Abstract
The influenza A virus NS1 protein, a virus-encoded alpha/beta interferon (IFN-α/β) antagonist, appears to be a key regulator of protein expression in infected cells. We now show that NS1 protein expression results in enhancement of reporter gene activity from transfected plasmids. This effect appears to be mediated at the translational level, and it is reminiscent of the activity of the adenoviral virus-associated I (VAI) RNA, a known inhibitor of the antiviral, IFN-induced, PKR protein. To study the effects of the NS1 protein on viral and cellular protein synthesis during influenza A virus infection, we used recombinant influenza viruses lacking the NS1 gene (delNS1) or expressing truncated NS1 proteins. Our results demonstrate that the NS1 protein is required for efficient viral protein synthesis in COS-7 cells. This activity maps to the amino-terminal domain of the NS1 protein, since cells infected with wild-type virus or with a mutant virus expressing a truncated NS1 protein—lacking approximately half of its carboxy-terminal end—showed similar kinetics of viral and cellular protein expression. Interestingly, no major differences in host cell protein synthesis shutoff or in viral protein expression were found among NS1 mutant viruses in Vero cells. Thus, another viral component(s) different from the NS1 protein is responsible for the inhibition of host protein synthesis during viral infection. In contrast to the earlier proposal suggesting that the NS1 protein regulates the levels of spliced M2 mRNA, no effects on M2 protein accumulation were seen in Vero cells infected with delNS1 virus.
Protein expression in eukaryotic cells is controlled by complex regulatory mechanisms, and viral infection usually results in an intricate interplay between viral and cellular products leading to significant changes in protein expression patterns. On one hand, cells encode several antiviral pathways dedicated to shutting off protein synthesis when infected by viruses. On the other hand, many viruses, including influenza A viruses, have developed mechanisms favoring the translation of viral over cellular mRNAs and leading to a switch from cellular to viral protein synthesis, while inhibiting the cell-encoded antiviral pathways (12). A key role in the modulation of expression of cellular and viral proteins during influenza virus infection has been attributed to the viral NS1 protein. This protein has been described to enhance viral protein expression by promoting translation of viral mRNAs over cellular ones (1, 6, 8, 9). Moreover, the NS1 protein has been postulated to inhibit host protein synthesis by blocking cellular mRNA polyadenylation and nuclear export (4, 10, 23, 27). The latter effects on host mRNA processing have been suggested to be a major cause of shutoff of host cellular protein synthesis during influenza virus infection (for reviews, see references 3 and 20). Expression of the NS1 protein in tissue culture cells was also reported to inhibit mRNA splicing (10, 17). This activity of the NS1 protein might also contribute to the regulation of host and viral protein expression in infected cells. In addition, it has been suggested that the NS1 protein enhances transcription of specific viral genes, contributing to higher viral protein expression (21). A recently described function of the NS1 protein is that of antagonizing the alpha/beta interferon (IFN-α/β) system (for a review, see reference 13). The anti-IFN properties of the NS1 protein map to its double-stranded (dsRNA)-binding domain, which is able to inhibit dsRNA activated antiviral pathways (30, 33). By this mechanism the NS1 protein prevents the synthesis of IFN-α/β, as well as the activation of the antiviral enzymes PKR (2, 15, 18, 32) and 2′-5′-oligoadenylate synthetases (N. Donelan and A. García-Sastre, unpublished observations).
To directly study the effects of the NS1 protein on viral protein synthesis and on host cell protein expression inhibition during influenza virus infection, we took advantage of a recombinant influenza virus lacking the NS1 gene (delNS1 virus) (14). Since this virus lacks the IFN antagonist activity of the NS1 protein, it is attenuated and does not replicate efficiently in IFN-competent systems. However, delNS1 virus can replicate in IFN-deficient systems to levels only slightly lower than those of wild-type virus. We also used recombinant viruses NS1-99 and NS1-126 (31, 33) expressing carboxy-terminal(ly) truncated NS1 proteins of 99 and 126 amino acids, respectively. These truncated NS1 proteins contain a functional RNA-binding domain but lack the effector domain that has been suggested to be involved in the inhibition of mRNA splicing, polyadenylation, and transport (3, 26). In the present study, the effects of the NS1 protein on protein expression were investigated in cells infected with these recombinant viruses, as well as in cells transfected with NS1-expressing plasmids. Our results indicate that the NS1 protein of influenza A virus is a general translational enhancer, most likely through its PKR-inhibitory activity, and that this protein is not required for the shutoff of host cell protein expression during virus infection.
MATERIALS AND METHODS
Cells and viruses.
