Abstract
Human cytomegalovirus (HCMV) has a coding capacity for glycoproteins which far exceeds that of other herpesviruses. Few of these proteins have been characterized. We have investigated the gene product(s) of reading frame 10, which is present in both the internal and terminal repeat regions of HCMV strain AD169 and only once in clinical isolates. The putative protein product is a 171-amino-acid glycoprotein with a theoretical mass of 20.5 kDa. We characterized the protein encoded by this reading frame in the laboratory strain AD169 and a recent isolate, TB40E. The results from both strains were comparable. Northern blot analyses showed that the gene was transcribed with early/late kinetics. Two proteins of 22 and 23.5-kDa were detected in virus-infected cells and in cells transiently expressing recombinant TRL10. Both forms contained only high-mannose-linked carbohydrate modifications. In addition, virus-infected cells expressed small amounts of the protein modified with complex N-linked sugars. Image analysis localized transiently expressed TRL10 to the endoplasmic reticulum. Immunoblot analyses as well as immunoelectron microscopy of purified virions demonstrated that TRL10 represents a structural component of the virus particle. Immunoblot analysis in the absence of reducing agents indicated that TRL10, like the other HCMV envelope glycoproteins, is present in a disulfide-linked complex. Sequence analysis of the TRL10 coding region in nine low-passage clinical isolates revealed strain-specific variation. In summary, the protein product of the TRL10 open reading frame represents a novel structural glycoprotein of HCMV and was termed gpTRL10.
Human cytomegalovirus (HCMV), the largest of the human herpesviruses, has a glycoprotein coding capacity which far exceeds that of other herpesviruses. The genome of the laboratory-adapted strain AD169 encodes at least 57 open reading frames (ORFs) with predicted characteristics of glycoproteins or glycoprotein exons (8). Low-passage clinical isolates contain an additional 13 ORFs that could potentially code for glycoproteins (5). The proteins are likely to have important functions in replication or pathogenicity of HCMV. Remarkably, fewer than 10 glycoprotein gene products have been characterized with respect to biosynthesis, function, and incorporation into the virion.
The envelope glycoproteins of herpesviruses have multiple roles in the viral replication cycle, including essential functions such as attachment, penetration, cell-to-cell spread, envelopment, and maturation of nascent virus particles (29). To understand these processes, it is necessary to have detailed knowledge of the structure and function of envelope glycoproteins. In addition, envelope glycoproteins can elicit a protective virus-neutralizing humoral immune response and thus have been proposed as important components of vaccines.
Among the HCMV structural glycoproteins, three major complexes (designated gCI to -III) have been identified to date (13). The gCI complex is formed by disulfide-linked homodimers of glycoprotein B (gB; also called gpUL55) (2, 3). The gCII complex is composed of gM (IMP; also called gpUL100) and gN (gpUL73), and large parts of this complex are also held together by disulfide bonds (28). Glycoproteins H (gH; also called gpUL75), L (gL; also called gpUL115) and O (gO; also called gpUL74), the constituents of gCIII, form a heterotrimeric, disulfide-linked complex (17, 18, 26). Thus, a hallmark of the structural glycoproteins of HCMV seems to be their formation of disulfide-linked high-molecular-weight complexes. Proteins within the gCI to -III complexes represent a group of structural glycoproteins which are conserved within the herpesvirus family, indicating important or even essential functions for the replication of these viruses (4, 29, 33).
In addition to the glycoproteins, such as gB, gH, gL, and gM, which have identifiable protein counterparts in all herpesviruses (family common), herpesviruses also code for a number of glycoproteins which are expressed only by individual genera of a herpesvirus subfamily (29). These “private” glycoproteins could contribute to the specific properties of individual herpesviruses, such as cell tropism or pathogenicity. For HCMV, very little is known about private structural glycoproteins. To our knowledge, the only reported structural glycoprotein that is not conserved throughout the herpesvirus family is gp48, the product of the UL4 reading frame (7). Interest in this glycoprotein has been limited almost exclusively to the complex transcriptional regulation of its expression (9, 15, 16, 36). Based on the large coding capacity of HCMV for glycoproteins, it can be postulated that HCMV virions will contain a number of glycoproteins which do not have counterparts in other herpesviruses.
We have studied the gene product(s) of reading frame 10, located within the repeat sequences flanking the long unique segment of the viral genome (terminal repeat long [TRL] and internal repeat long [IRL]). In the laboratory-adapted HCMV strain AD169, the reading frame is present twice and termed TRL10 and IRL10, respectively (8). In contrast, low-passage clinical isolates contain only the TRL10 reading frame, since large parts of IRL, including IRL10, are replaced by unique coding sequences (5).
Our data reveal that ORFs IRL10 and TRL10 of HCMV strain AD169 as well as TRL10 of the clinical isolate TB40E are transcribed with early/late kinetics. Using a rabbit antiserum, glycosylated proteins of 22 and 23.5-kDa were detected in cells infected with either virus. The proteins were also detected in purified HCMV virions by immunoblotting as well as immunoelectron microscopy. These results indicated that the product of the TRL10 reading frame was a structural protein. Analysis of the protein in the presence and absence of reducing agents indicated that TRL10, like the other previously described HCMV envelope glycoproteins, forms higher-molecular-weight disulfide-linked complexes.
MATERIALS AND METHODS
Cells and viruses.
HCMV strains AD169 and TB40E were propagated in primary human foreskin fibroblasts grown in minimal essential medium (Gibco-BRL, Glasgow, Scotland) supplemented with 5% fetal calf serum (FCS), glutamine (100 mg/liter), and gentamicin (350 mg/liter). Virions were isolated by glycerol-tartrate gradient centrifugation as described (39). 293T cells were cultured in Dulbecco‘s modified Eagle‘s medium (DMEM) (Gibco-BRL) supplemented with 10% FCS, glutamine, gentamicin, and 50 μg of G418 (Geneticin; Gibco-BRL) per ml. Cos7 cells were passaged in DMEM supplemented with 10% FCS, glutamine, and gentamicin. For immunofluorescence, Cos7 cells were grown on 13-mm glass cover slips.
Preparation of GST-TRL10 fusion proteins and antisera.
The TRL10 carboxyl-terminal region was amplified from AD169 DNA by PCR with the following primers containing either BamHI or SalI sites (underlined): TRL10.124/171s (5′-GGACGGATCCACACGCAAAAAGCTGGAACAA-3′ ) and TRL10.124/171as (5′-ATTAGTCGACGACGTTGTCGTCCTCGTCCTC-3′). The PCR product was digested with both endonucleases and inserted into the expression vector pGEX-1P (Pharmacia Biotech, Freiburg, Germany). The resulting plasmid, pGST/TRL10c, contained nucleotides (nt) 370 to 513 of TRL10AD169 fused to the glutathione S-transferase (GST) gene. Correct insertion was confirmed by DNA sequencing.
Plasmid DNA was used to transform Escherichia coli BL21 for expression of GST fusion proteins. The fusion protein was induced and purified from E. coli lysates according to the manufacturer’s instructions. One half of the purified glutathione-Sepharose 4B (Pharmacia Biotech)-coupled fusion protein was treated with PreScission protease (Pharmacia Biotech) to release the viral peptide. A mixture of pGST/TRL10c, the 34-kDa fusion, and pTRL10cut, the 6-kDa cleaved viral peptide, was used to raise an antiserum in rabbits. The resulting serum was affinity purified on the GST/TRL10c fusion protein by standard methods.
