Abstract
We have recently isolated a transmissible gastroenteritis virus (TGEV) infectious construct designated TGEV 1000 (B. Yount, K. M. Curtis, and R. S. Baric, J. Virol. 74:10600–10611, 2000). Using this construct, a recombinant TGEV was constructed that replaced open reading frame (ORF) 3A with a heterologous gene encoding green fluorescent protein (GFP). Following transfection of baby hamster kidney (BHK) cells, a recombinant TGEV (TGEV-GFP2) was isolated that replicated efficiently and expressed GFP. Replicon constructs were constructed that lacked either the ORF 3B and E genes or the ORF 3B, E, and M genes [TGEV-Rep(AvrII) and TGEV-Rep(EcoNI), respectively]. As the E and M proteins are essential for TGEV virion budding, these replicon RNAs should replicate but not result in the production of infectious virus. Following cotransfection of BHK cells with the replicon RNAs carrying gfp, GFP expression was evident by fluorescent microscopy and leader-containing transcripts carrying gfp were detected by reverse transcription-PCR (RT-PCR). Subsequent passage of cell culture supernatants onto permissive swine testicular (ST) cells did not result in the virus, GFP expression, or the presence of leader-containing subgenomic transcripts, demonstrating the single-hit nature of the TGEV replicon RNAs. To prepare a packaging system to assemble TGEV replicon particles (TGEV VRP), the TGEV E gene was cloned into a Venezuelan equine encephalitis (VEE) replicon expression vector and VEE replicon particles encoding the TGEV E protein were isolated [VEE-TGEV(E)]. BHK cells were either cotransfected with TGEV-Rep(AvrII) (E gene deletion) and VEE-TGEV(E) RNA transcripts or transfected with TGEV-Rep(AvrII) RNA transcripts and subsequently infected with VEE VRPs carrying the TGEV E gene. In both cases, GFP expression and leader-containing GFP transcripts were detected in transfected cells. Cell culture supernatants, collected ∼36 h posttransfection, were passed onto fresh ST cells where GFP expression was evident ∼18 h postinfection. Leader-containing GFP transcripts containing the ORF 3B and E gene deletions were detected by RT-PCR. Recombinant TGEV was not released from these cultures. Under identical conditions, TGEV-GFP2 spread throughout ST cell cultures, expressed GFP, and formed viral plaques. The development of infectious TGEV replicon particles should assist studies of TGEV replication and assembly as well as facilitate the production of novel swine candidate vaccines.
Transmissible gastroenteritis (TGE) is an economically important, acute enteric disease of swine, which is often 100% fatal in newborn piglets (23, 24, 46). TGE virus (TGEV), the causative agent of TGE, is a member of the Coronaviridae family and the order Nidovirales. In addition to the Coronaviridae, the order Nidovirales also includes the Arteriviridae family, of which the swine pathogen porcine reproductive and respiratory syndrome virus is a member (12, 18, 70). Despite significant size differences (∼13 to 32 kb), the polycistronic genome organization and regulation of gene expression from a nested set of subgenomic mRNAs are similar for all members of the order (18, 71).
TGEV possesses a single-stranded, positive-sense ∼28.5-kb RNA genome enclosed in a helical nucleocapsid structure that is surrounded by an envelope containing three viral proteins, including the S glycoprotein, the membrane (M) glycoprotein and a small envelope (E) protein (22, 25, 60, 61). Remarkably, only the E and M proteins are absolutely required for particle formation, defining a novel model for virion budding (27, 77). The TGEV genome contains eight large open reading frames (ORFs), which are expressed from full-length or subgenomic-length mRNAs during infection (22, 68, 69). The 5′-most ∼20 kb contains the replicase genes in two ORFs, 1A and 1B, the latter of which is expressed by ribosomal frameshifting (2, 22). The 3′-most ∼9 kb of the TGEV genome contains the structural genes, each preceded by a highly conserved intergenic sequence (IS) of 7 to 17 nucleotides (nt) in length that functions in the synthesis of each of the subgenomic RNAs (14, 22, 25, 73). The subgenomic mRNAs are arranged in a coterminal nested set structure from the 3′ end of the genome, and each contains a leader RNA sequence derived from the 5′ end of the genome. Although each mRNA is polycistronic, the 5′-most ORF is preferentially translated, necessitating the synthesis of a distinct mRNA species for each ORF (45, 49, 68, 69). Both full-length and subgenomic-length negative-strand RNAs are also produced and have been implicated in mRNA synthesis (8, 64, 66, 68, 69). Subgenomic RNA synthesis occurs by a method of discontinuous transcription, most likely by transcription attenuation during negative-strand synthesis (8, 64).
The coronavirus E and M proteins function in virion assembly and release, which involve the constitutive secretory pathway of infected cells. Coexpression of the E and M proteins results in virus-like particle formation in cells, defining a novel, nucleocapsid-independent mechanism of enveloped-virus assembly (77). The role of the E protein in virus assembly was further confirmed by reverse genetic analysis using targeted recombination (27). The TGEV M protein may serve to initiate the viral particle assembly process through interactions with genomic RNA and nucleoprotein in pre-Golgi compartments (52). The precise role of E in the assembly and release of coronavirus particles is not clear. Although an interaction between the E and M proteins has not yet been demonstrated, such an interaction likely occurs and would serve to facilitate the budding of viral particles. Additionally, E protein has been suggested to “pinch off the neck” of the assembled viral particles during the final stages of budding (77).
We have recently described a simple and rapid approach for systematically assembling a full-length cDNA copy of the TGEV genomic RNA from which infectious transcripts can be produced (80). Our approach, as well as that of Almazan et al. (2), will facilitate reverse genetic methods that impact all aspects of coronavirology. In this report, we describe the production of recombinant TGEV expressing green fluorescent protein (GFP) and the assembly of replicon RNAs encoding GFP that contain deletions within the two structural genes required for virion assembly and release (E and M). In addition, we have prepared a packaging system for TGEV replicons using Venezuelan equine encephalitis (VEE) virus replicon particles (VRPs) to express the TGEV E structural gene in trans, which will assist studies of virion assembly and maturation by allowing for the precise mapping of critical structural protein residues and/or motifs. This TGEV replicon system should prove useful for future studies of TGEV replication and assembly as well as provide for novel candidate mucosal vaccines against important swine pathogens.
MATERIALS AND METHODS
Virus and cells.
The Purdue strain of TGEV (ATCC VR-763) was obtained from the American Type Culture Collection and passaged once in swine testicular (ST) cells. ST cells were obtained from the American Type Culture Collection (ATCC 1746-CRL) and maintained in minimal essential medium containing 10% fetal clone II (HyClone Laboratories, Inc.) and supplemented with 0.5% lactalbumin hydrolysate, 1× nonessential amino acids, 1 mM sodium pyruvate, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). Baby hamster kidney (BHK) cells (BHK-21 [ATCC CCL10]) were maintained in alpha-minimal essential medium containing 10% fetal calf serum supplemented with 10% tryptose phosphate broth, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). To determine the effect of coinfection with TGEV and VEE VRPs on TGEV growth rate, cultures of ST cells (5 × 105) were infected with wild-type TGEV alone or with wild-type TGEV and VEE VRPs encoding a Norwalk-like virus (VEE-NCFL) capsid antigen (ORF 2) at a multiplicity of infection (MOI) of 5 for 1 h (33). The cells were washed twice with phosphate-buffered saline (PBS) to remove residual virus and VEE VRPs, and the cells were subsequently incubated at 37°C in complete medium. At different times postinfection, progeny virions were harvested and assayed by plaque assay in ST cells, as previously described (80).
Recombinant DNA manipulations of TGEV F subclone.
Plasmid DNAs were amplified in Escherichia coli DH5α and purified with the QIAprep Miniprep kit (Qiagen Inc., Valencia, Calif.). All enzymes were purchased from New England BioLabs (Beverly, Mass.) and used according to the manufacturer’s directions. DNA fragments were isolated from Tris-acetate-EDTA agarose gels (0.8%) with the QIAEX II gel extraction kit (Qiagen Inc.). All DNA was visualized using Dark Reader technology (Clare Chemical Research, Denver, Colo.) to prevent UV-induced DNA damage that could impact subsequent manipulations, including in vitro transcription. In our hands, increased concentrations of full-length transcripts and increased transfection efficiencies were achieved after Dark Reader technology was used to isolate the full-length TGEV cDNAs.