Influenza A/PR/8/34 virus (wild type, PR8) and transfectant NS1-126, NS1-99, and delNS1 viruses were grown in 7-day embryonated chicken eggs. All transfectant viruses are in a PR8 background, and their generation has been described elsewhere (14, 31, 33). NS1-99 and NS1-126 viruses encode truncated NS1 proteins containing the first 99 and 126 amino acids of wild-type (230 amino acids) NS1, respectively. DelNS1 virus does not encode an NS1 protein due to a deletion in its NS gene. Viral titers were obtained by plaque assay on MDCK cells as described previously (33). MDCK cells were maintained in minimal essential medium with 10% fetal calf serum (FCS) plus antibiotics. Vero, COS-7 and 293 cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% FCS plus antibiotics. U3A cells, a human cell line defective in STAT1 expression (22), were also maintained in DMEM−10% FCS plus antibiotics.
Expression plasmids.
pCAGGS-NS1(SAM) and pCAGGS-NS1-R38AK41A(SAM) contain the open reading frame of NS1 of PR8 virus in an expression vector under the control of a chicken β-actin promoter. They encode wild-type NS1 protein and an NS1 protein with two amino acid mutations (R38A and K41A) (30). pCAGGS-hPKR expresses human PKR with the same chicken β-actin promoter. This plasmid was constructed as follows. First, the PKR open reading frame from pcDNAI/Neo-PKR (kindly provided by Michael Katze) was amplified by PCR with the oligonucleotides 5′-GGCCGAGCTCACCATGGCTGGTGATCTTTCAGCAGG-3′ and 5′-GCGCCTCGAGCTAACATGTGTGTCGTTCATTTTTCTC-3′. The generated PCR product was digested by using SacI and XhoI restriction enzymes and cloned into SacI-XhoI digested pCAGGS. PKR expression from this plasmid was confirmed by Western blot analysis in pCAGGS-hPKR-transfected 293 cells by using the PKR-specific antibody K-17 (Santa Cruz Biotechnology) (data not shown). STAT1 was expressed in vector pRcCMV (Invitrogen) as previously described (25). pAdvantage (Promega) expresses the adenoviral virus-associated I RNA (VAI RNA) under the control of an RNA polymerase III promoter. The pCAT-control and pGL2-control plasmids (Promega) contain the coding regions for chloramphenicol acetyltransferase (CAT) and firefly luciferase, respectively, under the control of the simian virus 40 promoter and enhancer sequences. The pEGFP-N1 expression plasmid (Clontech) contains the EGFP cDNA under the control of the cytomegalovirus promoter.
Protein labeling experiments.
Confluent cell monolayers of Vero or COS-7 cells in 12-well plates were infected at a multiplicity of infection (MOI) of 5 with wild-type or transfectant influenza viruses. At various intervals postinfection, cells were labeled with l-[35S]cysteine and l-[35S]methionine for 30 min. Labeled cells were washed with phosphate-buffered saline and lysed in radioimmunoprecipitation assay buffer. Labeled proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Western blots.
The ratio of M1 and M2 proteins in virus-infected cells was determined by Western blotting. Confluent cell monolayers of Vero cells in 35-mm dishes were infected at an MOI of 5 with wild-type or delNS1 viruses. At 15 h postinfection, cells were washed with phosphate-buffered saline and lysed in 200 μl of SDS loading buffer. One-tenth portions of the cell extracts were blotted onto a nitrocellulose membrane after SDS−12.5% PAGE. The membrane was incubated in buffer solution containing first a 1:100 dilution of the mouse monoclonal antibody E10 (0.35 mg/ml) and then a 1:500 dilution of peroxidase-coupled secondary anti-mouse antibody (Boehringer Mannheim). The E10 antibody recognizes an amino-terminal peptide that is common to the M1 and M2 proteins. Bands corresponding to the M1 and M2 proteins were visualized by chemiluminescence (NEN Life Science Products). The viral nucleoprotein (NP) was visualized by Western blotting as previously described (31). Western blotting to detect the levels of green fluorescent protein (GFP) was also performed in U3A cells transfected with the GFP expression plasmid by using specific peptide antiserum from Clontech, according to the manufacturer’s protocol.
CAT assays.
293 cells were transfected with pCAT-control plasmid or pGL2-control plasmid in combination with the indicated expression plasmids. At 48 h postransfection, cell extracts were made and assayed for CAT activity as described previously (19) or for luciferase activity. Luciferase assays were performed by using the Promega Luciferase Assay System. All experiments with CAT and luciferase reporter genes were repeated at least three times, and the results from one representative experiment were represented.
RT-PCR of GFP mRNA.
