Abstract
A suppressor tRNATyr and mutant tyrosyl-tRNA synthetase (TyrRS) pair was developed to incorporate 3-iodo-l-tyrosine into proteins in mammalian cells. First, the Escherichia coli suppressor tRNATyr gene was mutated, at three positions in the D arm, to generate the internal promoter for expression. However, this tRNA, together with the cognate TyrRS, failed to exhibit suppressor activity in mammalian cells. Then, we found that amber suppression can occur with the heterologous pair of E.coli TyrRS and Bacillus stearothermophilus suppressor tRNATyr, which naturally contains the promoter sequence. Furthermore, the efficiency of this suppression was significantly improved when the suppressor tRNA was expressed from a gene cluster, in which the tRNA gene was tandemly repeated nine times in the same direction. For incorporation of 3-iodo-l-tyrosine, its specific E.coli TyrRS variant, TyrRS(V37C195), which we recently created, was expressed in mammalian cells, together with the B.stearothermophilus suppressor tRNATyr, while 3-iodo-l-tyrosine was supplied in the growth medium. 3-Iodo-l-tyrosine was thus incorporated into the proteins at amber positions, with an occupancy of >95%. Finally, we demonstrated conditional 3-iodo-l-tyrosine incorporation, regulated by inducible expression of the TyrRS(V37C195) gene from a tetracycline-regulated promoter.
INTRODUCTION
The incorporation of unnatural chemical groups into proteins has increasing importance in protein science and cell biology. Biophysical probes or structural modifications have been introduced into proteins by chemical modifications of amino acid residues (1) or by semi-synthetic methods involving protein ligations (2). The biosynthesis of proteins containing unnatural amino acids, alloproteins (3), is also a promising way of expanding the structural and chemical diversity in proteins (3–11). The site-specific incorporation of unnatural amino acids has been employed to study membrane proteins expressed in Xenopus oocytes (6,7). The utility of ‘site-specific alloproteins’ for regulating the interactions between cell signaling proteins has been shown in experiments in vitro (12). However, the only eukaryotic in vivo system available for alloprotein synthesis has been confined to Xenopus oocytes, which has severely limited the yields of alloproteins (7). The availability of alloproteins in mammalian cells should be extended for further applications in cell biology.
Unnatural amino acids have been attached to the specific adaptor tRNAs corresponding to amber codons (4,10,11), four base codons (13) or artificial codons with unnatural bases (14,15). One approach for site-specific-alloprotein synthesis involves the synthesis of the aminoacylated forms of the tRNAs, by chemical acylation (16) or by using an aminoacyl-tRNA synthetase (aaRS) (8,15), before their use in alloprotein synthesis. The adaptor tRNA cannot be reacylated during translation without the specific aaRS for the unnatural amino acid. The use of the tRNA in the aminoacylated form is prohibitive to the large-scale synthesis of alloproteins in vitro or in vivo, or to the conditional incorporation of unnatural amino acids regulated by certain signals, such as a signal for the expression of the specific tRNA and/or aaRS.
Another approach involves the use of specific tRNA·aaRS pairs for the unnatural amino acids, allowing the reacylation of the tRNA. Wang et al. have created two variants of Methanococcus jannaschii tyrosyl-tRNA synthetase (TyrRS) that are highly specific to O-methyl-l-tyrosine (17) and l-3-(2-naphthyl)alanine (18). When these amino acids were supplied in the growth medium, they were actually incorporated at amber positions by these variant enzymes and the cognate suppressor tRNATyr in Escherichia coli cells. The M.jannaschii TyrRS does not recognize E.coli tRNAs, while the E.coli aaRSs do not recognize the suppressor tRNATyr (17). Thus, this archaebacterial tRNATyr·TyrRS pair is ‘orthogonal’ to the E.coli translation system.
To extend this approach to eukaryotic systems, we recently created a 3-iodo-l-tyrosine-specific variant of E.coli TyrRS, with the two amino acid replacements Y37V and Q195C (19). This variant enzyme, TyrRS(V37C195), together with the E.coli suppressor tRNATyr, incorporates 3-iodo-l-tyrosine at amber positions in a wheatgerm cell-free translation (19). To use this system in a mammalian cell, the E.coli TyrRS and the cognate suppressor tRNATyr should be expressed and functional in the cell.