Six subgenomic cDNA clones (A to F) spanning the entire TGEV genome were isolated using standard molecular techniques as previously described (80). The 3′ end of the TGEV genome, carrying the S, ORF 3A, ORF 3B, E, M, N, and ORF S genes, is contained within the TGEV F subclone. nt 24828 to 25073 (GenBank accession no. AJ271965), corresponding to ORF 3A, were removed and replaced with the ClaI and PflMI restriction sites using conventional recombinant DNA techniques such that the adjacent ORFs (S and 3B) were not disrupted (62). The ClaI site was inserted just downstream of the 3A IS start, while the PflMI site is located just upstream of the ORF 3B IS (Fig. 1a). The mammalian codon-optimized version of the GFP gene was isolated from the noncytopathic Sindbis virus vector pSINrep19/GFP (kindly provided by Charlie Rice, Columbia University) and was inserted with a 5′ 20-nt N gene IS using the ClaI/PflMI cloning site by standard recombinant DNA techniques (Fig. 1a) (62). Clones were identified by DNA sequencing using an ABI model 377 automated sequencer, and the resulting construct, TGEV pFiGFP2(PflMI), was subsequently used in the assembly of recombinant TGEV viral cDNA and as the backbone for the construction of structural gene deletions (Fig. 1b).
FIG. 1.
Construction of TGEV replicon cDNAs. (a) TGEV F fragments. Structural genes are contained in the TGEV F fragment. FiGFP2(PflMI) (∼5.6 kb) was constructed from the wild-type TGEV F fragment (∼5.1 kb) by the deletion of ORF 3A (nt 24828 to 25073) and the insertion of GFP with a 5′ 20-nt N gene IS. Using this construct, we introduced deletions extending from the unique PflMI site at the very 3′ end of GFP to the unique AvrII (nucleotide position 25866) and EcoNI (nucleotide position 26935) sites present within the TGEV E and M genes, respectively. (b) Sequence organization of GFP in FiGFP2(PflMI). GFP was inserted just downstream of the ORF 3A IS. The TGEV sequence originating at the 3′ end of the S gene (nt 24693) through the start of the GFP gene is shown, and the important IS and restriction sites are labeled. (c) Strategy for assembling recombinant TGEV and replicon cDNAs. The six cDNA subclones (TGEV A, B1, B2, C, DE1, and F deletion fragments) spanning the genome are flanked by unique interconnecting BglI and BstXI sites, allowing for directional assembly into a full-length replicon cDNA by in vitro ligation (80). TGEV A contained a unique T7 start site at its 5′ end, and the F deletion fragments (FiGFP2-AvrII and FiGFP2-EcoNI) contain GFP and a 25-nt T tail, allowing for the synthesis of capped T7, polyadenylated transcripts in vitro.
Serial deletions within the TGEV structural gene region were generated from the unique PflMI site at the very 3′ end of the GFP gene and extended for various distances toward the 3′ end of the genome (Fig. 1b). TGEV pFiGFP2(PflMI) was digested with PflMI and AvrII or EcoNI, treated with T4 DNA polymerase under conditions in which the 5′→3′ exonuclease activity generated blunt ends (according to the manufacturer’s directions) (New England BioLabs), and religated using T4 DNA ligase. The unique AvrII site is located at nucleotide position 25866 within the E protein gene, and the unique EcoNI site is located at nucleotide position 26624 within the M protein gene (2). Clones containing each of the deletions were identified by restriction digestion analysis and confirmed by DNA sequencing using an ABI model 377 automated sequencer. The new TGEV F fragments (FiGFP2-AvrII and FiGFP2-EcoNI) were subsequently utilized in the assembly of the TGEV replicon constructs.
Assembly of full-length TGEV cDNAs.
The six cDNA subclones spanning the entire TGEV genome, including the FiGFP2(PflMI) and FiGFP2(PflMI) deletion subclones, were used to assemble TGEV viral and replicon constructs, respectively, as previously described (Fig. 1c) (80). The TGEV A fragment contains a T7 promoter while the TGEV FiGFP2(PflMI), FiGFP2-AvrII, and FiGFP2-EcoNI fragments terminate in a 25-nt poly(T) tract and a unique NotI site at the 3′ end, allowing for in vitro T7 transcription of capped, polyadenylated transcripts. To assemble full-length TGEV recombinant virus and subgenomic replicon cDNAs, plasmids were digested with BglI, BstXI, or NotI, and the appropriately sized inserts were isolated from agarose gels. The TGEV A-B1, B2-C, and DE-1-FiGFP2 fragments were ligated overnight at 4°C in the presence of T4 DNA ligase, according to the manufacturer’s directions. Systematically assembled products were isolated and extracted from agarose gels, and the TGEV A-B1, B2-C, and DE-1-FiGFP2 fragments were religated overnight. The final ligation products were purified by phenol-chloroform-isoamyl alcohol and chloroform extraction, precipitated under isopropanol, and washed with 70 and 90% ethanol. Purified TGEV full-length viral and replicon cDNA constructs, designated TGEV-GFP2, TGEV-Rep(AvrII), and TGEV-Rep(EcoNI), were subsequently used for T7 in vitro transcription.
TGEV in vitro transcription and transfection.
Capped runoff T7 transcripts were synthesized in vitro from assembled TGEV and replicon cDNAs using the mMessage mMachine kit as described by the manufacturer (Ambion, Austin, Tex.), with certain modifications. TGEV RNA transcription reaction mixtures (50-μl volume) were prepared containing 7.5 μl of a 30 mM GTP stock and incubated at 37°C for 2 h. Similar reactions were performed using 1 μl of PCR amplicons carrying the TGEV N gene sequence and 1 μg of pVR21-E1, each containing a 2:1 ratio of cap analog to GTP. A portion of the RNA transcripts (5 μl of the 50-μl reaction volume) were treated with DNase I, denatured, and separated in 0.5% agarose gels in Tris-acetate-EDTA buffer containing 0.1% sodium dodecyl sulfate. The remaining RNA transcripts were directly electroporated into BHK cells. As a control, separate transcription reaction mixtures were treated with RNase A for 15 min at room temperature prior to transfection.
BHK cells were grown to subconfluence (∼70%), trypsinized, washed twice with PBS, and resuspended in PBS at a concentration of 107 cells/ml. RNA transcripts were added to 800 μl of the cell suspension (8 × 106 cells) in an electroporation cuvette, and three electrical pulses of 850 V at 25 μF were given with a Bio-Rad Gene Pulser II electroporator. N gene transcripts (lacking the TGEV leader sequence) were included in all electroporations, as these transcripts may enhance the recovery of infectious TGEV virions derived from the full-length cDNA construct, TGEV 1000 (80). The BHK cells were either seeded alone or, in some instances, mixed with 106 ST cells in a 75-cm2 flask and incubated at 37°C in 5% CO2. Aliquots of cell culture supernatants were harvested ∼36 h postelectroporation, and fresh cultures of ST cells were infected for 1 h at room temperature and subsequently incubated at 37°C in complete medium.
Analysis of GFP expression and RT-PCR to detect leader-containing subgenomic transcripts.