Total RNA was prepared from transfected U3A cells at 48 h postransfection by using Trizol reagent (Gibco-BRL). The RNA was treated with DNase I and subjected to reverse transcription-PCR (RT-PCR) analysis as previously described (30). RNA was first reverse transcribed by using random hexamer primers and Moloney murine leukemia virus reverse transcriptase. The resulting cDNA was used as a template for 25 cycles of PCR in the presence of [32P]dATP with the following primers specific for GFP mRNA: 5′-ACGTAAACGGCCACAAGTTC-3′ and 5′-AAGTCGTGCTTCATGTG-3′. To rule out potential plasmid contamination, PCRs were also performed by using non-reverse-transcribed samples as a template. No products were observed in the absence of RT.
RESULTS
Enhancement of reporter gene activity by cotransfection with an NS1 expression plasmid.
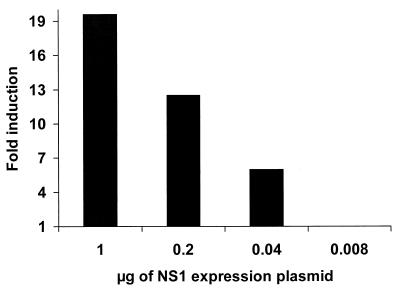
The expression of the NS1 protein of influenza A virus has been associated with the inhibition of host mRNA polyadenylation and transport (4, 10, 23, 27). These NS1 activities may lead to the inhibition of cell gene expression in NS1 protein-expressing cells. In order to investigate the effects of the NS1 protein on gene expression, we cotransfected the luciferase reporter plasmid pGL2-control with increasing concentrations of pCAGGS-NS1(SAM), encoding the NS1 protein of influenza A/PR/8/34 virus. Surprisingly, we found that expression of the NS1 protein resulted in an enhancement and not a decrease of reporter gene activity. This increase in reporter gene expression was NS1 dose dependent, and luciferase activity was increased almost 20-fold when pGL2-control was cotransfected with 1 μg of pCAGGS-NS1(SAM) (Fig. 1).
FIG. 1.
NS1 protein expression enhances reporter gene activity. 293T cells were transfected with 0.5 μg of the luciferase-expressing plasmid pGL2-control, together with the indicated amounts of the NS1-expressing plasmid pCAGGS-NS1(SAM). The total amount of DNA remained constant among transfections by adding pCAGGS empty plasmid to a total of 4 μg. At 48 h posttransfection, cell extracts were made and the luciferase activity was measured. The y axis represents the relative luciferase activity compared to that in cells transfected with pGL2-control plasmid in the absence of pCAGGS-NS1(SAM).
Enhancement of reporter gene activity by expression of influenza A NS1 protein and adenovirus VAI RNA.
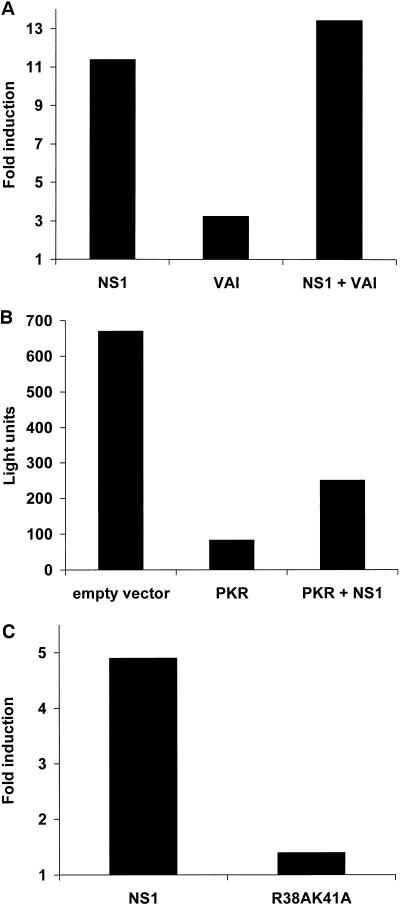
The previously described inhibitory properties of the NS1 protein on PKR activation (2, 15, 18, 32) may explain the NS1-mediated enhancement of plasmid gene expression. Transfection of mammalian cells with an expression plasmid often results in suboptimal protein expression due to the activation after transfection of the translation inhibitory protein PKR. pAdvantage is a commercial plasmid which enhances transient protein expression after cotransfection with protein expression plasmids by increasing translation initiation through the expression of adenoviral VAI RNA, a known PKR inhibitor (16, 24, 29). We therefore compared the abilities of pCAGGS-NS1(SAM) and pAdvantage plasmids to mediate enhancement of reporter gene expression. When pCAGGS-NS1(SAM) was cotransfected with the reporter plasmid pCAT-control in 293 cells, we observed an approximately 10-fold induction in CAT expression (Fig.2A). Cotransfection of pCAT-control with pAdvantage also resulted in enhancement of CAT expression, although the results were less dramatic (ca. threefold induction). Interestingly, when both pAdvantage and pCAGGS-NS1(SAM) were cotransfected with pCAT-control, there were no cooperative effects on CAT expression. This result suggests that the NS1 protein and the VAI RNA might enhance reporter gene expression through a common pathway and not by two independent mechanisms. However, it is still possible that these two viral products act through separate pathways that do not synergize.