Preparation of eukaryotic expression constructs and DNA transfection.
The entire TRL10 reading frame was amplified from AD169 cosmid pCM1052 (11) with the following primers that contain XhoI and HindIII restriction sites (underlined): TRL10 forward (5′-GGATCTCGAGATGTATCCGCGTGTAATG-3′) and TRL10 reverse (5′-ATACAAGCTTGACGTTGTCGTCCTCGTC-3′). The PCR product was cleaved and inserted into the expression vector pcDNA3.1myc/His (Invitrogen, Carlsbad, Calif.). The resulting plasmid, pcTRL10myc/his, encoded full-length pTRL10 fused to a C-terminal Myc/His tag. The integrity of pcTRL10myc/his was confirmed by DNA sequencing.
293T cells were transfected with the respective DNA using Lipofectamine Plus reagent (Gibco-BRL) according to the manufacturer’s suggestion except that the transfection mixture consisted of 1 μg of DNA, 95 μl of DMEM and 6 μl of Lipofectamine Plus reagent. The mixture was added to a cell culture dish (3.5-cm diameter, seeded with 2 × 105 cells 1 day before). After 48 h, cells were harvested, washed three times with phosphate-buffered saline (PBS), and stored at −20°C until used.
Image analysis.
Cos7 cells grown on glass cover slips in 24-well plates were transfected with 3 to 5 μg of plasmid pcTRL10myc/his using calcium chloride as described (35). Two days later, the cover slips were washed and fixed in 2.0% paraformaldehyde in PBS. The fixed cells were permeabilized with NP-40-containing buffer and then blocked with 20% normal goat serum (28). Primary antibodies, including an anti-Myc monoclonal antibody, rabbit anticalreticulin (endoplasmic reticulum [ER] marker), and rabbit anti-gm130 (Golgi marker) were then added. Following a wash, antibody binding was detected with a Texas red-conjugated anti-mouse immunoglobulin G (IgG) or fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (Southern Biotechnology Associates, Birmingham, Ala). The anticalreticulin serum was purchased from Affinity Bioreagents, Boulder, Colo. and the gm130 antiserum was kindly provided by Elizabeth Sztul (University of Alabama, Birmingham, Ala.). Images were collected using a Leitz Dialux fluorescence microscope fitted with a Photometrics charge-coupled device camera. Images were processed using Image Pro software and Adobe Photoshop as previously described (28).
DNA sequence analysis.
To establish the coding sequences for TRL10 in clinical isolates, DNA was extracted from lysates of infected fibroblasts and amplified by PCR using primers TRL10forward and TRL10SEQAS (5′-GACGGCGTTCGATGAACTTCC-3′). Nucleotide sequences were determined on an ABI 377 using the dye terminator sequencing kit according to the manufacturer’s instructions (PE Applied Biosystems, Foster City, Calif.). DNA and amino acid sequence evaluation was performed by use of the University of Wisconsin Genetics Computer Group software.
SDS-PAGE and immunoblotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10 to 15% polyacrylamide gels under standard conditions. Proteins were transferred to nitrocellulose membranes, and membranes were blocked with PBS containing 0.1% Tween 20 and 5% powdered milk. Antibodies and sera were diluted in PBS containing 0.1% Tween 20. For detection of primary antibody binding, horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antibody and the enhanced chemiluminescence detection system (Pharmacia Biotech) were used according to the manufacturer’s instructions. Removal of N-linked oligosaccharides was carried out using recombinant peptide:N-glycosidase F (PNGase F) and endoglycosidase H (Endo H) (New England Biolabs, Beverly, Mass.) according to the manufacturer’s specification.
Membrane topology was investigated via protease digestion. Gradient-purified virions were resuspended in MSB buffer containing 150 mM potassium acetate, 5 mM magnesium acetate, 50 mM HEPES (pH 7.6), and 200 mM sucrose. For solubilization of virions, Triton X-100 was added to a final concentration of 1% (wt/vol). Protease digestion was performed with 20 μg of proteinase K per ml for 2 h at 37°C. All samples were subjected to PAGE and immunoblot analysis immediately after incubation.
RNA methods.
For Northern blot analysis, whole-cell RNA was isolated from noninfected or infected cells at 7, 24, 48, and 72 h postinfection using an RNA isolation kit (Qiagen, Hilden, Germany). For gel electrophoresis, 20 μg of each RNA preparation was denatured with glyoxal and separated in 1% agarose gels containing 40% formaldehyde. Northern blot analysis was carried out according to standard procedures. The TRL10-specific antisense probe was 32P labeled in a single-strand PCR using full-length TRL10 as the template. As an internal control, the filter was hybridized with a glyceraldehyde-3-phosphate dehydrogenase-specific probe.
For reverse transcription (RT)-PCR, infection was blocked with either 150 μg/ml cycloheximide or phosphonoformic acid at a concentration of 185 μg/ml. Total RNA was isolated using the High Pure RNA isolation kit (Roche Diagnostics, Indianapolis, Ind.), and RT-PCR was carried out with the Titan One Tube RT-PCR System (Roche Diagnostics) following the manufacturer‘s specifications.
Immunoelectron microscopy.
For immunoelectron microscopy, purified virions were labeled with an antibody directed against pTRL10 as well as with preimmune sera and a control antibody against gB, monoclonal antibody 27-287. Detection of specific antibody binding was achieved by using protein A-gold. Stability tests at high ionic strength (14) ensured that the gold particles had been quantitatively covered by protein A, thus minimizing the possibility of nonspecific binding. The samples were subjected to labeling prior to fixation with 1% (vol/vol) glutaraldehyde and negative staining with 4% (wt/vol) uranyl acetate following published procedures (14). The specimens, which were mounted on 400-mesh, carbon-coated copper grids, were observed at an accelerating voltage of 80 kV.
Nucleotide sequence accession numbers.
The nucleotide sequences for the TRL10 coding sequences from clinical isolates have been assigned the following accession numbers from GenBank: AF432083, AF432084, AF432085, AF432086, AF432087, AF432088, AF432089, AF432090, AF432091, and AF432092.
RESULTS
Reading frames TRL10 and IRL10.