At ∼18 h postelectroporation, transfected cultures were analyzed for GFP expression by fluorescent microscopy using an Olympus model inverted microscope. Total intracellular RNA was also harvested from transfected cell cultures (pass 0) with Trizol reagent (Gibco BRL) and from cultures inoculated with pass 0 supernatants (pass 1) at ∼36 h postinfection. The intracellular RNA was used as a template for reverse transcription-PCR (RT-PCR) using primer sets to detect leader-containing transcripts encoding GFP. RT reactions were performed using Superscript II reverse transcriptase for 1 h at 42°C (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 0.1 M dithiothreitol), as described by the manufacturer (Gibco BRL), prior to PCR amplification using Taq polymerase (Expand Long kit; Roche Biochemical). To detect leader-containing GFP transcripts in TGEV-GFP2-infected cells, the 5′ leader-specific primer (nt 1 to 25) TGEV-L (5′-CAC TAT AGA CTT TTA AAG TAA AGT GAG TGT AGC-3′) was used with the 3′ primer F:5010(−) (5′-ATT AAG ATG CCG ACA CAC GTC-3′), located within ORF 3B at position 24828. To detect leader-containing GFP transcripts derived from replicon RNAs, two different primer sets were utilized. The 5′ leader-specific primer, TGEV-L, was used with the 3′ primer (−)E5546 (5′-GTT AAT GAC CAT TCC ATT GTC-3′), located just downstream of the AvrII site within TGEV E at nucleotide position 25866, to amplify across the PflMI-AvrII deletion. To amplify across the PlfMI-EcoNI deletion, the same 5′ leader-specific primer (TGEV-L) was used with the 3′ primer M6400(−) (5′-CAA GTG TGT AGA CAA TAG TCC-3′), located just downstream of the EcoNI site within TGEV M at nucleotide position 26624. Following 30 cycles of amplification (94°C for 25 s, 60°C for 25 s, 68°C for 90 s), PCR products were separated on agarose gels and visualized by using Dark Reader technology (Clare Chemical Research). All images were digitalized and assembled by using Adobe Photoshop 5.5 (Adobe Systems, Inc.). Products were subsequently isolated and subcloned directly into Topo XL TA cloning vectors (Invitrogen) as described by the manufacturer. Colonies were isolated on Luria-Bertani plates containing kanamycin (50 μg/ml), and plasmid DNAs were sequenced using an ABI model 377 automated sequencer.
Recombinant VEE replicon construct.
TGEV replicon helper constructs were generated using pVR21, a VEE replicon vector kindly provided by Nancy Davis and Robert Johnston (3). Using overlapping extension PCR, the TGEV E gene was inserted just downstream of the subgenomic 26S promoter within the poly cloning site of pVR21. Using the Expand Long Template PCR system (Roche Molecular Biochemicals), the TGEV (Purdue strain) E gene was amplified from the TGEV F fragment by 30 cycles of PCR (94°C for 25 s, 60°C for 25 s, 72°C for 1 min) by using the TGEV E(V+) 5′ primer (5′-AGT CTA GTC CGC CAA GAT GAC GTT TCC TAG GGC ATT G-3′) and the AscI site-containing TGEV E(V−) 3′ primer (5′-GGC GCG CCT CAA GCA AGG AGT GCT CCA TC-3′). In addition, a segment of the pVR21 vector containing a unique SwaI site followed by the 26S subgenomic promoter was amplified by PCR by using the 6198V primer (5′-GCA AAG CTG CGC AGC TTT CC) with the (−)7564V primer (5′-CAT CTT GGC GGA CTA GAC TAT GTC GTA GTC CAT TCA GGT TAG CCG). Appropriately sized amplicons were isolated on agarose gels and extracted as previously described (80).
The 5′-most 19 nt of primer (−)7546V were complementary to the 5′-most 19 nt of the 5′ TGEV E(V) primer, allowing for the adjoining of the two amplicons by overlapping PCR. Using the Expand Long Template PCR system, reactions were performed and consisted of 30 cycles of 94°C for 20 s, 58°C for 30 s, and 68°C for 2 min, with the first 5 cycles done in the absence of primers. The resulting amplicon, containing unique SwaI and AscI restriction sites at its 5′ and 3′ ends, respectively, was isolated and purified as previously described. Following AscI and SwaI restriction digest (unique to both the TGEV E amplicon and pVR21), the TGEV E gene was inserted into the pVR21 vector. The resulting recombinant VEE replicon vector (pVR21-E1) was cloned, and the sequence was confirmed using an ABI model 377 automated sequencer.
A bipartite helper system consisting of two helper RNAs derived from the V3014Δ520-7505 monopartite helper was used for the construction of VEE replicon particles (59). These helper RNAs express the individual capsid and glycoprotein genes of VEE, thereby supplying the structural genes in trans.
Recombinant VEE VRP production.
The recombinant VEE replicon construct (pVR21-E1) was linearized at a site downstream of the VEE cDNA sequence by NotI digestion, and T7 capped runoff transcripts were generated in vitro by using the T7 mMessage mMachine kit as described by the manufacturer (Ambion). Recombinant VEE replicon and helper RNAs were coelectroporated into BHK cells and incubated at 37°C in 5% CO2 for ∼24 to 27 h. Cell culture supernatants were harvested and clarified by centrifugation at 12,000 × g for 15 min. Recombinant VEE VRPs (VEE-TGEV[E]) were partially purified, concentrated, and resuspended in PBS as previously described (15). Although we were unable to quantitatively identify the presence of VEE-TGEV(E) VRPs due to our lack of anti-E antibody, a qualitative analysis was performed. BHK cells were infected with purified VEE-TGEV(E) VRPs for 1 h at room temperature. VRP titers were high as cytopathic effects were evident in 100% of the transfected cultures, suggesting titers of >108 VRP/ml, and transcripts encoding TGEV E were present in infected cells (data not shown).
Packaging of TGEV replicon RNA.
Two methods were used to supply TGEV E in trans, allowing for the packaging of TGEV-Rep(AvrII) replicon RNA. In the first method, TGEV-Rep(AvrII) replicon RNA and the helper RNA derived from pVR21-E1 were coelectroporated into BHK cells. In the second method, BHK cells were first electroporated with in vitro-transcribed TGEV-Rep(AvrII) RNA, seeded onto 75-cm2 flasks of ST cells, and at 3 h postelectroporation, subsequently infected with recombinant VEE-TGEV(E) VRPs for 1 h at room temperature. Cultures were visualized for GFP expression by fluorescent microscopy at ∼18 h postelectroporation. Culture fluids were harvested ∼36 h postelectroporation, and undiluted aliquots were inoculated onto fresh ST cell cultures (75-cm2 flasks) for 1 h at room temperature. Successful packaging and passing of TGEV-Rep(AvrII) replicon RNA were determined by GFP expression, and RT-PCR analysis was performed to detect leader-containing GFP transcripts, as described above. In addition, RT-PCR was performed to detect TGEV M and N gene leader-containing transcripts by using the TGEV-L 5′ primer and the 3′ primers TGEV-Mg (5′-AGA AGT TTA GTT ATA CCA TAG GCC TTT ATA CCA TAT GTA ATA ATT TTT CTT GCT CAC TC-3′) located at position 26870 within the M gene and TGEV-Ng (5′-CCA CGC TTT GGT TTA GTT CGT TAC CTC ATC AAT TAT CTC-3′) located at position 28038 within the N gene, respectively. Briefly, RT reactions were performed by using Superscript II reverse transcriptase for 1 h at 42°C (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 0.1 M dithiothreitol), as described by the manufacturer (Gibco BRL), prior to 30 cycles of PCR amplification using Taq polymerase (Expand Long kit; Roche Biochemical) (94°C for 25 s, 58°C for 25 s, 68°C for 1 min and 40 s). PCR products were separated on agarose gels, cloned, and sequenced as previously described.
To verify the presence of single-hit TGEV-Rep(AvrII) particles and the lack of recombinant TGEV with the wild-type phenotype, 60-mm2 cultures of ST cells were infected for 1 h at room temperature with TGEV VRPs obtained from previous TGEV-Rep(AvrII) packaging experiments (clarified and concentrated by high-speed centrifugation as previously described [15]), overlaid with fresh media, incubated at 37°C, and subsequently examined over a 72-h time period for GFP expression by fluorescent microscopy as well as virus production by plaque assay in ST cells. Under identical conditions, supernatants obtained from cell cultures transfected with TGEV-GFP2 transcripts were passaged onto fresh ST cells and examined for virus replication by GFP expression, for cytopathic effects, and by plaque assay, as previously described.
RESULTS
Recombinant TGEV expressing GFP.