FIG. 2.
Analysis of the NS1 protein-mediated enhancement of reporter gene expression. (A) 293 cells were transfected with 0.5 μg of the CAT expressing plasmid pCAT-control, together with 1 μg of NS1 protein expressing plasmid pCAGGS-NS1(SAM), pAdvantage (expressing VAI RNA), or a combination of both plasmids. The total amount of DNA remained constant among transfections by adding pCAGGS empty plasmid to a total of 2.5 μg. At 48 h posttransfection, cell extracts were made and the CAT activity was measured. The y axis represents the relative CAT activity compared to that in cells transfected only with pCAT-control plasmid. (B) 293 cells were transfected with 1 μg of the luciferase-expressing plasmid pGL2-control, together with 2 μg of pCAGGS empty plasmid, pCAGGS-hPKR, or a combination of pCAGGS-hPKR and pCAGGS-NS1(SAM). The total amount of DNA remained constant among transfections by adding pCAGGS empty plasmid to a total of 5 μg. At 48 h posttransfection, cell extracts were made and the luciferase activity was measured. (C) 293 cells were transfected with 1 μg of pGL2-control, together with pCAGGS-NS1(SAM) or pCAGGS-NS1-R38AK41A(SAM). At 48 h posttransfection, cell extracts were made and the luciferase activity was measured. The y axis represents the relative luciferase activity compared to that in cells transfected with pGL2-control and pCAGGS-empty plasmids.
Expression of NS1 partly overcomes the inhibition of translation by PKR.
In order to directly test whether the NS1 protein would prevent the inhibitory effects of PKR on gene expression, we investigated the effects of NS1 expression on reporter gene activity in 293 cells overexpressing PKR. Overexpression of PKR resulted in a decrease in luciferase activity after cotransfection of the reporter plasmid pGL2-control and pCAGGS-hPKR. These effects are known to be mediated by increased PKR kinase levels due to overexpression, resulting in phosphorylation of eIF-2α and in inhibition of translation. Coexpression of NS1 with PKR partly restored luciferase activity in pGL2-control transfected cells (Fig. 2B). These results are in agreement with the described NS1-mediated inhibition of eIF-2α phosphorylation by PKR (15, 18).
Enhancement of gene expression by the NS1 protein requires its RNA-binding activity.
We next investigated whether the dsRNA-binding properties of the NS1 protein are required for its reporter gene expression enhancement. The luciferase reporter gene pGL2-control was cotransfected into 293 cells with pCAGGS-NS1(SAM) or with pCAGGS-NS1-R38AK41A(SAM), the latter expressing a mutant form of NS1 with impaired dsRNA-binding activity (5). In contrast to wild-type NS1 protein, the mutant NS1 protein was unable to enhance reporter gene expression (Fig. 2C). Interestingly, the anti-PKR activity of the NS1 protein also requires an intact NS1 dsRNA-binding domain (18). Consequently, when we cotransfected pCAGGS-NS1-R38AK41A(SAM) together with the reporter plasmids pGL2-control and pCAGGS-hPKR, expression of this RNA-binding-defective NS1 mutant protein, in contrast to that of wild-type NS1 protein, did not result in increased luciferase activity (data not shown).
Enhancement of gene expression by the NS1 protein is mediated at a translational level.
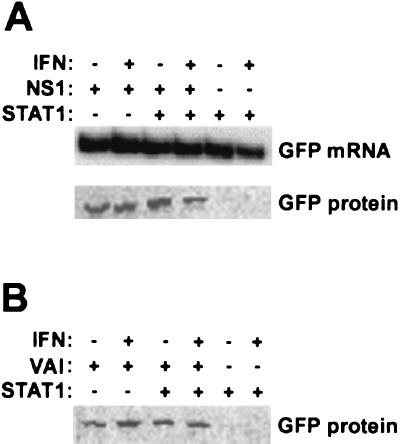
The above-described experiments support a model whereby the NS1 protein enhances gene expression due to an increase in translation through the inhibition of PKR and possibly of other dsRNA-activated antiviral pathways. In order to prove that the NS1 protein functions as a translational enhancer, we determined GFP-specific mRNA and protein levels in cells cotransfected with pCAGGS-NS1(SAM) and a GFP reporter plasmid. As shown in Fig. 3A, no major differences were found in GFP mRNA levels in cells expressing or not expressing the NS1 protein. However, coexpression of NS1 and GFP resulted in the appearance of green cells which were brighter than those transfected with GFP reporter plasmid alone (data not shown). Measurement of total amounts of GFP protein by Western blotting confirmed higher GFP expression in cells transfected with the NS1 expression plasmid (Fig. 3A). In these experiments we used the STAT1-deficient U3A cell line. Transfection of these cells with an STAT1 expression plasmid in the presence or absence of IFN treatment allowed us to determine whether the translational enhancement properties of the NS1 protein were affected by the presence of STAT1 and/or IFN. Regardless of STAT1 expression, the NS1-mediated translational enhancement was not influenced by IFN treatment. The same results were obtained when the NS1 expression plasmid was replaced by the PKR inhibitor VAI expression plasmid (Fig. 3B), suggesting again that both VAI RNA and NS1 protein enhance translation through a common mechanism.