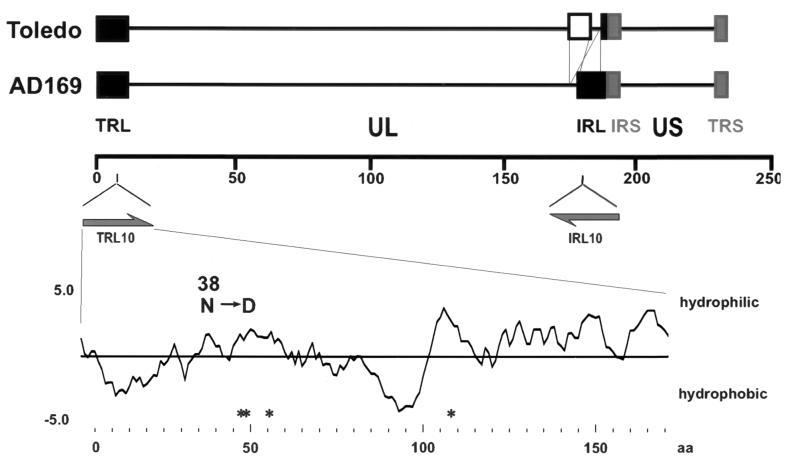
The genome of HCMV strain AD169 contains two large inverted repeats (IRL and TRL) of approximately 12 kb each. These sequences flank the unique long part of the HCMV AD169 genome (Fig. 1). The repeats show nearly complete homology and contain at least 14 ORFs (8). In strain AD169, reading frame 10 (termed TRL10 and IRL10; nucleotides 8101 to 8694 and 180773 to 181336, respectively) codes for a putative glycoprotein of 171 amino acids with a theoretical molecular mass of 20.5 kDa. TRL10/IRL10 is predicted to be a type I glycoprotein containing a cleavable signal sequence between amino acids 1 and 23 and a hydrophobic membrane anchor sequence between amino acids 86 and 102 (Fig. 1) (predicted by PSORTII server [http://psort.nibb.ac.jp/]) (32).
FIG. 1.
Genomic localization of ORFs TRL10/IRL10 and structural predictions of the protein product. The upper part of the figure indicates the localization of TRL/IRL10 within the genome of HCMV strains AD169 and Toledo. The lower part represents a computer prediction of the hydrophilicity and potential N-linked glycosylation sites (*) of the predicted TRL10 protein. The single amino acid divergence at position 38 between TRL10 and IRL10 in strain AD169 is indicated.
According to the nucleotide sequence of HCMV strain AD169, TRL10 and IRL10 are not completely identical. IRL10 differs from TRL10 by a D→N exchange at position 38 (8). Low-passage HCMV isolates, in contrast to the laboratory-adapted strains, have a different genetic organization with respect to the TRL and IRL regions. In these isolates, reading frames IRL14 to IRL1 of the IRL sequence are replaced by a unique sequence containing at least 19 genes not found in the laboratory-adapted strains (5). Thus, in contrast to the laboratory-adapted strains, recent clinical isolates contain only a single TRL10 allele (see Fig. 1).
The presence of two ORFs which are part of repeat sequences in strain AD169 as opposed to a single unique reading frame in clinical isolates poses a problem in nomenclature, since the coding sequence in clinical isolates corresponds to IRL10AD169 rather than TRL10AD169 (see below). Nevertheless, to avoid confusion and to facilitate consistency throughout the paper, we will designate the gene and its product TRL10 and gpTRL10, respectively, being aware that in low-passage isolates TRL10 is not part of repeat sequences and that in AD169 transcripts and protein products cannot be unequivocally assigned to TRL10 or IRL10.
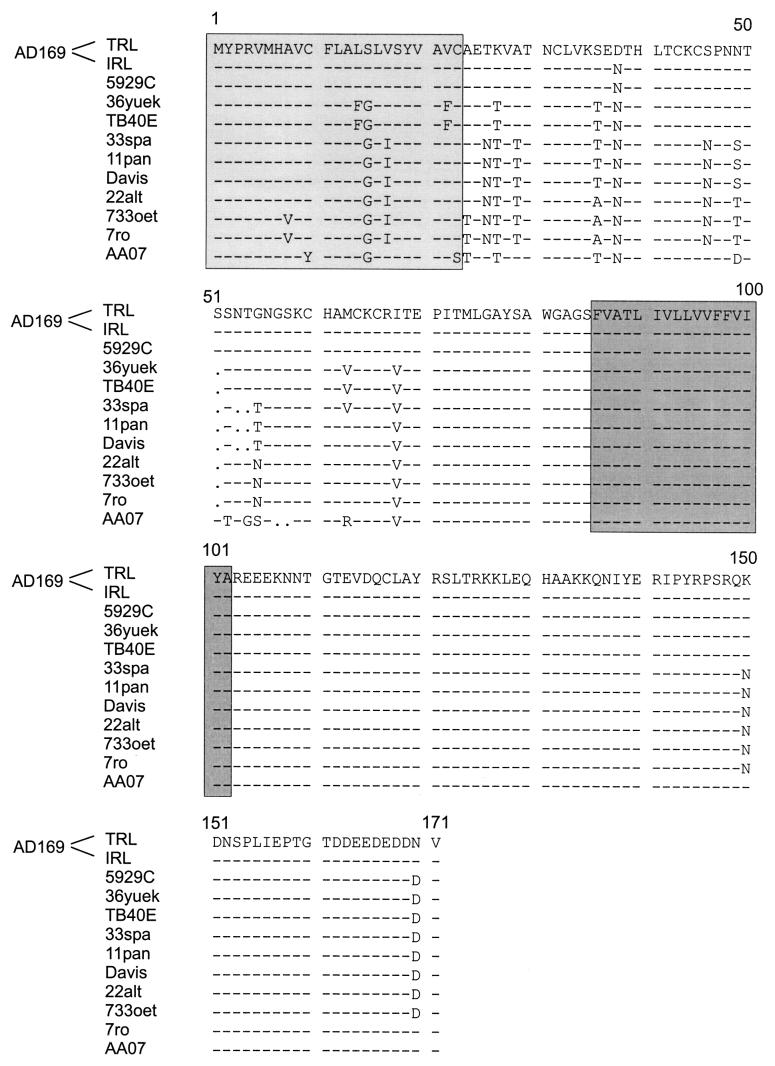
Since we wanted to study the protein product of the TRL10/IRL10 reading frames, we included both the laboratory-adapted strain AD169 and the clinical isolate TB40E (37) in our studies to account for the different number of alleles. To verify that TB40E carried only a single TRL10 coding sequence, the genome was cloned into overlapping cosmids. Partial DNA sequencing revealed that the genome arrangement of TB40E was homologous to that of HCMV strain Toledo. Sequence analysis also confirmed the presence of an ORF in strain TB40E with close homology to TRL10 from strain AD169 (see Fig. 7). It should be noted that TRL10TB40E contained the D→N exchange at position 38 which is characteristic of the IRL10AD169 sequence (see Fig. 7).
FIG. 7.
Alignment of TRL10 amino acid sequences of clinical isolates with AD169 as the reference. Numbers above the sequences represent amino acid residues. Dashes indicate amino acid identity, and dots indicate gaps. Predicted signal and anchor sequences are highlighted in light and dark grey, respectively.
RNA and protein product(s) of the TRL10 gene.
To analyze transcripts originating from the TRL10 ORFs, total RNA from human fibroblasts infected with HCMV strains AD169 and TB40E was isolated at different time points postinfection. The RNA was analyzed in Northern blots using the entire TRL10AD169 antisense sequence as a probe. Signals corresponding to RNA sizes of 4.2 kb, 3.9 kb, and 3.4 kb were detected in samples obtained 48 and 72 h postinfection in AD169-infected cells (data not shown). The signal intensity of the 4.2-kb and 3.4-kb RNAs remained constant in both samples, whereas the signal from the 3.9-kb RNA increased during this interval. The 3.4-kb and 3.9-kb RNA species were also detected in TB40E-infected cells. The kinetics of expression were identical for the 3.9-kb RNA (increasing signal intensities between 48 h and 72 h), whereas the 3.4-kb RNA was already detected at 24 h postinfection.