Our method of developing recombinant TGEV and replicon constructs was based on the same strategy previously used to assemble the full-length cDNA construct TGEV 1000 (80). In this strategy, a full-length cDNA construct is assembled from a series of six cDNA subclones that span the entire TGEV genome (fragments TGEV A, B1, B2, C, D/E, F), each flanked by restriction sites that leave unique interconnecting junctions of 3 or 4 nt in length (BglI and BstXI). These sticky ends are not complementary to most other sticky ends generated with the same enzyme at other sites in the DNA, allowing for the systematic assembly of TGEV cDNAs by in vitro ligation.
Six cDNA subclones that span the entire TGEV genome were used in the assembly of recombinant TGEV cDNA (TGEV-GFP2) (Fig. 1c). The 3′ end of the TGEV genome, including the S, ORF 3A, ORF 3B, E, M, N, and ORF S genes, is carried by the 5.1-kb F fragment. To generate TGEV cDNA constructs containing a reporter gene, the TGEV ORF 3A gene was replaced with the GFP gene, as previous studies suggest that TGEV ORFs 3A and 3B may encode luxury functions unnecessary for virus replication (46, 49, 78). To potentially enhance GFP expression, we inserted a 20-nt N gene IS (TGGTATAACTAAACTTCTAA) just upstream of the GFP gene (Fig. 1a), as the transcripts initiated from this IS are the most abundant throughout the TGEV replication cycle (35, 38). This new TGEV F fragment encoding GFP was designated FiGFP2(PflMI).
Appropriately sized fragments [TGEV A-FiGFP2(PflMI)] were isolated by BglI and either BstXI or NotI digestion. Following a series of in vitro ligations (80), full-length TGEV-GFP2 cDNA was isolated and subsequently used for in vitro transcription. The TGEV A fragment contains a T7 promoter while the TGEV FiGFP2(PflMI) fragment has a poly(T) tract at its very 3′ end, allowing for in vitro T7 transcription of capped, polyadenylated mRNAs. Following in vitro transcription, full-length TGEV-GFP2 transcripts were directly electroporated into 8 × 106 BHK cells, and recombinant TGEV-GFP2 was harvested at 31 h posttransfection. TGEV-GFP2 displayed growth kinetics similar to those of wild-type TGEV generated from the TGEV 1000 infectious construct, with both viruses growing to over 108 PFU/ml in ∼20 h (Fig. 2a). In addition, high levels of GFP expression from TGEV-GFP2, but not from the wild-type virus, were evident by fluorescence microscopy (Fig. 2b). Recombinant TGEV-GFP2 is stable for at least 10 high-titer passages in ST cells, as demonstrated by high levels of GFP expression (data not shown).
FIG. 2.
Recombinant TGEV virus expressing GFP. (a) Cultures of ST cells were infected with either recombinant TGEV-GFP2 virus or the TGEV infectious construct (icTGEV) (80) at an MOI of 5 for 1 h at room temperature. The inocula were removed, and the cultures were incubated in complete medium at 37°C. Samples were harvested at the indicated times and assayed by plaque assay in ST cells. (b) Cultures of ST cells were infected as described for panel a. At ∼12 h postinfection, GFP expression was observed by fluorescent microscopy in TGEV-GFP2-infected cultures (B) but not in icTGEV-infected cultures (A).
Assembly of TGEV replicons encoding GFP.
Our data demonstrate that an ORF 3A deletion is not detrimental to stable replication and passage of recombinant TGEV expressing GFP. Consequently, replicon constructs were generated by deleting the E and M structural genes from the previously constructed FiGFP2(PflMI) F fragment (Fig. 1b). In the first construct (pFiGFP2-AvrII), TGEV ORF 3B and the very 5′ end of the E gene (first 10 nt), including the E gene IS and ATG start codon, were removed by PflMI-AvrII digestion, resulting in an ∼800-nt deletion from pFiGFP2(PflMI). In the second construct (pFiGFP2-EcoNI), ORF 3B, E, and the 5′-most 508 nt of the M gene, including the M gene IS, were removed by PflMI-EcoNI digestion, resulting in an ∼1.5-kb deletion. These constructs were then used to assemble full-length TGEV replicon cDNAs (Fig. 1c).
Replication competence and heterologous gene expression from TGEV replicon RNAs.
As with the construction of TGEV-GFP2, six cDNA subclones that span the entire TGEV genome were used in the assembly of TGEV-Rep constructs (Fig. 1c). Appropriately sized fragments were isolated by BglI and either BstXI or NotI digestion. Following a series of in vitro ligations (80), full-length TGEV subgenomic replicons [TGEV-Rep(AvrII) and TGEV-Rep(EcoNI)] were isolated and subsequently used for in vitro transcription of TGEV replicon RNAs (∼29.1 kb and ∼28.4 kb, respectively). Using transcripts driven from various pSin replicons as a control, we predict that these transcripts were likely of the appropriate lengths (data not shown). Replicon RNA transcripts were directly electroporated into 8 × 106 BHK cells or treated with RNase A prior to transfection, and the cultures were visualized for GFP expression at ∼18 h posttransfection (Fig. 3a).
FIG. 3.
Heterologous gene expression from TGEV replicon RNAs. (a) Cultures of BHK cells were transfected with TGEV replicon RNAs encoding GFP that contained ORF 3B and E (A) [TGEV-Rep(AvrII) transcripts] or ORF 3B, E, and M gene deletions (C). Alternatively, transcripts were treated with RNase prior to transfection into BHK cells (B and D) [TGEV-Rep(AvrII) DNA and TGEV-Rep(EcoNI) DNA only, respectively]. At ∼18 h posttransfection, GFP expression was observed by fluorescent microscopy. (b) Intracellular RNA was isolated from transfected cell cultures and used as a template for RT-PCR. Leader-containing GFP subgenomic transcripts were detected using a 5′ leader primer (TGEV-L) and 3′ primers located just downstream of the AvrII [(−)E5546] or EcoNI [M6400(−)] sites. Appropriately sized amplicons of ∼850 bp were generated, corresponding to transcripts encoding GFP. Cells transfected with TGEV-Rep(AvrII) (A) or TGEV-Rep(EcoNI) (B) RNA are shown. A 1-kb ladder is shown in both panels (lanes 1). Arrows indicate leader-containing GFP amplicons (lanes 2).
GFP expression was observed in <1% of the cells transfected with TGEV-Rep(AvrII) and TGEV-Rep(EcoNI) RNAs (Fig. 3a), demonstrating that TGEV ORF 3A, ORF 3B, E, and M were not required for subgenomic mRNA synthesis and GFP expression. Transcripts of TGEV-Rep(AvrII) and TGEV-Rep(EcoNI) treated with RNase A prior to transfection did not result in observable GFP expression. Leader-containing subgenomic transcripts should be present that encode GFP and contain the appropriate deletions that were introduced into the replicon cDNAs. Consequently, intracellular RNA was isolated at ∼36 h postelectroporation and analyzed by RT-PCR with primer pairs located within the leader RNA sequence and downstream of the deletions in the TGEV-Rep constructs. Appropriately sized amplicons of ∼850 bp were generated from the TGEV-Rep(AvrII)- and TGEV-Rep(EcoNI)-transfected cells (Fig. 3b), corresponding to leader-containing GFP transcripts. Subsequently, the TGEV-Rep(EcoNI) amplicon was gel extracted, cloned, and sequenced, confirming the synthesis of leader-containing GFP transcripts with the PflMI-EcoNI deletion that originated from the ORF 3A IS (see Fig. 7a and 7c). Identical results were seen following TGEV-GFP2 infection, indicating that the 20-nt N gene IS was silent in this configuration and that the natural ORF 3A IS was preferentially used in subgenomic mRNA synthesis. In contrast, wild-type TGEV-infected cells yielded multiple amplicons corresponding to leader-containing transcripts carrying TGEV ORF 3A, ORF 3B, E, and M (data not shown). These transcripts were not detected in TGEV-Rep(AvrII)- and TGEV-Rep(EcoNI)-electroporated cells, respectively, as these genes were completely or partially deleted in the TGEV-Rep constructs (Fig. 1b). Taken together, these data demonstrate the synthesis of subgenomic mRNA and heterologous gene expression from the TGEV-Rep(AvrII) and TGEV-Rep(EcoNI) subgenomic replicon RNAs.