FIG. 3.
NS1 protein expression enhances translation. (A) STAT1-deficient U3A cells were transfected with a GFP expression plasmid, together with NS1 and/or STAT1 expression plasmids as indicated. When indicated, cells were treated at 42 h posttransfection for 6 h with 1,000 U of IFN/ml. Cell extracts were made at 48 h posttransfection, and the levels of GFP mRNA and of GFP protein were determined in parallel samples by RT-PCR and Western blot analysis, respectively. (B) GFP Western blotting was done in parallel in U3A cells transfected under the same conditions, except that the NS1 expression plasmid was replaced by a VAI RNA expression plasmid.
The NS1 protein is not required for shutoff of host protein expression by influenza A virus.
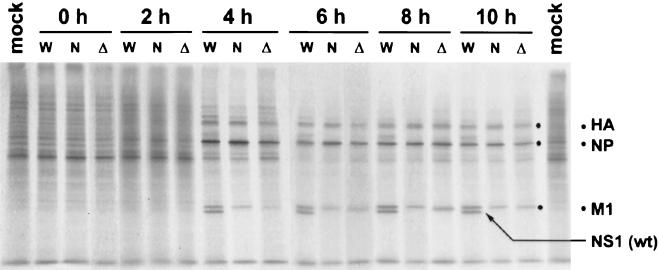
Despite the proposed role of the NS1 protein of influenza A virus in inhibiting host mRNA processing, no inhibition of reporter gene mRNA or protein expression was observed in cells expressing NS1 (Fig. 3A). However, the NS1 protein may still inhibit host protein expression during influenza virus infection, resulting in the observed shutoff of cell protein expression in virus-infected cells. Therefore, we compared levels of protein expression at different times postinfection in cells infected with wild-type PR8 virus, with NS1-99 virus (expressing a truncated NS1 protein lacking the NS1-effector domain postulated to be associated with the inhibition of cell mRNA processing and transport), and with delNS1 virus (lacking the entire NS1 gene). In this experiment we used IFN-deficient Vero cells, which previously were found to be highly permissive for delNS1 virus replication (14). As shown in Fig. 4, host protein synthesis shutoff was evident at 4 h postinfection. No significant differences were found in the levels of host protein synthesis shutoff among the three viruses. Therefore, the NS1 protein does not seem to play a role in regulating decreased host cell protein expression during infection of IFN-deficient Vero cells. In addition, levels of viral protein synthesis, with the exception of the NS1 protein, were comparable among the three viruses, although expression of the M1 protein appeared to be slightly decreased in delNS1- and NS1-99-infected cells.
FIG. 4.
Patterns of protein expression in Vero cells infected with wild-type (W), NS1-99 (N), and delNS1 (Δ) viruses. Vero cells infected at an MOI of 5 were 35S labeled for 30 min at the indicated times postinfection. Labeled proteins were analyzed by SDS−12% PAGE. Labeled mock-infected cells are also shown. The positions of the major viral proteins HA, NP, M1, and NS1 are indicated on the right.
The NS1 protein is not involved in the regulation of M1/M2 protein levels in influenza A virus-infected cells.
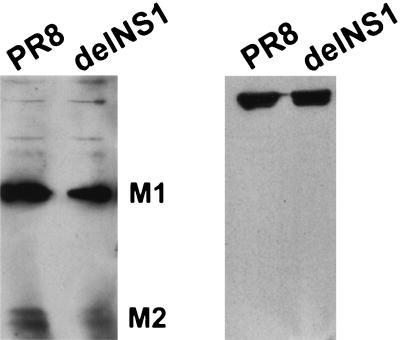
It has previously been reported that expression of NS1 protein results in the inhibition of splicing of the M segment-derived mRNA (17). This inhibition would result in a higher ratio of M1 protein (encoded by the unspliced M-specific mRNA) versus M2 protein (encoded by the spliced M-specific mRNA) in the presence of NS1. To investigate the role of the NS1 protein in the regulation of expression of the M gene products, we determined relative M1/M2 protein levels in Vero cells infected with wild-type virus or delNS1 mutant virus by using a monoclonal antibody which recognizes both proteins. As shown in Fig. 5, no differences were found for either virus, suggesting that the splicing of the M-derived mRNA in Vero cells is not affected by the NS1 protein during influenza virus infection.
