vector | |
| Proliferating | 1 | 1 | 1 |
| 7-day-arrested G0 | 0.18 ± 0.05 | 0.14 ± 0.08 | 0.002 ± 0.001 |
| 14-day-arrested G0 | 0.11 ± 0.05 | 0.09 ± 0.06 | <0.001 |
| Stimulated 7-day-arrested G0a | 0.66 ± 0.06 | 0.87 ± 0.04 | 0.03 ± 0.02 |
Initiation of cell proliferation 4 days posttransduction of 7-day-arrested G0 cells.
We investigated whether EIAV vectors, like HIV vectors (13), could be maintained as a stable transduction intermediate in quiescent cells. Quiescent cells were transduced with the various vectors at an MOI of 5. Seven days later, the quiescent transduced cells were stimulated to proliferate by the addition of serum-containing medium and subjected to flow cytometric analysis 2 days later. Gene transfer levels increased four- to sixfold with the EIAV and HIV vectors compared to those of nonstimulated quiescent cells, whereas MLV vectors were inefficient at transducing the stimulated cells (Table 1). These data suggest that, like HIV, EIAV vectors form stable transduction intermediates in quiescent cells, which may be a characteristic of all lentiviral vector systems.
EGFP protein levels are lower in EIAV vectors than in HIV vectors.
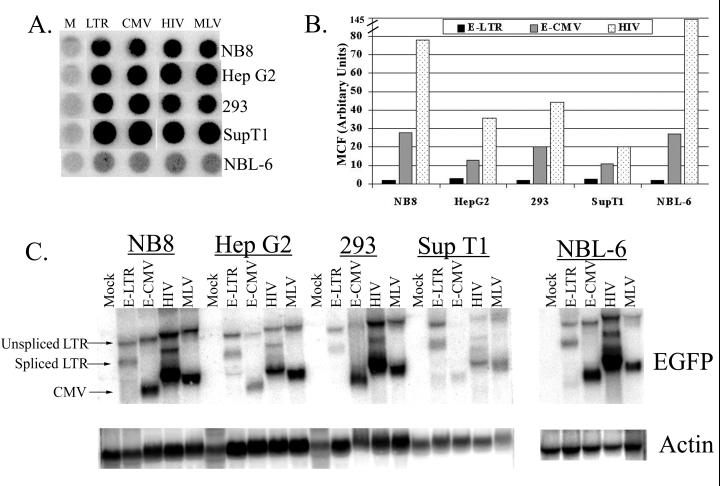
To directly compare transgene expression levels from EIAV and HIV vectors, cells were transduced at an MOI of 0.5 to prevent multiple vector integration events per cell, expanded, and sorted with a fluorescence-activated cell sorter for EGFP expression 14 days later to ensure vector integration. The EIAV long terminal repeat (E-LTR) vector was created by excising the internal CMV promoter from the parental EIAV vector (E-CMV), which allows transcription of the EGFP gene to be driven solely by the E-LTR. Gene expression was measured by flow cytometric analysis of the EGFP protein and by Northern blot analysis. Dot blot analysis of genomic DNA isolated from the cells sorted with a fluorescence-activated cell sorter showed that similar vector genome copy numbers existed in the EIAV and HIV vector-transduced cell populations (Fig. 1A). In all cell lines, the EGFP protein levels were two- to fivefold lower in the E-CMV-transduced cells than those in the HIV-transduced cells (Fig. 1B). The EGFP protein levels obtained with the E-LTR vectors were 4- to 14-fold lower than those obtained with the E-CMV vectors, demonstrating the necessity of internal promoters in EIAV-based vectors.
FIG. 1.