FIG. 7.
Sequence analysis of leader-containing amplicons encoding GFP. TGEV-Rep(AvrII) and TGEV-Rep(EcoNI) leader-containing amplicons were isolated from agarose gels and subcloned into Topo II TA cloning vectors. Inserts were sequenced using universal primers and an automated sequencer. (A) Sequence of the 5′ end of GFP amplicons generated from TGEV-Rep(EcoNI)-transfected cells, indicating that the leader-containing transcripts initiated from the TGEV ORF 3A IS. (B) Leader-containing GFP transcripts with the PflMI-AvrII deletion derived from TGEV-Rep(AvrII) VRP-infected cells. (C) Leader-containing GFP transcripts with the PflMI-EcoNI deletion derived from TGEV-Rep(EcoNI)-transfected cells. Underlined bases correspond to the GFP stop codon, and the shaded bases correspond to the deletion and blunt-end ligation sites.
TGEV-Rep(AvrII) lacks all of ORF 3B and a portion of the E gene and therefore should not produce infectious virions. Successful assembly of infectious TGEV from this replicon should be prevented on at least two levels. First, the E gene IS and flanking sequences have been deleted in this replicon, which should preclude the synthesis of E gene subgenomic mRNA transcripts. Secondly, the E gene start codon has been deleted, and the next possible ATG start codon is out of the E gene reading frame at nucleotide position 25888 and would potentially encode an irrelevant 14-amino-acid protein. However, one possible in-frame E gene start codon is located 33 bp downstream of the PflMI-AvrII deletion, at nucleotide position 25899, and expression from this site would result in a truncated E protein, with about a 17% (14 of 83 amino acids) deletion from the N terminus, including residues within a putative membrane anchor. Although unlikely, the expression of a biologically active, truncated E protein may result via read-through from other TGEV mRNAs or from cryptic IS sites that drive expression of a subgenomic mRNA encoding the E protein, allowing for the assembly of infectious virions. We think that this is unlikely, as cryptic subgenomic leader-containing E transcripts were not detected by RT-PCR that would encode this E protein truncation. To address whether small amounts of truncated E are produced which function in virus assembly and release, aliquots of cell culture supernatants were harvested ∼36 h postelectroporation and passed onto fresh cultures of ST cells (pass 1). By RT-PCR, there was no evidence of virus replication. In addition, virus-induced cytopathology and GFP expression were not apparent in these cultures (data not shown).
Packaging of TGEV replicon RNA in virus particles.
Previous data indicate that the PflMI-AvrII deletion prevented E protein function and the assembly of infectious virus. Consequently, E protein provided in trans should complement the E gene deletion and result in infectious TGEV VRPs. The VEE replicon system has been used previously for the high-level expression of a number of heterologous genes (3, 11, 34, 58, 59, 67) and was used as an efficient means for expressing the TGEV E protein in trans. We hypothesized that VEE VRPs expressing TGEV E would supply sufficient concentrations of E protein in trans to allow for efficient assembly and release of packaged TGEV-Rep(AvrII) VRPs (Fig. 4a). To determine the effect of VEE VRPs on TGEV replication, cultures of ST cells were either infected with wild-type TGEV alone or coinfected with VEE VRPs containing a G1 VEE-NCFL capsid gene (33) and wild-type TGEV. Progeny TGEV virions were harvested at different times postinfection and quantified by plaque assay in ST cells (Fig. 4b). Clearly, the TGEV growth rate was not adversely affected by coinfection with VEE VRPs. Similar results have been shown with another alphavirus, Sindbis virus, and the murine coronavirus mouse hepatitis virus (MHV) (7).
FIG. 4.
Strategy to assemble TGEV-Rep(AvrII) VRPs. (a) In the full-length TGEV-Rep(AvrII) cDNA construct, ORF 3A has been replaced with GFP, and ORF 3B and the 5′ end of the E gene have been deleted. To produce packaged replicon particles, replicon RNA-transfected cells were infected with VEE VRPs expressing the TGEV E protein [VEE-TGEV(E)]. Alternatively, TGEV-Rep(AvrII) replicon RNAs can be coelectroporated with pVR21-E1-derived transcripts. TGEV VRPs should be released from cells that can be used as single-hit expression vectors. (b) Cultures of ST cells were infected with either wild-type (wt) TGEV alone or with TGEV and VEE VRPs expressing a G1 Norwalk-like virus capsid (wt + VEE) at an MOI of 5 for 1 h at room temperature. The inocula were removed, and the cultures were incubated in complete medium at 37°C. Samples were harvested at the indicated times and assayed by plaque assay in ST cells.
The TGEV E gene was inserted into the VEE replicon vector pVR21, just downstream of the subgenomic 26S promoter. The resulting construct, designated pVR21-E1, was subsequently used for the production of VEE VRPs expressing the TGEV E protein [VEE-TGEV(E)]. Capped runoff TGEV-Rep(AvrII) transcripts were electroporated into 8 × 106 BHK cells (pass 0), seeded onto cultures of ST cells in 75-cm2 flasks and then infected with VEE-TGEV(E) VRPs. Alternatively, BHK cells were coelectroporated with TGEV-Rep(AvrII)- and pVR21-E1-derived transcripts. In both experiments, GFP expression was evident by fluorescent microscopy by ∼18 h posttransfection, demonstrating the subgenomic transcription and heterologous gene expression from the TGEV-Rep(AvrII) genome in the presence of VEE replicon RNAs (Fig. 5). Cell culture supernatants were harvested ∼36 h posttransfection and used to inoculate fresh cultures of ST cells (pass 1) to determine if the TGEV-Rep(AvrII) replicon RNA had been packaged into TGEV VRPs. By ∼18 h postinfection, GFP expression was observed in these pass 1 cultures (Fig. 5), confirming that replicon RNAs had been packaged into infectious TGEV VRPs. However, TGEV VRP titers were low, estimated to be 103 to 105 infectious units/ml by fluorescent microscopy, depending on the experiment.
FIG. 5.
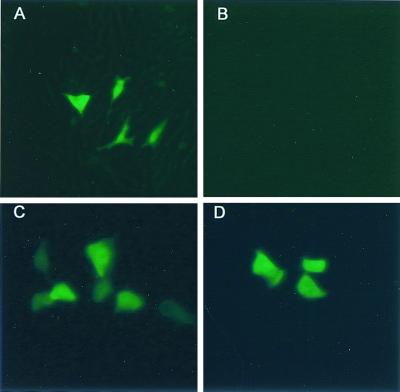
TGEV-Rep(AvrII) VRPs. Transfected cultures displayed GFP expression at ∼18 h posttransfection (A) [TGEV-Rep(AvrII) pass 0]. Supernatants were harvested ∼36 h posttransfection and used to infect fresh cultures of ST cells for 1 h at room temperature. At ∼18 h postinfection, GFP expression was observed by fluorescent microscopy in these pass 1 cultures (C and D). (C) TGEV VRP production following coelectroporation with VEE-E RNAs [TGEV-Rep(AvrII) plus E protein transcripts pass 1]; (D) TGEV VRP production following infection with VEE VRPs encoding the E protein [TGEV-Rep(AvrII) plus VEE-TGEV(E) pass 1]. Passage of supernatants from cells transfected with TGEV-Rep(AvrII) transcripts without expression of the E protein in trans did not result in detectable GFP expression (B) [TGEV-Rep(AvrII) without E protein pass 1].