RT-PCR analysis of TRL10 transcription following treatment of infected cells with cycloheximide or phosphonoformic acid were consistent with classification of the TRL10 transcripts in the early/late kinetic class (data not shown). Thus, although we cannot definitively identify transcripts encoding the TRL10 RNA in either strain, the data are consistent with the interpretation that TRL10 RNA was expressed with early/late kinetics.
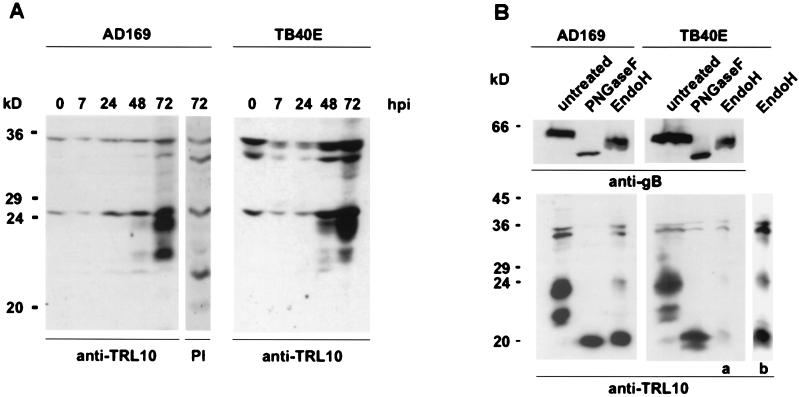
In order to characterize the protein product of ORF TRL10, fibroblasts were infected with AD169 or TB40E, cell lysates from virus-infected cells were prepared at different time points after infection and subjected to immunoblot analysis. The protein was detected using a polyclonal rabbit antiserum raised against a bacterial fusion protein comprising amino acids 124 to 171 of TRL10AD169. Two proteins of 22-kDa and 23.5-kDa molecular mass were detected with the antiserum beginning 48 h after infection (Fig. 2A). These proteins were not detected in uninfected cells or by the preimmune serum. The signal intensity increased considerably between 48 h and 72 h.
FIG. 2.
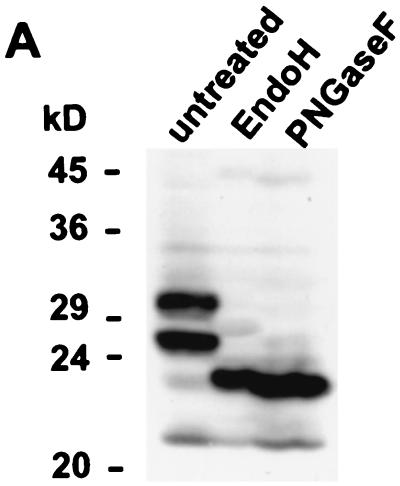
Synthesis and glycosylation of gpTRL10 in infected cells. Human foreskin fibroblasts were infected with HCMV strains AD169 and TB40E. (A) Lysates were prepared at the indicated time points after infection (hours postinfection) and subjected to immunoblotting. pTRL10 was detected using a TRL10-specific rabbit serum (anti-TRL10). The lane designated PI indicates a lysate that was analyzed using the preimmune serum. (B) Infected cells were harvested after 72 h. Cell lysates were digested with either PNGase F or Endo H or left untreated. As shown in the lower part, the membrane was probed with the anti-TRL10 rabbit serum (anti-TRL10) to demonstrate the effect of Endo H on gpTRL10 expressed in TB40E-infected cells. On the extreme right, lane b represents an overexposure of lane a. To control for the efficiency of endoglycosidase digestion, the same blot was stripped and reprobed using monoclonal antibody 27-287 (anti-gB), directed against gp58 of glycoprotein B. This is shown in the upper panel.
In repeated experiments, we observed a difference in the relative abundance of the 22-kDa and the 23.5-kDa proteins expressed in cells infected with the two different virus isolates. In lysates from AD169-infected cells, the proteins appeared to be expressed in nearly equivalent quantities, whereas in lysates from TB40E-infected cells, the 23.5-kDa protein consistently appeared to be overexpressed compared to the 22-kDa protein. In addition, the 22-kDa signal appeared as a doublet in TB40E-infected cells (Fig. 2A).
The predicted primary amino acid sequence of both TRL10AD169 and TRL10TB40E contains two motifs for N-linked glycosylation in the extraluminal portion of the molecule. To determine whether the protein was in fact glycosylated, lysates from infected cells 72 h postinfection were treated with endoglycosidases and subsequently analyzed by immunoblotting. Treatment of lysates from AD169-infected cells with 1PNGase F, which removes complex as well as high-mannose N-linked sugars, resulted in the loss of both the 23.5-kDa and the 22-kDa proteins. Instead a single protein of 20 kDa was detected in these lysates (Fig. 2B).
Treatment with Endo H, which removes only high-mannose carbohydrates, resulted in the nearly complete loss of signal from the 23.5-kDa protein and the disappearance of the signal from the 22-kDa protein. Again, a new species of 20 kDa was detected in these samples. Similar results were obtained with lysates from TB40E-infected cells except that the 22-kDa and 20-kDa signals appeared as doublets (Fig. 2B). The proteins of 32 kDa and 34 kDa that were also detected in this analysis represent nonspecific staining, since they were also detected with the preimmune serum (Fig. 2A). To control for possible proteolytic activity which may have occurred during the enzymatic digestion, blots were stripped and redeveloped using antibody 27-287, which is specific for the gp58 part of glycoprotein B (Fig. 2B). In agreement with published data, we observed an 8-kDa reduction with PNGase F and a 3-kDa reduction with Endo H (3). Partial digests were not seen.
These findings were most consistent with the interpretation that the 22-kDa protein contained exclusively high-mannose N-linked carbohydrates, whereas the signal at 23.5 kDa was produced by two proteins, of which the vast majority contained high-mannose forms, whereas a small fraction was modified by complex sugars. In any case, our results clearly show that the protein product of the TRL10 reading frame was a glycoprotein, and therefore we have designated it gpTRL10.
gpTRL10 is a structural glycoprotein.
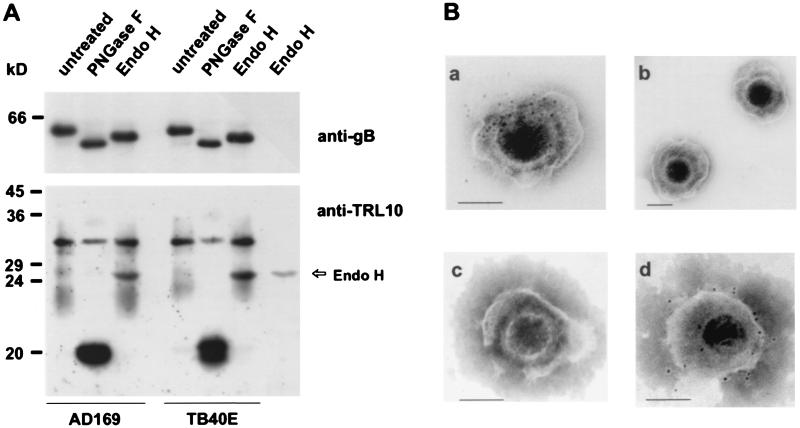
To analyze whether gpTRL10 represents a structural glycoprotein, HCMV virions were purified in glycerol-tartrate gradients and analyzed for the presence of gpTRL10 in immunoblots. A diffusely migrating band at 22 to 34 kDa was detected using the TRL10-specific rabbit antiserum (Fig. 3A). Upon treatment of the viral lysates with PNGase F, the diffusely migrating proteins disappeared and a protein of 20 kDa was generated. Digestion of viral lysates with Endo H did not alter the migration of the virion 22- to 34-kDa protein (Fig. 3A). Identical results were obtained using dense bodies as the antigen (data not shown). The 25-kDa protein detected in the virion lysate after Endo H digestion was also demonstrated in a control lane which contained only Endo H, indicating nonspecific reactivity of the rabbit serum with Endo H (Fig. 3A) These data indicated that gpTRL10 was a virion structural protein which, in contrast to the intracellular forms of this protein, contained only complex N-linked sugars.