The pair of a suppressor tRNAGln and glutaminyl-tRNA synthetase (GlnRS) from E.coli, which is orthogonal to the eukaryotic translation system, has been expressed in mammalian cells and caused amber suppression (20). The cytidine at position 9 (C9) in this tRNA was replaced by A to generate the internal promoter for expression in mammalian cells. On the other hand, the corresponding engineering for the E.coli tRNATyr would require three base substitutions in the positions involved in the tertiary interactions that support the L-shaped structure, which could impair tRNA function. This difficulty has been circumvented by importing the aminoacylated form of this suppressor tRNATyr, with no such substitutions, into mammalian cells (21).
In the present study, the Bacillus stearothermophilus suppressor tRNATyr was expressed in mammalian cells together with E.coli TyrRS(V37C195), for the incorporation of 3-iodo-l-tyrosine into proteins (Fig. 1); the Bacillus tRNATyr contains the internal promoters in its native sequence (Fig. 2), and can be recognized by E.coli TyrRS (22). Finally, we created a cell line that stably maintains this variant TyrRS gene expressed from a tetracycline-regulated promoter, for conditional incorporation of 3-iodo-l-tyrosine in the presence of this inducer.
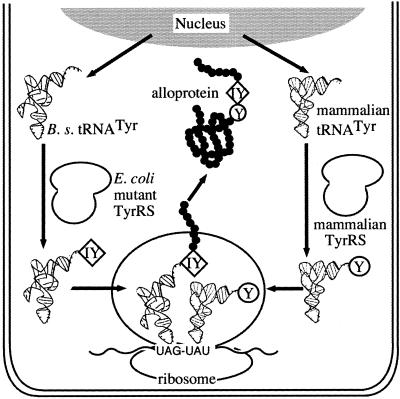
Figure 1.
The mammalian cell system for incorporating 3-iodo-l-tyrosine into proteins in response to amber codons. 3-Iodo-l-tyrosine (IY), present together with l-tyrosine (Y) in the growth medium, is taken up into the cell and is then attached, by its specific E.coli mutant TyrRS, to the B.stearothermophilus (B. s.) suppressor tRNATyr. This tRNA carries this unnatural amino acid to the amber codon on the mRNA and incorporates it into a protein (alloprotein). On the other hand, the endogenous, mammalian tRNATyr·TyrRS pair incorporates l-tyrosine into the proteins at the corresponding tyrosine codon.
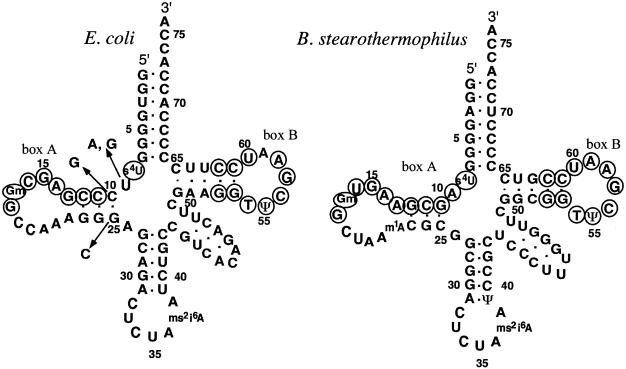
Figure 2.
The suppressor tRNATyr derived from the E.coli tRNA2Tyr (left) and the B.stearothermophilus suppressor tRNATyr (right). The nucleotides corresponding to the consensus internal promoter sequences (box A, TRGCNNAGY for positions 8–16 and G18G19; box B, GGTTCGANTCC for positions 52–62) (25) are circled. The base substitutions made in the E.coli tRNA to generate box A are shown by the arrows. Modified nucleotides found in these tRNAs, prepared from their native organisms, are 4-thiouridine (s4U), 2′-O-methylguanosine (Gm), 2-methylthio-N6-isopentenyladenosine (ms2i6A), 5-methyluridine (T), pseudouridine (ψ) and 1-methyladenosine (m1A) (22).