TGEV VRPs should express leader-containing subgenomic mRNAs encoding GFP and the various downstream ORFs, including M and N. Following TGEV VRP infection, intracellular RNA was isolated and subjected to RT-PCR by using primer pairs in the leader RNA and downstream of the GFP, M, and N genes. As in the previous TGEV-Rep(AvrII) experiments, a leader-containing GFP amplicon of ∼850 bp was generated (Fig. 6a) and sequenced to confirm the presence of leader-containing GFP transcripts with the PflMI-AvrII deletion (Fig. 7b). Leader-containing subgenomic transcripts were also detected that contained the TGEV M and N genes, ∼900 bp and ∼1.2 kb, respectively (Fig. 6b), demonstrating that transcripts for at least two of the structural genes were expressed in TGEV VRP-infected cells. Larger leader-containing amplicons were also observed and likely corresponded to cryptic start sites noted within GFP as well as the larger GFP leader-containing amplicons (Fig. 6b). These data demonstrate the replication competence and heterologous gene expression from packaged TGEV-Rep(AvrII) RNAs.
FIG. 6.
Leader-containing subgenomic transcripts are present in cultures infected with TGEV VRPs. Intracellular RNA was isolated from cell cultures infected with pass 0 supernatants and used as a template for RT-PCR to detect leader-containing GFP, M, and N subgenomic transcripts using a 5′ leader primer (TGEV-L) and 3′ primers located just downstream of each respective gene. (A) An appropriately sized amplicon of ∼850 bp was generated (indicated by an arrow), corresponding to leader-containing transcripts encoding GFP (lane 1). Lane 2 shows a 1-kb ladder. (B) In addition, amplicons of ∼900 bp and ∼1.2 kb were generated, corresponding to leader-containing M and N gene mRNA transcripts, respectively. The arrows correspond to leader-containing M (lane 2) or N (lane 3) transcripts. A 1-kb ladder is shown in lane 1.
TGEV replicon particles function as single-hit virus vectors.
An important aspect of viral replicon particle systems, in terms of future use as an expression vector for vaccine development, is the lack of recombinant virus production. It is possible that mutations may evolve which restore E protein expression and function or recombinant TGEVs emerge following mixed TGEV-Rep(AvrII) and VEE-TGEV(E) infection. To conclusively demonstrate the lack of recombinant virus production from the E deletion replicon RNA, ST cells were infected with concentrated TGEV VRPs and examined for GFP expression as well as virus production by plaque assay over a 72-h time period (Fig. 8). Under identical conditions, parallel ST cell cultures were infected with pass 0 supernatant (obtained from transfected cell cultures) containing TGEV-GFP2. Expansion of GFP expression was clearly observed in TGEV-GFP2-infected cells (Fig. 8a, panels A to E) while no expansion was noted in TGEV VRP-infected cells (Fig. 8a, panels F to J). In fact, GFP expression in these TGEV VRP-infected cells eventually decreased after the 24-h time point and eventually disappeared. Additionally, infectious TGEV particles were not detected by plaque assay in TGEV VRP-infected cultures during this same 72-h period (Fig. 8b), while infectious TGEV-GFP2 virus rapidly reached titers of 2 × 106 PFU/ml by 48 h postinfection under identical conditions. These data clearly demonstrate the lack of revertant wild-type and recombinant virus production from the TGEV-Rep(AvrII) VRP stocks.
FIG. 8.
TGEV replicon particles function as single-hit virus vectors. Cultures of ST cells were infected for 1 h at room temperature with either concentrated TGEV VRPs or supernatant from TGEV-GFP2-transfected cells. (a) Cultures were then examined over a 72-h time period for GFP expression by fluorescent microscopy. Expansion of GFP expression was observed in TGEV-GFP2-infected cells (panels A to E) while no expansion was noted in TGEV VRP-infected cells (panels F to J). (b) In addition, supernatants were harvested from the TGEV-GFP2- and TGEV VRP-infected cultures and assayed by plaque assay in ST cells. Symbols: ⧫, TGEV-GFP2; ▪, TGEV-VRP.
DISCUSSION
In this report, we describe the assembly of recombinant TGEVs and replicons that express GFP, which should prove useful in furthering our understanding of coronavirus biology. The successful expression of heterologous genes from recombinant TGEVs and VRPs may also facilitate future vaccine development against homologous and heterologous pathogens (1, 3, 9, 10, 19–21, 34, 39, 41, 42, 47, 56–58, 67, 74, 75). Importantly, the use of replicons as a vaccine delivery system offers a number of important advantages over the use of live attenuated virus vaccines, which are capable of independent spread and recombination with wild-type virus populations. Replicon vectors are an inherently safer alternative to the use of live attenuated virus vaccines due to the lack of progeny virus production. In addition, high-level expression of heterologous genes should result in the use of a relatively low dose of VRPs for vaccination and immune induction.
TGEV vectors should provide an optional system for the incorporation and expression of one or more foreign genes, as coronaviruses contain a polycistronic genome organization and synthesize multiple subgenomic-length mRNAs (25). It has been shown that TGEV ORFs 3A and 3B likely encode luxury functions that can be deleted without affecting infectivity or replication in vitro and may serve as appropriate sites for the insertion of heterologous genes into the TGEV genome (25, 46, 49, 76, 78). In contrast to arteriviruses, the TGEV IS sites rarely overlap (or overlap slightly) with upstream ORFs, simplifying the design and expression of foreign genes from downstream IS sites (14, 22, 73). The helical nucleocapsid structure of TGEV will also likely minimize packaging constraints and allow for the expression of multiple large genes from a single construct (25, 45, 61). Importantly, recombinant TGEV VRPs could be readily targeted to other mucosal surfaces in swine or to other species by simple replacements in the S glycoprotein gene, which has been shown to determine tissue and species tropism (16, 44, 63, 72). For these reasons, TGEV VRPs may provide a novel approach for the development of combination vaccines in a variety of mammalian hosts.
We have successfully inserted and expressed GFP from the TGEV genome, demonstrating the feasibility of using TGEV-based replicon vectors for heterologous gene expression. Efficient self-replication of in vitro-transcribed recombinant TGEV and replicon RNAs was demonstrated by GFP expression, the presence of leader-containing subgenomic transcripts, and the production of infectious recombinant virus and VRPs. These data support previous results suggesting that the TGEV ORFs 3A and 3B were not required for virus replication in vitro, although these genes may confer subtle fitness advantages that cannot be detected by these assays (46, 49, 78). It should also be possible to determine the minimal TGEV replicon size by serially deleting each of the downstream ORFs. However, this may be complicated by the requirement of RNA secondary structures that may be essential for genome replication or subgenomic RNA synthesis (36, 37, 79). As has been previously reported, efficient TGEV RNA transfection and expression in cells were dependent upon the coelectroporation of N transcripts (78). Because the nucleocapsid protein interacts with leader and negative-strand RNA and colocalizes with the viral polymerase sites of RNA synthesis, it is possible that the N protein may function as part of the transcription complex in some undetermined manner (6, 17). Replicon constructs containing N gene deletions may facilitate studies of the possible N gene function(s) in TGEV replication.
Using synthetic defective-interfering (DI) RNA genomes, some groups have reported that downstream ISs suppress transcription from upstream ISs (40, 43). In addition, data suggest that the N gene IS is the strongest initiator of TGEV subgenomic RNA transcription (35, 38). In our recombinant virus and replicon RNAs, GFP subgenomic mRNA synthesis was initiated from the normal ORF 3A IS rather than from the 20-nt N gene IS that has been duplicated just upstream of gfp (Fig. 7a). Our results do not necessarily contradict earlier reports, as in this context the N gene IS function was silent and would not display the reported phenotypes. Also, fundamental differences exist between the two systems used in these analyses (DI versus nearly full-length replicon), including the rapid replication of small ∼2- to 3-kb DI RNAs and IS presentation, compared with 28.5-kb genome-length RNAs. For example, experiments utilizing DI systems involved IS elements within primary and secondary flanking genome sequence contexts that were not authentic, while the ISs in our recombinant viruses and replicons closely approximate the wild-type TGEV genome. Our data suggest that IS location and flanking sequences likely have an impact on gene expression, especially in promoter proximal locations, which are fundamentally different in the two systems. The simplest interpretation of our data is that the random insertion of a large ∼20-nt IS element is not sufficient to initiate TGEV subgenomic mRNA synthesis unless this IS is provided to the viral transcriptional machinery in an appropriate context that as of yet remains unknown. Our data, as well as that obtained concerning bovine coronavirus (54), lead us to hypothesize that TGEV subgenomic RNA transcription may be mediated by long-range RNA and/or ribonucleoprotein interactions, which are most definitely dependent on higher orders of genome structure.