FIG. 3.
Detection of gpTRL10 in extracellular virions. (A) Immunoblot analysis of virion proteins. Lysates of extracellular virions of strains AD169 and TB40E were digested with endoglycosidases (PNGase F and Endo H) or left untreated and then subjected to SDS-PAGE and transferred to nylon membranes. Membranes were developed using the anti-TRL10 rabbit serum (anti-TRL10) or the anti-gB monoclonal antibody 27-287. The rightmost lane contains Endo H in an amount equivalent to that used for digestion. The signal at 26 kDa is therefore considered a nonspecific reaction of the rabbit serum with the enzyme. (B) Immunoelectron microscopy. Extracellular virions were incubated with the anti-TRL10 serum (panels a and b), preimmune serum (panel c), or the gB-specific monoclonal antibody 27-287 (panel d) prior to Immunoelectron microscopy analysis. Bound antibodies were detected using protein A-colloidal gold particles. The bar in the lower right-hand corner of each photograph represents 0.1 μm.
In order to directly confirm the virion association of gpTRL10, immunoelectron microscopy was performed. Virions were incubated with the TRL10 rabbit antiserum or preimmune serum, and binding of antibodies was detected using protein A-gold particles (5 nm). The samples were subjected to labeling prior to fixation with glutaraldehyde and negative staining. Virions incubated with preimmune serum were not labeled. Using the TRL10 antiserum, gold particles only labeled virions with damaged envelopes (Fig. 3B). Intact virions were negative (Fig. 3B). In contrast, when a monoclonal antibody against gB was used under the same conditions, staining was observed on intact as well as damaged virions (Fig. 3B). The data are consistent with the fact that the TRL10 antiserum was raised against amino acid sequences which, according to computer predictions, are localized in intraluminal parts of the protein. In intact virions, these epitopes should be buried beneath the virion membrane and therefore not accessible to antibodies.
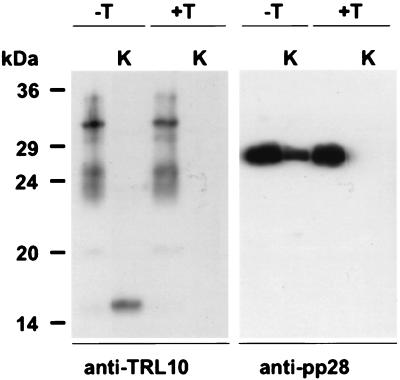
To further define the virion association of gpTRL10, we treated extracellular virions with detergent to disrupt the virion envelope and then digested solubilized virions with proteinase K. Neither a known tegument protein, pp28 (UL99), nor gpTRL10 could be detected in virions which were initially solubilized with detergent and then digested with proteinase K (Fig. 4). In the absence of detergent, the virion pp28 in the tegument remained intact in the face of proteinase K digestion. In contrast, when untreated virions were digested with proteinase K, the forms of gpTRL10 migrating between 22 and 34 kDa were no longer present, but an immunoreactive form of gpTRL10 of approximately 16 kDa was detected (Fig. 4). Because the gpTRL10 antiserum is directed at the carboxyl terminus, this finding provided evidence that gpTRL10 was exposed on the envelope of HCMV and its topology was most consistent with that of a type I glycoprotein.
FIG. 4.
Membrane association of virion gpTRL10. Extracellular virions (AD169) were treated with Triton X-100 (+T) or left untreated (−T). One half of each virus preparation was subsequently digested with proteinase K (K). The preparations were subjected to immunoblot analysis, developed with either anti-TRL10 serum or anti-pp28 monoclonal antibody 41-18 (35).
Recombinant expression of gpTRL10 and intracellular distribution of the protein.
The rabbit serum which was raised against prokaryotically expressed gpTRL10 sequences failed to react with gpTRL10 in infected cells when assayed by immunofluorescence. In order to obtain more information about the intracellular distribution and trafficking of the protein, we constructed the expression plasmid TRL10myc by fusing a Myc/His tag to the carboxyl terminus of the TRL10 open reading frame. Expression of this protein could then be detected using a Myc-specific monoclonal antibody. Following transient transfection of the TRL10myc plasmid into 293T cells, lysates were prepared and analyzed by immunoblotting. Both the Myc-specific antibody and the gpTRL10-specific polyclonal rabbit antisera detected proteins of 26 kDa and 30 kDa (results using an anti-Myc monoclonal antibody are shown in Fig. 5A). Following treatment with either PNGase F or Endo H, a single protein of 23 kDa was observed, indicating that after transient expression gpTRL10myc was modified only by high-mannose carbohydrates (Fig. 5A). The difference in migration between gpTRL10 in infected cells compared to transfected cells was the result of the additional amino acid sequences present in the Myc/His tag (≈3 kDa).
FIG. 5.
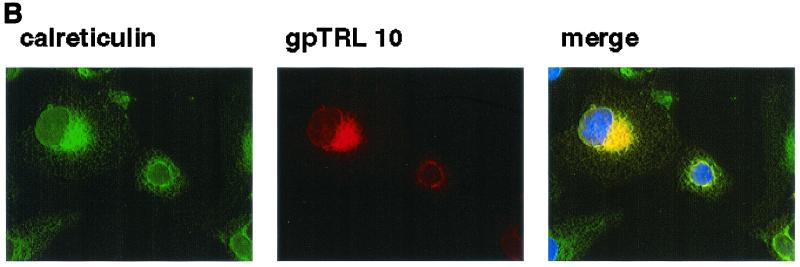
Recombinant expression and cellular localization of gpTRL10. (A) 293T cells were transfected with plasmid pcTRL10myc/his, expressing full-length gpTRL10 as a Myc/His-tagged fusion protein. Cell lysates were prepared and subjected to immunoblot analysis following digestion with endoglycosidases. Blots were developed using an anti-Myc monoclonal antibody. (B) Cos7 cells grown on glass cover slips were transfected with plasmid pcTRL10myc/his as described above. Two days after transfection, the cells were fixed in 4% paraformaldehyde in PBS and then permeabilized in PBS containing NP-40. The cover slips were then incubated with an anti-Myc antibody or a rabbit serum reactive with an ER resident protein, calreticulin. Following extensive washing, antibody binding was detected with FITC-labeled anti-rabbit IgG or Texas Red-conjugated anti-mouse IgG. Cell nuclei were stained with Hoechst 33258 to demarcate the nucleus. Images were collected as described in Materials and Methods. Yellow indicates colocalization of the signal.