FIG. 5.
The NS1 protein does not affect the relative levels of M1 and M2 proteins in influenza virus-infected cells. Vero cells were infected at an MOI of 5 with wild-type PR8 or delNS1 virus. At 15 h postinfection, cell extracts were made and analyzed by Western blotting with an antibody (E10) recognizing both M1 and M2 proteins (left) and NP-specific antibody (right).
The NS1 protein is required for efficient viral protein expression in COS-7 cells.
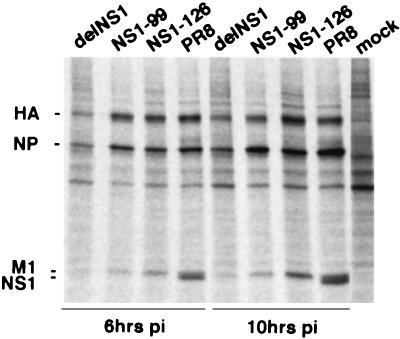
In order to investigate the NS1-mediated effects on protein expression in cells not deficient in IFN-α/β, we performed protein-labeling experiments in COS-7 cells. No differences in protein expression were found in COS-7 cells infected with wild-type virus or with the mutant virus NS1-126 expressing an NS1 protein lacking the effector domain (Fig. 6). In contrast, a general reduction of both cellular and viral protein expression was found in COS-7 cells infected with delNS1 virus, demonstrating a requirement of the RNA-binding domain of the NS1 protein for efficient protein expression in COS-7 cells. Interestingly, cells infected with the NS1-99 virus showed an intermediate phenotype and displayed reduced levels of hemagglutinin (HA) and M1 protein synthesis but not of NP.
FIG. 6.
Patterns of protein expression in COS-7 cells infected with delNS1, NS1-99, NS1-126, and wild-type PR8 viruses. COS-7 infected cells at an MOI of 5 were 35S labeled for 30 min at the indicated times postinfection. Labeled proteins were analyzed by SDS−10% PAGE. Labeled mock-infected cells are also shown. The positions of the major viral proteins HA, NP, M1, and NS1 are indicated on the left.
DISCUSSION
The NS1 protein of influenza A virus has been implicated in a plethora of functions related to the regulation of gene expression in infected cells. Some of these described functions, such as the inhibition of mRNA polyadenylation, splicing, and nucleocytoplasmic transport, would result in inhibition of gene expression and possibly in shutoff of host protein synthesis (4, 10, 17, 23, 27). Some others, such as the inhibition of PKR (2, 15, 18, 32) and of IFN-α/β-related pathways (28, 30, 33), would result in sustained protein synthesis during viral infection, which otherwise should induce the host IFN-α/β antiviral system. Thus, in the absence of NS1 protein expression, activation of PKR in virus-infected cells would lead to the phosphorylation and inactivation of the translational factor eIF-2α, resulting in protein synthesis inhibition (11). We have investigated here the overall effects of the NS1 protein on cellular and viral gene expression. Transfection experiments resulted in the enhancement of reporter gene expression when reporter plasmids were cotransfected with an NS1 expression plasmid (Fig. 1 to 3). This effect did not appear to be promoter specific, since it was seen with both simian virus 40- and cytomegalovirus-based reporter plasmids.
The NS1-mediated enhancement of reporter plasmid expression was found to occur at a posttranscriptional level (Fig. 3). Interestingly, a translational enhancement activity of the NS1 protein has already been proposed (1, 6, 8). However, it was believed that this effect is specific for a subset of influenza virus mRNAs (8). While we cannot exclude the possibility that the NS1 protein preferentially enhances the translation of some or of all viral mRNAs, our results show that the NS1 protein, in the absence of any other influenza virus protein, functions as a general enhancer of protein translation. In fact, the effects of NS1 expression in reporter gene activity are comparable to or even more dramatic than the effects of a known translational enhancer molecule, the adenoviral VAI RNA (Fig. 2A and 3). Further experiments will be required to completely understand the effects of NS1 in viral and cellular translation, especially as relates to its ability to interact with the eukaryotic initiation factor 4GI (1).