Analysis of gene expression from vector-transduced cells. Cells were transduced with the various vectors at an MOI of 0.5, expanded for 14 days, and sorted for EGFP expression. (A) Dot blot analysis of vector copies. DNA (30 μg) from the sorted EGFP cells (10 μg of DNA from NBL-6 cells) was spotted onto a nylon filter and probed with 32P-labeled EGFP cDNA. Lane M, mock-transduced. (B) Flow cytometric analysis of EGFP protein levels in sorted EGFP-positive cells transduced with the different vectors. Results are the mean levels of channel fluorescence (MCF) of EGFP-expressing cells and were normalized to the minimum gate setting. (C) Northern blot analysis of sorted EGFP-positive cells. Total RNAs were isolated from the sorted cells, and the blot was sequentially probed with 32P-labeled EGFP and β-actin. β-Actin was used to normalize for RNA loading. Arrows indicate the unspliced and spliced messages originating from viral LTRs and the transcripts originating from the internal CMV promoter. Mock, mock transduced. Data shown are representative of the gene expression levels observed from multiple experiments.
Northern blot analysis was used to quantitate the steady-state RNA levels in the sorted EGFP-positive cells (Fig. 1C). We detected three RNA bands that represented the full-length and spliced transcripts originating from the viral LTRs and an RNA transcript originating from the internal CMV promoter. Similar to what occurred with the protein levels, there was a three- to fivefold decrease in the total amount of vector-derived RNA (represented by the addition of transcripts from the LTR [spliced and unspliced] and the internal CMV promoter) from E-CMV-transduced cells compared to the amount from the HIV-transduced cells. The steady-state levels of EGFP RNA obtained with the E-LTR vectors were two- to ninefold lower than those obtained with the E-CMV vectors. The level of EGFP RNA generated by the MLV vectors was comparable to that generated by the HIV vectors.
To assess the influence of vector sequences on promoter activity, we analyzed the amount of EGFP RNA originating from the internal CMV promoter only. Surprisingly, the amount of EGFP RNA originating from the CMV promoter in E-CMV-transduced cells was two- to eightfold less than that originating from the HIV-transduced cells, suggesting that the endogenous sequence element(s) found in the EIAV vector backbone may contribute to the decreased amount of steady-state RNAs in transduced cells. Taken together, these data demonstrate that the differences in EGFP protein levels detected by flow cytometry obtained with E-CMV and HIV vectors were not due to differing translational efficiencies between the vector systems but were the direct result of decreased amounts of steady-state RNA produced from the EIAV-based vectors.
Decreased stability of RNA transcripts from EIAV vectors.
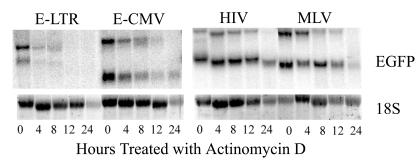
Since the levels of steady-state EGFP RNAs originating from identical internal CMV promoters from EIAV vectors were less than those originating from HIV vectors in all cell lines examined, we hypothesized that these differences may be due to differences in mRNA stability between the vector systems. Actinomycin D (10 μg/ml)-treated transduced NB8 cells were analyzed for their EGFP RNA half-life by Northern blot analysis (Fig. 2). The half-life of transcripts originating from the internal CMV promoter in the E-CMV vector was 6 h. In contrast, the half-life of transcripts originating from the internal CMV promoter in HIV vectors was 24 h, a finding consistent with the data from the study of Zufferey et al. (17). The half-lives of RNAs derived from both the EIAV and HIV vectors in HepG2 cells, 293 cells, and the horse fibroblast cell line NBL-6 were similar to those found in the NB8 cells (data not shown), demonstrating that the short RNA half-lives of EIAV vector transcripts do not vary markedly between cell types or in a species-specific manner. Surprisingly, transcripts from the E-LTR vector lacking the internal CMV promoter were dramatically more labile than transcripts from other lentiviral vectors, with half-lives of less than 4 h. These data suggest that the low levels of steady-state RNAs from EIAV vectors are due, in part, to the inherent instability of EIAV-derived transcripts.
FIG. 2.