To define the intracellular distribution of gpTRL10, we used transient expression of gpTRL10myc followed by fluorescence imaging with antibodies directed against the Myc-tagged viral protein or against cellular markers of the secretory pathway. In Cos7 cells, we observed complete colocalization of gpTRL10myc with markers staining the ER (Fig. 5B). The transiently expressed gpTRL10myc failed to colocalize with markers of the ER-Golgi intermediate compartment (ERGIC), Golgi, or trans-Golgi compartment (data not shown).
Complex formation of gpTRL10.
All but one of the structural glycoproteins of HCMV which have been characterized thus far have been shown to form disulfide-linked complexes (2, 3, 26, 28). It was therefore of interest to investigate potential complex formation of gpTRL10. Lysates of cells transfected with the plasmid encoding TRL10myc or gradient-purified virions were solubilized in the presence and absence of the reducing agent 2-mercaptoethanol and analyzed by immunoblotting.
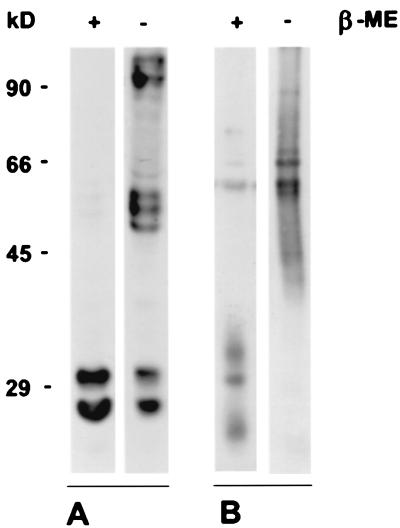
In the presence of 2-mercaptoethanol, protein bands of the expected sizes between 22 and 34 kDa were detected (Fig. 6). In the absence of 2-mercaptoethanol, higher-molecular-weight proteins were seen. In lysates from TRL10myc-transfected cells, proteins of 50 to 60 kDa and >90 kDa were detected by the Myc-specific antibody in the nonreduced lysates. However, it should be noted that a significant fraction of gpTRL10 remained in monomeric form (Fig. 6). In contrast, the 22- and 34-kDa forms of gpTRL10 present in viral lysates completely disappeared in the samples which were solubilized in the absence of 2-mercaptoethanol. Instead, a broadly migrating band between 40 and 90 kDa was detected by the TRL10 antiserum, demonstrating formation of a disuldfide-bonded complex which contained gpTRL10 as at least one protein component and likely a second and unidentified virion protein.
FIG. 6.
Complex formation of gpTRL10. (A) 293T cells were transfected with a plasmid (pcTRL10myc/his) expressing full-length gpTRL10 as a Myc/His-tagged fusion protein. Cell lysates were solubilized in the presence (+) and absence (−) of 2-mercaptoethanol (β-ME) and analyzed by immunoblotting with a Myc-specific monoclonal antibody. (B) Extracellular TB40E virions were subjected to immunoblot analysis in the presence (+) or absence (−) of 2-mercaptoethanol (β-ME) and developed with the rabbit TRL10-specific antiserum. Estimated molecular masses (in kilodaltons) are shown on the left.
Comparison of gpTRL10 coding sequences from different HCMV isolates.
Amino acid sequences of HCMV structural glycoproteins show variable degrees of homology between isolates (25, 27, 31). It was therefore of interest to determine the nucleotide and deduced amino acid sequences of the TRL10 gene from different isolates. Nucleotide sequences were determined for 11 HCMV strains, including nine low-passage clinical isolates and the laboratory-adapted strains AD169 and Davis. The sequences were aligned and compared to the published sequence of TRL10AD169 (Fig. 7).
On the nucleotide level, the gpTRL10 coding sequences showed overall homology of from 93.7 to 99.7%. On the amino acid level, the putative membrane anchor and carboxy-terminal regions between residues 86 and 171 were highly conserved among the isolates. The only differences in the predicted sequence were a K→N exchange at position 150 and an N→D exchange at position 170. The predicted extraluminal part of the protein showed a somewhat higher degree of variation (1.2 to 15.5% amino acid dissimilarities). However, the position and number of cysteine residues as well as potential N-linked glycosylation sites were perfectly conserved among the isolates. Note that the aspartic acid (D) in position 38 of TRL10AD169 seems to be the exception, since all other isolates have an asparagine (N) at the same position and thus resemble IRL10AD169-like sequences.
DISCUSSION
We have identified the product of the TRL10 ORF as a structural envelope glycoprotein, gpTRL10, of HCMV. The gene encoding gpTRL10 was interesting because it is present twice in laboratory-adapted strains such as AD169 and Towne, yet only once in clinical isolates such as Toledo and TB40E. In strain AD169, the nucleotide sequences of the duplicated gene as represented by TRL10 and IRL10 are not identical. The predicted amino acid sequence of the gpIRL10 differs from the gpTRL10 by a D→N exchange at position 38 (8). Whether similar divergence occurs in other laboratory strains is unknown.
The genomic area from both strains gave rise to numerous RNAs, making an assignment of the TRL10-specific transcripts via Northern blot analysis not possible even by using strand-specific probes. However, our results suggested that the TRL10 ORF was transcribed with early/late kinetics, which is in agreement with results obtained by microarray analysis (6).
Computer algorithms predict that gpTRL10 is a type I glycoprotein with a theoretical molecular mass of 20.5 kDa. The protein exhibits no amino acid homology to other herpesvirus proteins, including the closely related betaherpesviruses, murine cytomegalovirus (34), and human herpesvirus types 6 and 7 (12). In agreement with the predicted mass of the TRL10 protein, a polypeptide of 20 kDa was observed in infected cells after removal of N-linked carbohydrates.
Interestingly, in infected cells gpTRL10 existed in two predominant forms having molecular masses of 22 and 23.5 kDa. These molecules were modified by the addition of high-mannose sugars, as indicated by their sensitivity to Endo H. The most likely explanation for the expression of the two forms of gpTRL10 in infected cells was the presence of differentially glycosylated forms of gpTRL10. Potential sites for the addition of N-linked carbohydrates were present at amino acid positions 48/49 and 56 in the extraluminal domain of the gpTRL10. The presence of high-mannose carbohydrates on the vast majority of gpTRL10 present in virus-infected fibroblasts suggested that these molecules were likely localized to the ER. In addition to the predominant Endo H-sensitive forms, we also identified immunoreactive gpTRL10 which contained complex N-linked sugars, as reflected by their resistance to Endo H digestion and sensitivity to Endo F. Forms of TRL10 modified with complex carbohydrates were far less abundant in infected cells. In contrast, the gpTRL10 present in extracellular viral particles contained only complex N-linked sugars.
There are at least two possible explanations for the differences between the sugar modifications on the gpTRL10 in virus and that found in infected cells. The first is that the rabbit antiserum recognized predominantly forms of gpTRL10 modified with simple N-linked sugars. This possibility seems unlikely, since it was raised against an amino acid sequence at the C terminus of the molecule (amino acids 124 to 171) which is distal to the proposed N-linked glycosylation sites. Furthermore, transiently expressed gpTRL10 containing a C-terminal Myc epitope tag was modified nearly exclusively with simple N-linked sugars. The second and more likely possibility was that the gpTRL10 containing simple, high-mannose modifications was produced in excess during virus infection and its transport from proximal compartments of the secretory pathway was rate limiting. This resulted in overabundance of the gpTRL10 containing only simple, high-mannose sugar modifications within infected cells. Similar findings have been noted for other HCMV glycoproteins, including gB. Within infected cells, a significant amount of gB can be detected as an uncleaved polyprotein modified with simple, high-mannose sugars (3).