Three different lines of indirect evidence suggest that the NS1-mediated translational enhancement is at least partly a result of the inhibition of PKR activity in transfected cells. First, coexpression of VAI RNA and NS1 protein does not have a cooperative effect in reporter gene expression (Fig. 2A). Second, expression of NS1 partly overcomes the translational inhibition induced by overexpression of PKR (Fig. 2B). Third, a mutant NS1 impaired in its ability to inhibit PKR is also compromised in translational enhancement activity (Fig. 2C). These results are also in agreement with the known anti-PKR activity of the NS1 protein of influenza A virus (2, 15, 18, 32). However, at this point we cannot rule out the involvement of other cellular proteins besides PKR in the translational enhancement mediated by the NS1 protein.
Only recently has it become possible to study the effects of the NS1 protein on viral expression by using recombinant mutant viruses containing selected mutations in their NS1 genes (7, 9, 14, 34). We have taken advantage of different influenza A virus recombinants containing deletions in their NS genes to study the effects of the NS1 protein on protein expression during influenza A virus infection. A number of different viruses, including influenza A viruses, induce cellular protein synthesis shutoff while maintaining viral protein synthesis in infected cells. It has been postulated that the NS1 protein affects cellular mRNA processing and that this protein may be responsible for the influenza virus-induced protein synthesis shutoff (4, 23). Therefore, we investigated the overall levels of cellular protein synthesis in cells infected with wild-type virus, with NS1-99 virus lacking the NS1 carboxy-terminal effector domain postulated to be involved in inhibition of mRNA processing, and with delNS1 virus. In order to distinguish effects due to NS1 protein expression from those due to low levels of viral replication, we used IFN-deficient Vero cells, in which the levels of replication of wild-type and NS1 mutant viruses are comparable (7, 14). Since no major differences in overall protein expression were found among the three viruses, we conclude that the NS1 protein does not play a major role in host cell protein synthesis shutoff. Clearly, some other viral product(s) plays a more prominent role than the NS1 protein in the shutoff of cell protein expression in influenza virus-infected cells. Our results are in agreement with recent findings from Zürcher et al. (34) showing identical levels of protein synthesis shutoff in cells infected with mutant influenza A viruses expressing truncated NS1 proteins. These mutants express NS1 proteins of 81 and 156 amino acids which lack the effector domain. However, Enami and Enami (9) recently reported impaired protein synthesis shutoff in MDBK cells infected with an influenza A virus mutant expressing an NS1 protein of 110 amino acids. The reason for this discrepancy could be that this NS1 truncated protein has unusual characteristics in MDBK cells. Furthermore, this group did not have the benefit of working with a virus lacking the entire NS1 gene, and no experiments were done in IFN-deficient cell systems.
The NS1 protein has also been implicated in the inhibition of mRNA splicing during influenza A virus infection (10, 17). Specifically, it was shown that expression of the NS1 protein results in the inhibition of splicing of the M-specific mRNA in transfection assays, resulting in high levels of unspliced M1 mRNA and low levels of spliced M2 mRNA (17). However, in our experiments no changes in the levels of M1/M2 protein expression were found in cells infected with delNS1 virus, demonstrating that the NS1 protein does not have a major impact on the levels of M1 and M2 protein expression in virus-infected cells.
Despite all of the proposed functions of the carboxy-terminal domain of the NS1 protein in overall cellular mRNA processing, we were unable to detect major changes on overall protein expression in IFN-deficient Vero cells infected with delNS1 virus or with a recombinant virus expressing an NS1 protein lacking the effector domain. In fact, only when COS-7 cells were used did we find that the presence of the NS1 protein is required for preventing a general inhibition of viral protein expression in infected cells. These results are consistent with a role of NS1 in inhibiting antiviral responses in cells having an intact IFN system. Thus, no major effects on viral protein expression are expected when the NS1 protein is expressed in infected Vero cells unable to produce IFN and therefore unable to induce high levels of PKR and other antiviral proteins encoded by IFN-stimulated genes. However, infection of COS-7 cells with delNS1 virus most likely results in the stimulation of IFN synthesis, leading to the transcriptional induction of IFN-stimulated genes, such as PKR, and to the inhibition of protein synthesis. In fact, activation of PKR readily occurs when IFN-competent substrates are infected with delNS1 virus or with NS1 temperature-sensitive (ts) mutant viruses at a nonpermissive temperature (2, 15). The NS1-99 virus displayed an intermediate phenotype between delNS1 and wild-type viruses in COS-7-infected cells. Thus, while HA and M1 protein expression was reduced in cells infected with this virus, NP expression was comparable to that observed in cells infected with wild-type virus. Similar results have been reported with other NS1 mutant viruses (7, 9). Interestingly, we found that the NS1-99 protein was poorly expressed in virus-infected cells, suggesting that this truncated protein is unstable (data not shown). Therefore, we also used in these experiments NS1-126 virus, expressing a stable NS1 protein of 126 amino acids lacking an effector domain (33). The results clearly show that this mutant virus induces a wild-type pattern of viral and cellular protein expression in infected COS-7 cells. Thus, the effect seen in NS1-99 virus-infected cells most likely reflects low levels of NS1 protein expression. Nevertheless, we cannot exclude the possibility that a domain between amino acid residues 99 and 126 is required for efficient expression of the HA and M1 proteins. In any case, it is intriguing that the expression of the late HA and M1 proteins is more sensitive to NS1 regulation than the expression of the early NP protein. A slight defect in M1 protein expression was also observed in Vero cells infected with NS1-99 and delNS1 viruses (Fig. 4). These effects might be attributed to activation of basal levels of PKR in Vero cells and/or to decreased or lack of interaction between NS1 and eIF-4GI, resulting in a reduced translational rate for the M1 mRNA.