We noted smaller leader-containing RNAs, indicating the presence of cryptic transcription start sites within GFP. This phenomenon was previously observed following expression of GFP from the MHV genome (26, 65). Both the TGEV and MHV genomes contain a number of atypical start sites that result in the transcription of aberrant subgenomic RNAs (data not shown) (26). These data further substantiate our conclusion that the insertion of IS elements may not be enough to direct TGEV subgenomic mRNA synthesis and that genome location, flanking sequence, and secondary sequence likely function in this process.
The VEE, Kunjin, and Sindbis replicon packaging systems involve the cotransfection of replicon and helper RNAs that express the structural genes (59). In addition to this method, replicon RNAs have been packaged by the expression of structural genes in trans from wild-type or mutated virus (53, 56). An important concern with these types of replicon systems is the production of recombinant virus, especially when considering the development of a replicon particle vaccine. Recombinant viruses have been isolated from Sindbis virus and VEE virus replicon systems that may be the result of recombination between replicon and helper RNAs and/or copackaging of replicon and helper RNAs into the same VRP (28, 30, 59). Coronaviruses undergo homologous recombination at high frequencies during mixed infection, which presents a significant concern for the production and use of TGEV replicons (5, 29, 50). Our TGEV replicon packaging system involves the use of an unrelated virus vector (VEE). In our system, TGEV-Rep(AvrII) RNAs were packaged by the expression of TGEV E in trans from VEE VRPs as well as from pVR21-E1-derived helper RNA transcripts. Because the TGEV replicon packaging system involves the use of an unrelated virus vector (VEE), the possibility of a recombination event between replicon and helper RNAs may be reduced. In addition, the TGEV E gene carried in the VEE replicon construct lacks an appropriate IS, which is necessary for the initiation of subgenomic RNA transcription. As this same IS is deleted in the TGEV-Rep(AvrII) construct, expression of TGEV helper genes that are recombined into TGEV replicon RNAs should be minimized in our system. Although recombinant wild-type TGEV was not detected, this concern could also be reduced by the inclusion of attenuating mutations in the helper proteins and/or the engineering of a bipartite replicon and helper system (TGEV E and M structural proteins) (59). Nevertheless, the possibility of RNA recombination between the replicon RNA and wild-type virus cannot be eliminated and remains an important issue requiring additional analysis.
Another advantage to the use of VEE replicon vectors to express TGEV structural genes is the possibility of developing combination vaccines. Dendritic cells, which are professional antigen-presenting cells and potent inducers of T-cell responses to viral antigens, are preferred targets of VEE and VRP infection, while TGEV targets the mucosal surfaces of the respiratory and gastrointestinal tract (4, 23, 25, 48). As the VEE and TGEV replicon RNAs synergistically interact, two-vector vaccine systems are feasible that may result in increased immunogenicity when compared with either vector alone. Combination prime-boost vaccines (e.g., DNA immunization and vaccinia virus vectors) have dramatically enhanced the immune response (notably cellular responses) against target papillomavirus and lentivirus antigens compared to single-immunization regimens (13, 31, 32, 55). Using different recombinant viral vectors (influenza and vaccinia) to prime and boost may also synergistically enhance the immune response, sometimes by an order of magnitude or more (31).
Our strategy for the assembly of TGEV replicon constructs was based on the use of six cDNA subclones that span the entire length of the TGEV genome, designated fragments A to F, which can be systematically ligated in vitro into a full-length cDNA (80). This strategy is capable of circumventing problems associated with genome size constraints as well as regions of chromosomal instability, while allowing for simple reverse genetic applications. Full-length TGEV cDNA constructs must be synthesized de novo and do not exist intact in bacterial vectors, circumventing problems with sequence instability. However, this did not restrict the applicability of our approach. In fact, the separation of the replicon constructs into distinct fragments allows for genetic manipulation of independent subclones, thereby minimizing the occurrence of spurious mutations that arise during recombinant DNA manipulation. Because replicon cDNAs are consumed during in vitro transcription in this strategy, a weakness of our approach is that the full-length replicon cDNAs are consumed with use and must be continually rebuilt. However, replicon cDNAs will likely be stable in bacterial artificial chromosome vectors after reverse genetic manipulations, preventing the need for repetitive de novo synthesis by engineered constructs (2, 51).
The synthesis of large RNA transcripts (∼27 to 29 kb) in vitro is problematic, and the electroporation of such large RNA constructs has also proven difficult, resulting in a <1% transfection efficiency. This low efficiency of transfection will likely limit the production of TGEV VRPs and their use as vaccine vectors. However, transfecting cells with helper packaging constructs and subsequently passing the TGEV VRPs in the presence of VEE-TGEV(E) VRPs may address this issue. In this way, VRPs can be amplified and high concentrations may amplify replicon titers for future applications. In addition, the use of a DNA launch platform, such as with a cytomegalovirus promoter, may overcome the problems associated with our RNA launch system.
In summary, we have developed efficient RNA-based recombinant TGEV and replicon particles capable of expressing GFP in cultured cells. Our data are in agreement with other studies showing that the TGEV E protein is absolutely essential for coronavirus particle assembly (27, 77). This TGEV replicon system allows for the dissociation of RNA replication from virion assembly and should provide a powerful model system to independently address questions of coronavirus RNA synthesis, polymerase function, assembly, and release. In addition, recombinant TGEVs and VRPs may facilitate the production of novel swine candidate vaccines.
Acknowledgments
We are grateful to Robert E. Johnston and Nancy Davis for providing the VEE expression vector pVR21 and for their assistance with the VEE replicon system. In addition, we thank Bob Bagnell for assistance with fluorescent microscopy and Patrick Harrington for helpful discussions during the course of these studies.
This work was supported by research grants from the National Institutes of Health (AI23946 and GM63228).