The predominance of forms of gpTRL10 containing only simple sugars could result from either an inefficient folding program, as has been proposed for gB, or perhaps the lack of sufficient expression of associated transporter molecules required for its transport to more distal sites of the secretory pathway. Consistent with this latter explanation was the finding that gpTRL10 in extracellular virions was approximately 8 to 10 kDa larger than intracellular forms of gpTLR10. Because this difference in size was believed to be a result of the presence of heterogeneous complex carbohydrate modification on virion gpTRL10, we would argue that gpTRL10 must associate with a second viral protein to traffic to distal compartments of the secretory pathway for terminal carbohydrate modifications and entry into a cytoplasmic assembly compartment.
Our findings indicated that gpTRL10 was present in a higher-molecular-weight complex. Expression of the TRL10 ORF in transfected cells indicated that in the absence of a reducing agent, the gpTRL10 formed higher-molecular-weight complexes or aggregates; however, a significant amount of the protein remained in monomeric form. In contrast, when extracellular virions were disrupted in the absence of a reducing agent, only higher-molecular-weight immunoreactive forms between 40 and 90 kDa were detected and none of the lower-molecular-weight forms. Together, these findings indicated that in the absence of other viral proteins, gpTRL10 could form higher-molecular-weight species, perhaps as a result of aggregation or nonnative disulfide bond formation between unpaired cysteines. In contrast, the gpTRL10 found in extracellular virions was present almost entirely as a higher-molecular-weight complexes. This result provided additional support for the argument that gpTRL10 was complexed with a second viral protein.
Consistent with this argument was the lack of intracellular transport and carbohydrate processing of gpTRL10 when it was expressed in the absence of other viral proteins. Thus, it appeared that similar to better-described HCMV envelope glycoproteins, gpTRL10 was complexed with other viral proteins, most likely through disulfide bonds, and that intracellular transport and incorporation of the protein into the virion progeny required complex formation. We have not identified the viral protein(s) in the complex and therefore cannot definitively exclude the possibility that the larger, complex forms of gpTRL10 were homodimers or dimers of differentially processed forms of gpTRL10. However, the ER retention and the lack of terminally glycosylated forms of gpTRL10 in cells transfected with a plasmid encoding the TRL10 ORF suggested that a virus function was required for its correct intracellular localization. In this respect, gpTRL10 might be similar to other HCMV glycoproteins such as gH, gL, gO, gM, and gN, which also require complex formation for proper folding and transport to the cell surface (19, 28, 38).
Structural glycoproteins of HCMV have been studied in a number of previous investigations. Solubilization of envelope constituents and subsequent analysis by various chromatographic methods have led to the characterization of three glycoprotein complexes (gCI to -III) within the envelope of HCMV (13). The glycoprotein composition of these complexes has been defined by investigators in several laboratories. The gCI complex is formed by homodimeric molecules of gB (2, 3). Glycoprotein complex II is composed of gM and gN, whereas glycoproteins H, L, and O together constitute the gCIII complex (17, 18, 26, 28). A protein corresponding to the size of gpTRL10 was not detected during the course of these studies, suggesting that this polypeptide was not an additional component of one of these complexes. However, other studies have noted the existence of envelope constituents with molecular weights similar to that of gpTRL10, although at a low abundance (1, 10). Thus, it remains to be determined if gpTRL10 is associated with one of the previously defined gCI to -III complexes or with an as yet unidentified envelope protein.
Because of its location in the virion envelope, gpTRL10 is potentially capable of inducing neutralizing antibodies and thus could represent a target of the humoral immune response during natural infection. Several studies have investigated antibody specificities against structural proteins of HCMV in detail (21, 22, 23, 24). Proteins which could correspond to gpTRL10 were not well described. However, in SDS-PAGE, gpTRL10 migrated as a very diffuse band and could have escaped detection. Other envelope glycoproteins such as gp116 (the amino-terminal component of gB) and gN, which have been shown to be highly immunogenic are also not routinely detected during immunoblot analysis with human sera.
Induction of strain-specific antibodies is a well-known phenomenon in HCMV biology, and antigenic variation could have limited antibody recognition of gpTRL10 by human sera (20, 44, 46). A number of glycoproteins have been described as polymorphic and thus able to induce strain-specific antibodies (30, 42). Our analysis of gpTRL10 coding sequences from clinical isolates revealed some sequence polymorphism in the amino-terminal part of the molecule. Since the extraluminal part is rather small (62 amino acids after removal of the signal peptide) and therefore could contain only a limited number of antibody binding sites, even a limited degree of variability would be sufficient to result in induction of strain-specific antibodies. Interestingly, computer algorithms indicate that the most variable domain between amino acids 40 and 60 is also highly antigenic.
Several other interesting findings were revealed by the sequence analysis. All low-passage isolates contained the D→N substitution at position 38 of TRL10AD169 and thus corresponded to the IRL10AD169 sequence. This result suggested that the terminal repeats of AD169 may have originated as a duplication of the internal repeat sequences. Moreover, eight of the isolates that were analyzed contained an N→D substitution at position 170. This substitution resulted in the extension of the acidic cluster at the C terminus of gpTRL10. Acidic clusters of membrane-bound proteins are known to play important roles in protein trafficking to the trans-Golgi network and are a common motif in the carboxyl terminus of herpesvirus glycoproteins (43). Whether this additional acidic residue in the C terminus of TRL10 resulted in a change in the intracellular trafficking of gpTRL10 from these clinical isolates of HCMV is unknown; however, it was of some interest that the TRL10 nucleotide sequence from most clinical isolates had this amino acid substitution.
Structural glycoproteins of herpesviruses can be grouped into two general classes, those that are conserved in members of the same subgroup of viruses or even throughout the entire family of herpesviruses and those glycoproteins that are unique to members of a specific subgroup of viruses. The conserved glycoproteins which have homologs in most members of the herpesviruses have been studied extensively and have been shown in many cases to be required for herpesvirus replication, including essential roles in particle assembly and early events in virus infection (4, 27, 33). Much less is known about structural glycoproteins which are unique to members of subgroups of herpesviruses. These proteins likely play very critical roles in the in vivo replication and pathogenic behavior of the respective viruses. An obvious example is the gp350/220 of Epstein-Barr virus, which binds to the cellular receptor CD21 on B cells (40, 41). Homologs of this viral protein have not been described in other alpha-, beta-, and gammaherpesviruses and point to a unique role of this protein in the in vivo replication of Epstein-Barr virus. Other examples include gD of HSV, a glycoprotein which has been shown to have an important role in adsorption (45).
A gD homolog has not been identified in betaherpesviruses such as HCMV, a finding which underscores the requirements of different viral glycoproteins for the unique and often characteristic biological behavior of herpesviruses. Whether gpTRL10 plays an essential role in the biology of HCMV remains to be determined; however, the maintenance of this ORF in the genome of clinical isolates strongly argues for an essential role in the replication of this virus in the natural host. A more complete study of the function of envelope glycoproteins whose expression is restricted to individual members of a virus group could provide insight into the role of these virion envelope components in specific aspects of infection, such as cell tropism and disease induction, which in turn define the unique phenotypes of individual herpesviruses.