MATERIALS AND METHODS
tRNA genes for the expression in mammalian cells
The human, E.coli and B.stearothermophilus suppressor tRNATyr genes were constructed by annealing two oligodeoxynucleotides, commercially synthesized by Amersham Pharmacia Biotech; each gene consists of the corresponding tRNA sequence, lacking the 3′-CCA, and the 5′-flanking sequence (AGCGCTCCGGTTTTTCTGTGCTGAACCTCAGGGGACGCCGACACACGTACACGTC) of the human tRNATyr gene (23), linked to the 5′ end of the tRNA sequence. These genes were each cloned within the EcoRI and HindIII sites of pBR322. The gene cluster consisting of nine unidirectional copies of the B.stearothermophilus suppressor tRNATyr gene was constructed by the following two steps (Fig. 3). First, three PCRs, each with distinct sets of primers, were performed, using a GeneAmp PCR system 9700 (Applied Biosystems). The first reaction produced a primer-binding site (pbs1) at one end, upstream of the gene, and a BstXI site, CCAGCAGACTGG (designated BstXI-1), at the other end, downstream of the gene. The second reaction produced the BstXI-1 site and another type of BstXI site, CCAGCTTCCTGG (BstXI-2), at the ‘upstream’ and ‘downstream’ ends, respectively, while the third reaction produced the BstXI-2 site (upstream) and the other primer-binding site, pbs2 (downstream). These PCR products were ligated with each other to generate a sub-cluster consisting of three copies of the tRNA gene. This sub-cluster was amplified with the primers for pbs1 and pbs2, each with an additional sequence for a restriction site, to generate EcoRI and HindIII sites at the ‘pbs1’ and ‘pbs2’ ends, respectively. The HindIII–EcoRI and EcoRI–BamHI sites, each in the order of the pbs1 to pbs2 ends, were also generated similarly. Thus, three types of the sub-cluster with different combinations of the restriction sites were finally produced. These sub-clusters were cloned together within the EcoRI and BamHI sites of pBR322 to generate a plasmid carrying nine copies of the Bacillus suppressor tRNATyr gene.
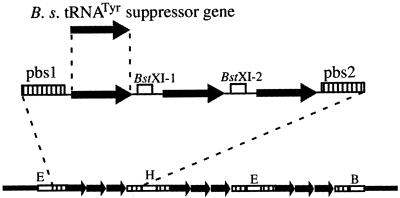
Figure 3.
Construction of a plasmid carrying nine tandem copies of the B.stearothermophilus suppressor tRNATyr. Three copies of this suppressor tRNATyr gene, represented by the arrow, were assembled, by ligation at the BstXI sites (BstXI-1 and BstXI-2), into a sub-cluster with the primer-binding sites (pbs1 and pbs2) at the ends. Three copies of this sub-cluster, each with the EcoRI (E), HindIII (H) or BamHI (B) sites, were then assembled and cloned within the EcoRI–BamHI sites of pBR322.
Construction of TyrRS genes and reporter genes
The FLAG tag (DYKDDDDK) was added to the C-termini of the wild-type TyrRS and TyrRS(V37C195) genes, the ras gene and the epidermal growth factor receptor gene (24) by amplifying these genes with appropriate PCR primers. The PCR products were each cloned into the vector pcDNA3.1/Zeo(+) (Invitrogen) for expression in mammalian cells. For TyrRS(V37C195), the PCR product was also cloned into the vector pcDNA4/TO (Invitrogen) to generate plasmid pEYSM1 for tetracycline-regulated expression. The site-directed mutagenesis of the ras gene was performed by PCR with mutagenic primers. Similarly, the tyrosine codon in position 1068 of the epidermal growth factor receptor was mutated to an amber codon. For the gene encoding a green fluorescent protein (the cyano-fluorescent variant) (Clontech), the first methionine residue was replaced by a short peptide, encoded by ATGGGAACTAGTCCATAGTGGTGGAATTCTGCAGATATCCAGCACAGTGGCGGCCGCGTC (the amber codon is underlined) and the FLAG tag was added to the C-terminus. The resulting gene was cloned into the vector pcDNA3.1/Zeo(+). The sequences of the constructed genes were confirmed using an ABI PRISM 377 DNA sequencer (Applied Biosystems).
Amber suppressions in mammalian cells
Transfections were carried out with 0.5–2 µg of DNA for each plasmid per 35 mm plate, according to the procedure for LipofectAMINE 2000 (Gibco BRL). Opti-MEM I (Gibco BRL) was used as the growth medium. Cell extracts were prepared 24 h after the transfection and were subjected to SDS–PAGE, followed by western blotting using the anti-FLAG M2 antibody (Sigma) and the ECL+ immunodetection system (Amersham Pharmacia Biotech). The band intensity was measured using an image analyzer, LAS-1000plus (Fuji Photo Film Co., Japan). The ras and ras(Am) products (0.5 µg each) were purified, using an anti-FLAG M2 antibody affinity gel (Sigma), from one and five culture plates (100 mm diameter), respectively. The LC-MS analysis and the tandem mass spectrometry sequencing were performed as previously described (15).