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Pragai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989–12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasuriya, U. B., H. W. Heidner, J. F. Hedges, J. C. Williams, N. L. Davis, R. E. Johnston, and N. J. MacLachlan. 2000. Expression of the two major envelope proteins of equine arteritis virus as a heterodimer is necessary for induction of neutralizing antibodies in mice immunized with recombinant Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:10623–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245–252. [DOI] [PubMed] [Google Scholar]
- 5.Baric, R. S., K. Fu, M. C. Schaad, and S. A. Stohlman. 1990. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology 177:646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baric, R. S., E. Sullivan, L. Hensley, B. Yount, and W. Chen. 1999. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73:638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund, P., C. Smerdou, M. N. Fleeton, I. Tubulekas, and P. Liljestrom. 1998. Enhancing immune responses using suicidal DNA vaccines. Nat. Biotechnol. 16:562–565. [DOI] [PubMed] [Google Scholar]
- 10.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caley, I. J., M. R. Betts, D. M. Irlbeck, N. L. Davis, R. Swanstrom, J. A. Frelinger, and R. E. Johnston. 1997. Humoral, mucosal, and cellular immunity in response to a human immunodeficiency virus type 1 immunogen expressed by a Venezuelan equine encephalitis virus vaccine vector. J. Virol. 71:3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavanagh, D., and M. C. Horzinek. 1993. Genus Torovirus assigned to the Coronaviridae. Arch. Virol. 128:395–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C., T. Wang, C. Hung, D. M. Pardoll, and T. Wu. 2000. Boosting with recombinant vaccinia increases HPV-16 E7-specific T cell precursor frequencies of HPV-16 E7-expressing DNA vaccines. Vaccine 18:2015–2022. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. M., D. Cavanagh, and P. Britton. 1995. Cloning and sequencing of a 8.4-kb region from the 3′-end of a Taiwanese virulent isolate of the coronavirus transmissible gastroenteritis virus. Virus Res. 38:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, N. L., I. J. Caley, K. W. Brown, M. R. Betts, D. M. Irlbeck, K. M. McGrath, M. J. Connell, D. C. Montefiori, J. A. Frelinger, R. Swanstrom, P. R. Johnson, and R. E. Johnston. 2000. Vaccination of macaques against pathogenic simian immunodeficiency virus with Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delmas, B., J. Gelfi, R. L’Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denison, M. R., W. J. M. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. I. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiCiommo, D. P., and R. Bremner. 1998. Rapid, high level protein production using DNA-based Semliki Forest virus vectors. J. Biol. Chem. 273:18060–18066. [DOI] [PubMed] [Google Scholar]
- 20.Dollenmaier, G., S. M. Mosier, F. Scholle, N. Sharma, K. L. McKnight, and S. M. Lemon. 2001. Membrane-associated respiratory syncytial virus f protein expressed from a human rhinovirus type 14 vector is immunogenic. Virology 281:216–230. [DOI] [PubMed] [Google Scholar]
- 21.Dubensky, T. W., D. A. Driver, J. M. Polo, B. A. Belli, E. M. Latham, C. E. Ibanez, S. Chada, D. Brumm, T. A. Banks, S. J. Mento, D. J. Jolly, and S. M. Chang. 1996. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J. Virol. 70:508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eleouet, J. F., D. Rasschaert, P. Lambert, L. Levy, P. Vende, and H. Laude. 1995. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology 206:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enjuanes, L., C. Sanchez, A. Mendez, and M. L. Ballesteros. 1995. Tropism and immunoprotection in transmissible gastroenteritis coronaviruses. Dev. Biol. Stand. 84:145–152. [PubMed] [Google Scholar]
- 24.Enjuanes, L., C. Smerdou, J. Castilla, I. M. Anton, J. M. Torres, I. Sola, J. Golvano, J. M. Sanchez, and B. Pintado. 1995. Development of protection against coronavirus induced diseases. A review. Adv. Exp. Med. Biol. 380:197–211. [DOI] [PubMed] [Google Scholar]
- 25.Enjuanes, L., and B. A. M. van der Zeijst. 1995. Molecular basis of transmissible gastroenteritis coronavirus (TGEV) epidemiology, p.337–376. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 26.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu, K., and R. S. Baric. 1992. Evidence for variable rates of recombination in the MHV genome. Virology 189:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geigenmuller-Gnirke, U., B. Weiss, R. Wright, and S. Schlesinger. 1991. Complementation between Sindbis viral RNAs produces infectious particles with a bipartite genome. Proc. Natl. Acad. Sci. USA 88:3253–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalo, R. M., D. Rodriguez, A. Garcia-Sastre, J. R. Rodriguez, P. Palese, and M. Esteban. 1999. Enhanced CD8+ T cell response to HIV-1 env by combined immunization with influenza and vaccinia virus recombinants. Vaccine 17:887–892. [DOI] [PubMed] [Google Scholar]
- 32.Hanke, T., T. J. Blanchard, J. Schneider, C. M. Hannan, M. Becker, S. C. Gilbert, A. V. Hill, G. L. Smith, and A. McMichael. 1998. Enhancement of MHC class 1-restricted peptide specific T cell induction by a DNA prime/MVA boost vaccination regime. Vaccine 16:439–445. [DOI] [PubMed] [Google Scholar]
- 33.Harrington, P. R., B. Yount, R. E. Johnston, N. Davis, C. Moe, and R. S. Baric. 2002. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 76:730–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hevey, M., D. Negley, P. Pushko, J. Smith, and A. Schmaljohn. 1998. Marburg virus vaccines based upon alphavirus replicons protect guinea pigs and nonhuman primates. Virology 251:28–37. [DOI] [PubMed] [Google Scholar]
- 35.Hiscox, J. A., D. Cavanagh, and P. Britton. 1995. Quantification of individual subgenomic mRNA species during replication of the coronavirus transmissible gastroenteritis virus. Virus Res. 36:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs, L., B. A. M. van der Zeijst, and M. C. Horzinek. 1986. Characterization and translation of transmissible gastroenteritis virus mRNAs. J. Virol. 57:1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johanning, F. W., R. M. Conry, A. F. LoBuglio, M. Wright, L. A. Sumerel, M. J. Pike, and D. T. Curiel. 1995. A Sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic Acids Res. 23:1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khromykh, A. A. 2000. Replicon-based vectors of positive strand RNA viruses. Curr. Opin. Mol. Ther. 2:555–569. [PubMed] [Google Scholar]
- 42.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laude, H., D. Rasschaert, B. Delmas, M. Godet, J. Gelfi, and B. Charley. 1990. Molecular biology of transmissible gastroenteritis virus. Vet. Microbiol. 23:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356–1361. [DOI] [PubMed] [Google Scholar]
- 48.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGoldrick, A., J. P. Lowings, and D. J. Paton. 1999. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 144:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendez, A., C. Smerdou, A. Izeta, F. Gebauer, and L. Enjuanes. 1996. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology 217:495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pancholi, P., Q. Liu, N. Tricoche, P. Zhang, M. E. Perkus, and A. M. Prince. 2000. DNA prime-canarypox boost with polycistronic hepatitis C virus (HCV) genes generates potent immune responses to HCV structural and nonstructural proteins. J. Infect. Dis. 182:18–27. [DOI] [PubMed] [Google Scholar]
- 56.Percy, N., W. S. Barclay, M. Sullivan, and J. W. Almond. 1992. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J. Virol. 66:5040–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porter, D. C., D. C. Ansardi, W. S. Choi, and C. D. Morrow. 1993. Encapsidation of genetically engineered poliovirus minireplicons which express human immunodeficiency virus type 1 Gag and Pol proteins upon infection. J. Virol. 67:3712–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pushko, P., M. Bray, G. V. Ludwig, M. Parker, A. Schmaljohn, A. Sanchez, P. B. Jahrling, and J. F. Smith. 2000. Recombinant RNA replicons derived from attenuated Venezuelan equine encephalitis virus protect guinea pigs and mice from Ebola hemorrhagic fever virus. Vaccine 19:142–153. [DOI] [PubMed] [Google Scholar]
- 59.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389–401. [DOI] [PubMed] [Google Scholar]
- 60.Rasschaert, D., and H. Laude. 1987. The predicted primary structure of the peplomer protein E2 of the porcine coronavirus transmissible gastroenteritis virus. J. Gen. Virol. 68:1883–1890. [DOI] [PubMed] [Google Scholar]
- 61.Risco, C., I. M. Anton, L. Enjuanes, and J. L. Carrascosa. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 63.Sánchez, C. M., A. Izeta, J. M. Sánchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Durán, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaad, M. C., and R. S. Baric. 1993. Evidence for new transcriptional units encoded at the 3′ end of the mouse hepatitis virus genome. Virology 196:190–198. [DOI] [PubMed] [Google Scholar]
- 66.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schultz-Cherry, S., J. K. Dybing, N. L. Davis, C. Williamson, D. L. Suarez, R. Johnston, and M. L. Perdue. 2000. Influenza virus (A/HK/156/97) hemagglutinin expressed by an alphavirus replicon system protects chickens against lethal infection with Hong Kong-origin H5N1 viruses. Virology 278:55–59. [DOI] [PubMed] [Google Scholar]
- 68.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siddell, S., H. Wege, and V. Ter Meulen. 1983. The biology of coronaviruses. J. Gen. Virol. 64:761–776. [DOI] [PubMed] [Google Scholar]
- 71.Snijder, E. J., and M. C. Horzinek. 1993. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 74:2305–2316. [DOI] [PubMed] [Google Scholar]
- 72.Tresnan, D. B., R. Levis, and K. V. Holmes. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 70:8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tung, F. Y., S. Abraham, M. Sethna, S. L. Hung, P. Sethna, B. G. Hogue, and D. A. Brian. 1992. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology 186:676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varnavski, A. N., and A. A. Khromykh. 1999. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology 255:366–375. [DOI] [PubMed] [Google Scholar]
- 75.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaughn, E. M., P. G. Halbur, and P. S. Paul. 1995. Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J. Virol. 69:3176–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wesley, R. D., R. D. Woods, and A. K. Cheung. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 65:3369–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams, G. D., R. Y. Chang, and D. A. Brian. 1999. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 73:8349–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]