The necessity for a virus to take over the cellular synthetic machinery in order to replicate its own genome is balanced by the necessity that the cell maintain a basal level of synthesis sufficient to permit its survival until the end of the replication cycle. The NS1 protein of influenza A virus appears to inhibit PKR and other dsRNA-activated pathways to maintain high levels of viral protein synthesis. This effect is mainly due to the amino-terminal, RNA-binding domain of this protein. Although it is still possible that the expression of selected cellular genes might be inhibited by the NS1 carboxy-terminal (effector) domain, we have disproved a major role of the NS1 protein and its effector domain in the overall inhibition of protein synthesis in cells transfected with an NS1 expression plasmid or in influenza A virus-infected cells. Therefore, we conclude that non-NS1-mediated mechanisms are responsible for the observed generalized host shutoff of cell protein synthesis during influenza virus infection.
Acknowledgments
We thank members of the laboratories of A.G.-S. and P.P. for critical discussions. We are also grateful to Michael G. Katze for providing pcDNAI/Neo-PKR plasmid and to George R. Stark for the U3A cell line.
This work was supported by NIH research grants to P.P. and A.G.-S. M.S. was partly supported by a fellowship from the “Istituto Superiore di Sanitàa” of Italy. C.F.B. was the recipient of an NIH NRSA postdoctoral fellowship.
REFERENCES
- 1.Aragón, T., S. de La Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Z., and R. M. Krug. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376–383. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, C. Y., R. Tejero, Y. Huang, D. E. Zimmerman, C. B. Rios, R. M. Krug, and G. T. Montelione. 1997. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 4:891–895. [DOI] [PubMed] [Google Scholar]
- 6.de la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enami, K., T. A. Sato, S. Nakada, and M. Enami. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enami, M., and K. Enami. 2000. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J. Virol. 74:5556–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortes, P., A. Beloso, and J. Ortín. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale, M. J., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29–46. [DOI] [PubMed] [Google Scholar]
- 12.Gale, M. J., S.-L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A virus and other negative strand RNA viruses. Virology 279:375–384. [DOI] [PubMed] [Google Scholar]
- 14.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. [DOI] [PubMed] [Google Scholar]
- 15.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45:195–200. [DOI] [PubMed] [Google Scholar]
- 17.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817–1828. [DOI] [PubMed] [Google Scholar]
- 18.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228. [DOI] [PubMed] [Google Scholar]
- 19.Luytjes, W., M. Krystal, M. Enami, J. D. Parvin, and P. Palese. 1989. Amplification, expression, and packaging of a foreign gene by influenza virus. Cell 59:1107–1113. [DOI] [PubMed] [Google Scholar]
- 20.Lyles, D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marión, R. M., T. Zürcher, S. de la Luna, and J. Ortín. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447–2451. [DOI] [PubMed] [Google Scholar]
- 22.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 88:11455–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000. [DOI] [PubMed] [Google Scholar]
- 24.O’Malley, R. P., T. M. Mariano, J. Siekierka, and M. B. Mathews. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 44:391–400. [DOI] [PubMed] [Google Scholar]
- 25.Ouchi, T., S. W. Lee, M. Ouchi, S. A. Aaronson, and C. M. Horvath. 2000. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-γ target genes. Proc. Natl. Acad. Sci. USA 97:5208–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, E. J., I. Marié, A. Prakash, A. García-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951–8957. [DOI] [PubMed] [Google Scholar]
- 29.Svensson, C., and G. Akusjarvi. 1985. Adenovirus VA RNAI mediates a translational stimulation which is not restricted to the viral mRNAs. EMBO J. 4:957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talon, J., M. Salvatore, R. E. O’Neill, Y. Nakaya, H. Zheng, T. Muster, A. García-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757–766. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NF-κB and induction of type I IFN. J. Virol. 74:11566–11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zürcher, T., R. M. Marión, and J. Ortín. 2000. Protein synthesis shut-off induced by influenza virus infection is independent of PKR activity. J. Virol. 74:8781–8784. [DOI] [PMC free article] [PubMed] [Google Scholar]