Analysis of half-lives of viral RNA from NB8 cells transduced with E-LTR, E-CMV, HIV, and MLV vectors. NB8 cells were transduced at an MOI of 0.5 with the indicated vector, expanded, and sorted for EGFP expression. Positive cells were cultured in medium containing 10 μg of actinomycin D/ml, and total RNA was isolated at the indicated time points. For Northern blot analysis, total RNA blots were probed with 32P-labeled EGFP probes from E-LTR-, E-CMV-, HIV-, or MLV-transduced cells. The EIAV blot was overexposed to more clearly visualize the decreasing RNA levels. An ethidium bromide-stained gel showing the 18S rRNA was used to assess RNA loading.
Long-term gene expression.
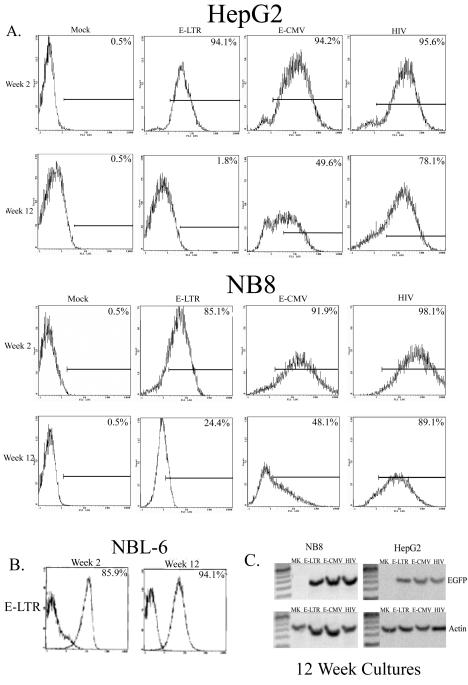
We next investigated the long-term expression of EGFP in sorted cells transduced with either the EIAV or HIV vector system. NB8 and HepG2 cells were assayed for EGFP expression by flow cytometry 2 and 12 weeks after the cell sorting. In NB8 and HepG2 cells transduced with the E-LTR vector, there were 5-fold and 47-fold decreases, respectively, in the numbers of EGFP-expressing cells after 12 weeks in culture (Fig. 3A). In cells transduced with the E-CMV vector, there were nearly twofold losses of EGFP-expressing cells in both cell lines after 12 weeks. In contrast, in NB8 and HepG2 cells transduced with the HIV vector, there were only slight decreases in the number of EGFP-positive cells after 12 weeks in culture. However, the horse fibroblast cell line NBL-6 transduced with E-LTR demonstrated no loss of EGFP-expressing cells over the same culture period (Fig. 3B). This finding suggests some species specificity in the sensitivity of the EIAV vectors to transgene-silencing mechanisms.
FIG. 3.
Long-term gene expression. Cells were transduced at an MOI of 0.5, expanded, and sorted for EGFP expression 14 days after transduction. The sorted cells were cultured, and the numbers of EGFP-positive cells were analyzed by flow cytometry 2 and 12 weeks later. The number of EGFP-positive cells is indicated in the corner of each histogram. (A) HepG2 and NB8 cells. Mock, mock transduced. (B) E-LTR-transduced NBL-6 horse fibroblast cells. (C) DNA-PCR analysis of EGFP levels in transduced cells. DNAs were isolated from HepG2- and NB8-transduced cells cultured for 12 weeks after transduction. Primers specific for EGFP (forward, 5′-CGAGCTGGACGGCGACGTAAAC-3′; reverse, 5′-GCGCTTCTCGTTGGGGTCTTTG-3′) or β-actin (forward, 5′-CATTGTGATGGACTCCGGAGACGG-3′; reverse, 5′-CATCTCCTGCTCGAAGTCTAGAGC-3′) were used to amplify 300 ng of DNA (30 cycles) from mock-transduced (lanes MK) or E-LTR-, E-CMV-, or HIV-transduced cells.