Acknowledgments
We thank S. Lang for providing anti-Myc monoclonal antibody.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung, and the National Institutes of Health.
REFERENCES
- 1.Benko, D. M., and W. Gibson. 1986. Primate cytomegalovirus glycoproteins: lectin-binding properties and sensitivities to glycosidases. J. Virol. 59:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britt, W. J. 1984. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology 135:369–378. [DOI] [PubMed] [Google Scholar]
- 3.Britt, W. J., and D. Auger. 1986. Synthesis and processing of the envelope gp55–116 complex of human cytomegalovirus. J. Virol. 58:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt, W. J., and M. Mach. 1996. Human cytomegalovirus glycoproteins. Intervirology 39:401–412. [DOI] [PubMed] [Google Scholar]
- 5.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C. P., D. H. Vesole, J. Nelson, M. B. Oldstone, and M. F. Stinski. 1989. Identification and expression of a human cytomegalovirus early glycoprotein. J. Virol. 63:3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., and M. F. Stinski. 2000. Activation of transcription of the human cytomegalovirus early UL4 promoter by the Ets transcription factor binding element. J. Virol. 74:9845–9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrar, G. H., and J. D. Oram. 1984. Characterization of the human cytomegalovirus envelope glycoproteins. J. Gen. Virol. 65:1991–2001. [DOI] [PubMed] [Google Scholar]
- 11.Fleckenstein, B., I. Müller, and J. Collins. 1982. Cloning of the complete human cytomegalovirus genome in cosmids. Gene 18:39–46. [DOI] [PubMed] [Google Scholar]
- 12.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51. [DOI] [PubMed] [Google Scholar]
- 13.Gretch, D. R., B. Kari, L. Rasmussen, R. C. Gehrz, and M. F. Stinski. 1988. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J. Virol. 62:875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppert, M., and A. Holzenburg. 1998. Electron microscopy in microbiology. BIOS Scientific Publishers Limited, Oxford, England.
- 15.Huang, L., C. L. Malone, and M. F. Stinski. 1994. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J. Virol. 68:2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 69:7612–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber, M. T., and T. Compton. 1997. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J. Virol. 71:5391–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye, J. F., U. A. Gompels, and A. C. Minson. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693–2698. [DOI] [PubMed] [Google Scholar]
- 20.Klein, M., K. Schoppel, N. Amvrossiadis, and M. Mach. 1999. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J. Virol. 73:878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landini, M. P., and M. La-Placa. 1991. Humoral immune response to human cytomegalovirus proteins: a brief review. Comp. Immunol. Microbiol. Infect. Dis. 14:97–105. [DOI] [PubMed] [Google Scholar]
- 22.Landini, M. P., G. Mirolo, M. C. Re, A. Ripalti, and M. La-Placa. 1989. Antibody reactivity to cytomegalovirus structural polypeptides in subclinical and clinical human immunodeficiency virus infections. Eur. J. Clin. Microbiol. Infect. Dis. 8:159–163. [DOI] [PubMed] [Google Scholar]
- 23.Landini, M. P., M. C. Re, G. Mirolo, B. Baldassarri, and M. La-Placa. 1985. Human immune response to cytomegalovirus structural polypeptides studied by immunoblotting. J. Med. Virol. 17:303–311. [DOI] [PubMed] [Google Scholar]
- 24.Landini, M. P., E. Rossier, and H. Schmitz. 1988. Antibodies to human cytomegalovirus structural polypeptides during primary infection. J. Virol. Methods 22:309–317. [DOI] [PubMed] [Google Scholar]
- 25.Lehner, R., T. Stamminger, and M. Mach. 1991. Comparative sequence analysis of human cytomegalovirus strains. J. Clin. Microbiol. 29:2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., J. A. Nelson, and W. J. Britt. 1997. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J. Virol. 71:3090–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lurain, N. S., K. S. Kapell, D. D. Huang, J. A. Short, J. Paintsil, E. Winkfield, C. A. Benedict, C. F. Ware, and J. W. Bremer. 1999. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J. Virol. 73:10040–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mach, M., B. Kropff, P. Dal-Monte, and W. J. Britt. 2000. Complex formation of human cytomegalovirus glycoprotein M (gpUL100) and glycoprotein N (gpUl73). J. Virol. 74:11881–11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manservigi, R., and E. Cassai. 1991. The glycoproteins of the human herpesviruses. Comp. Immunol. Microbiol. Infect. Dis. 14:81–95. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, H., V. A. Sundqvist, L. Pereira, and M. Mach. 1992. Glycoprotein gp116 of human cytomegalovirus contains epitopes for strain-common and strain-specific antibodies. J. Gen. Virol. 73:2375–2383. [DOI] [PubMed] [Google Scholar]
- 31.Monte, P. D., S. Pignatelli, M. Mach, and M. P. Landini. 2001. The product of human cytomegalovirus UL73 is a new polymorphic structural glycoprotein (gpUL73). J. Hum. Virol. 4:26–34. [PubMed] [Google Scholar]
- 32.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajcani, J., and A. Vojvodova. 1998. The role of herpes simplex virus glycoproteins in the virus replication cycle. Acta Virol. 42:103–118. [PubMed] [Google Scholar]
- 34.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schleiss, M. R., C. R. Degnin, and A. P. Geballe. 1991. Translational control of human cytomegalovirus gp48 expression. J. Virol. 65:6782–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinzger, C., K. Schmidt, J. Knapp, M. Kahl, R. Beck, J. Waldman, H. Hebart, H. Einsele, and G. Jahn. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 80:2867–2877. [DOI] [PubMed] [Google Scholar]
- 38.Spaete, R. R., K. Perot, P. I. Scott, J. A. Nelson, M. F. Stinski, and C. Pachl. 1993. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in transport of gH to the cell surface. Virology 193:853–861. [DOI] [PubMed] [Google Scholar]
- 39.Talbot, P., and J. D. Almeida. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol. 36:345–349. [DOI] [PubMed] [Google Scholar]
- 40.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203–213. [DOI] [PubMed] [Google Scholar]
- 41.Thorley-Lawson, D. A., and C. A. Poodry. 1982. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J. Virol. 43:730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urban, M., W. Britt, and M. Mach. 1992. The dominant linear neutralizing antibody-binding site of glycoprotein gp86 of human cytomegalovirus is strain specific. J. Virol. 66:1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan, L., S. S. Molloy, L. Thomas, G. Liu, Y. Xiang, S. L. Rybak, and G. Thomas. 1998. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94:205–216. [DOI] [PubMed] [Google Scholar]
- 44.Waner, J. L., and T. H. Weller. 1978. Analysis of antigenic diversity among human cytomegaloviruses by kinetic neutralization tests with high-titered rabbit antisera. Infect. Immun. 21:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, d. L. Ponce, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zablotney, S. L., B. B. Wentworth, and E. R. Alexander. 1978. Antigenic relatedness of 17 strains of human cytomegalovirus. Am. J. Epidemiol. 107:336–343. [DOI] [PubMed] [Google Scholar]