To ensure that the decreases in the numbers of EGFP-expressing cells were not due to the loss of integrated EIAV provirus in transduced cells, PCR was performed on genomic DNAs isolated from the 12-week cultures (Fig 3C). In NB8- and HepG2-transduced cells, there were no differences in gene transfer levels among E-LTR-, E-CMV-, and HIV-sorted cell lines, demonstrating that the loss of EGFP expression in EIAV-transduced human cells was not due to loss of the provirus but to gene-silencing mechanisms.
Our data demonstrate that EIAV-based vectors are effective gene transfer vehicles in dividing and nondividing human cells, a finding in direct contrast to those for two other nonprimate lentivirus vectors based on caprine arthritis encephalitis virus and visna virus (3, 12). However, other nonprimate lentiviral vectors, such as those based on feline immunodeficiency virus, can also effectively transduce human cells. To increase gene transfer to G0-arrested cells, transfer vectors containing the EIAV central polypurine tract are being developed. In contrast to the similarities in the levels of gene transfer obtained with EIAV- and HIV-based vectors, gene expression from EIAV-based vectors was significantly less than that from HIV-based vectors, due to the relative instability of EIAV-derived mRNA transcripts. It is important to note that during the preparation of this paper, Yamada et al. reported lower levels of EIAV-derived mRNA than those derived from HIV vectors in a transduced human B-cell line (16). However, the mechanism for this difference in mRNA levels was not investigated.
To increase the duration and levels of gene expression from EIAV vectors, modifications of EIAV vectors are necessary. Preliminary experiments using either the woodchuck hepatitis posttranscriptional element or the constitutive transport element from the Mason-Pfizer monkey virus, both of which increase gene expression from HIV vectors, also showed increased gene expression from EIAV vectors in vitro and in vivo (data not shown). Alterations in the EIAV core poly(A) site, which reside in the 3′ LTR, may increase gene expression by increasing the mRNA stability of EIAV-derived vectors. Removing polyadenylation sequences from HIV vectors and replacing them with a bovine growth hormone polyadenylation signal increased the viral titer twofold, suggesting a possible increase in viral RNA stability (8). Further developments in EIAV vector design similar to improvements made in HIV and MLV vectors, such as the creation of self-inactivating vectors to minimize promoter interference and the elimination of possible methylation-sensitive and transcriptional repressor sequences found in the E-LTR, are under way.
Finally, improvements in EIAV vector design should address the problem of the transgene silencing which was observed in long-term cultures of all human cell lines (Fig. 3). The nature of the gene-silencing mechanisms occurring in the EIAV vectors is unclear; however, it is hypothesized that gene silencing with retroviral vectors occurs through a two-step mechanism, where methylation of transcriptional elements occurs first, followed by repressive alterations in chromatin structure (4). However, culture of the EIAV-transduced silenced cell lines in the presence of the DNA methyltransferase inhibitor 5-azacytidine or the histone deacetylase inhibitor trichostatin A failed to reactivate transgene expression (data not shown). McInerney et al. reported that these drugs were ineffective in reversing retroviral vector gene silencing when administered 3 months after vector administration, which is similar to our findings with the EIAV vectors, suggesting that there is a limited time period in which it is possible to reactivate silenced transgenes after vector administration before silencing becomes refractive to methylase or deacetylase inhibitors (9). Alternatively, the presence of high levels of EIAV mRNA in human cells may impede cell growth or induce cell death. Therefore, over the culture period, a cell population expressing little or no EIAV mRNA would outgrow a cell population expressing a large amount of mRNA, resulting in an apparent loss of EGFP-expressing cells over time.
In summary, as gene transfer agents, EIAV vectors are as effective as HIV-derived vectors. Limitations in the duration and levels of gene expression from EIAV vectors necessitate ongoing improvements in EIAV vector design. These modifications are needed to make EIAV-derived vectors a viable alternative to HIV-derived vectors.
Acknowledgments
The work in this report was supported by grants AI47693 (B.A.B.) and HL10430 (J.P.O.) from the National Institutes of Health.
REFERENCES
- 1.Arya, S. K., M. Zamani, and P. Kundra. 1998. Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Hum. Gene Ther. 9:1371–1380. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., H. Ilves, W. Y. Lin, K. Eckert, A. Coward, S. Tamaki, G. Veres, and I. Plavec. 2001. Construction and molecular analysis of gene transfer systems derived from bovine immunodeficiency virus. J. Virol. 75:3371–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz, R. D., H. Ilves, I. Plavec, and G. Veres. 2001. Gene transfer systems derived from Visna virus: analysis of virus production and infectivity. Virology 279:116–129. [DOI] [PubMed] [Google Scholar]
- 4.Bestor, T. H. 1998. Gene silencing. Methylation meets acetylation. Nature 393:311–312. [DOI] [PubMed] [Google Scholar]
- 5.Curran, M. A., S. M. Kaiser, P. L. Achacoso, and G. P. Nolan. 2000. Efficient transduction of nondividing cells by optimized feline immunodeficiency virus vectors. Mol. Ther. 1:31–38. [DOI] [PubMed] [Google Scholar]
- 6.D’Costa, J., H. Brown, P. Kundra, A. Davis-Warren, and S. Arya. 2001. Human immunodeficiency virus type 2 lentiviral vectors: packaging signal and splice donor in expression and encapsidation. J. Gen. Virol. 82:425–434. [DOI] [PubMed] [Google Scholar]
- 7.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217–222. [DOI] [PubMed] [Google Scholar]
- 8.Iwakuma, T., Y. Cui, and L. J. Chang. 1999. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology 261:120–132. [DOI] [PubMed] [Google Scholar]
- 9.McInerney, J. M., J. R. Nawrocki, and C. H. Lowrey. 2000. Long-term silencing of retroviral vectors is resistant to reversal by trichostatin A and 5-azacytidine. Gene Ther. 7:653–663. [DOI] [PubMed] [Google Scholar]
- 10.Metharom, P., S. Takyar, H. H. Xia, K. A. Ellem, J. Macmillan, R. W. Shepherd, G. E. Wilcox, and M. Q. Wei. 2000. Novel bovine lentiviral vectors based on Jembrana disease virus. J. Gene Med. 2:176–185. [DOI] [PubMed] [Google Scholar]
- 11.Mitrophanous, K., S. Yoon, J. Rohll, D. Patil, F. Wilkes, V. Kim, S. Kingsman, A. Kingsman, and N. Mazarakis. 1999. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 6:1808–1818. [DOI] [PubMed] [Google Scholar]
- 12.Mselli-Lakhal, L., C. Favier, M. F. Da Silva Teixeira, K. Chettab, C. Legras, C. Ronfort, G. Verdier, J. F. Mornex, and Y. Chebloune. 1998. Defective RNA packaging is responsible for low transduction efficiency of CAEV-based vectors. Arch Virol. 143:681–695. [DOI] [PubMed] [Google Scholar]
- 13.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. [DOI] [PubMed] [Google Scholar]
- 14.Olsen, J. C. 1998. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 5:1481–1487. [DOI] [PubMed] [Google Scholar]
- 15.Tarantal, A. F., J. P. O’Rourke, S. S. Case, G. C. Newbound, J. Li, C. I. Lee, C. R. Baskin, D. B. Kohn, and B. A. Bunnell. 2001. Rhesus monkey model for fetal gene transfer: studies with retroviral-based vector systems. Mol. Ther. 3:128–138. [DOI] [PubMed] [Google Scholar]
- 16.Yamada, K., J. C. Olsen, M. Patel, K. W. Rao, and C. E. Walsh. 2001. Functional correction of fanconi anemia group C hematopoietic cells by the use of a novel lentiviral vector. Mol. Ther. 3:485–490. [DOI] [PubMed] [Google Scholar]
- 17.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73:2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]