The CHO-Y cells
To create the CHO-Y cells, which stably maintain the TyrRS(V37C195) gene, T-REX-CHO cells (Invitrogen), constitutively producing the tetracycline repressor protein, were transfected with the plasmid pEYSM1. The transfectants were selected in growth medium containing 25 µg/ml Zeocin (Invitrogen) and the selected cells were examined for expression of TyrRS(V37C195) in the presence of 1 µg/ml tetracycline, to obtain the CHO-Y cells. For alloprotein synthesis, CHO-Y cells were transfected with the plasmids containing the ras(Am) and Bacillus suppressor tRNATyr genes. Tetracycline (1 µg/ml) and 3-iodo-l-tyrosine (0.3 mM) were added to the medium 24 h after transfection and the cell extracts were prepared after a further 24 h.
RESULTS
Amber suppression in mammalian cells dependent on the expression of a prokaryotic tRNATyr·TyrRS pair
The expression of tRNA in eukaryotic cells requires two internal promoters (boxes A and B) in the tRNA-encoding sequence (25). Since the E.coli tRNATyr sequence contains only box B, U9 and C10 were replaced by A and G, respectively, to generate box A (Fig. 2). The resulting mismatched base pair, G10·G25, was repaired by the substitution G25C. Positions 9, 10 and 25 are involved in the tertiary interactions supporting the L-shaped structure, but are not involved in the identity of E.coli tRNATyr (26). The sequence of tRNATyr(A9G10C25) with a CUA anticodon was linked to the 5′-flanking sequence of the human tRNATyr gene. The human suppressor tRNATyr gene was similarly constructed as a control of amber suppression. To analyze the amber suppression, the tyrosine codon in position 32 in a truncated, synthetic ras gene (27) for the wild-type c-Ha-Ras was mutated to an amber codon. To detect expression, a FLAG peptide tag was added to the C-termini of the ras gene, ras(WT), and its amber derivative, ras(Am), as well as to the C-termini of the E.coli TyrRSs.
These ras genes were introduced into CHO cells and their products were detected by western blotting of the cell extracts with an anti-FLAG antibody (Fig. 4A). In the absence of the suppressor tRNAs, expression of the ras(WT) gene was detected (lane 1), whereas that of ras(Am) was not detected (lane 2), indicating the lack of an intrinsic suppressor activity within the cell. The human suppressor tRNATyr suppressed the amber mutation in ras(Am), with an efficiency of 26%, as determined by the band intensity (lane 3). On the other hand, the E.coli tRNATyr(A9G10C25) with a CUA anticodon, together with the wild-type TyrRS from E.coli, did not cause suppression (lane 4). Then, we tested another E.coli suppressor tRNATyr variant, with the other possible set of nucleotides to generate box A (G9G10C25); this tRNA also failed to cause suppression (data not shown). Thus, the generation of box A in the E.coli suppressor tRNATyr impaired suppression activity, probably through a distortion in the tertiary structure, which may lead to inhibition of the maturation or the aminoacylation of the tRNA.
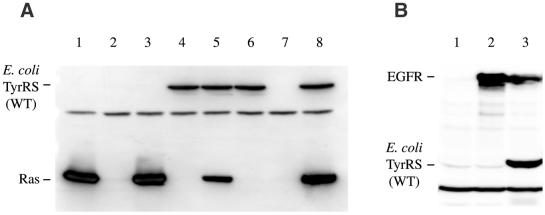
Figure 4.
Western blots with the anti-FLAG antibody for the detection of amber suppression in CHO cells. (A) The ras gene (lane 1) or the ras(Am) gene (lanes 2–8), each with the FLAG tag, was introduced into the cells. The human suppressor tRNATyr (lane 3) or the pair E.coli TyrRS and E.coli suppressor tRNATyr (lane 4) was expressed in the cells. This enzyme also has the FLAG tag. The B.stearothermophilus suppressor tRNATyr was expressed, together with the E.coli TyrRS (lanes 5 and 8) or without the enzyme (lane 7), or the E.coli TyrRS alone was expressed in the cells (lane 6). The B.stearothermophilus suppressor tRNATyr was expressed from the plasmid carrying a single copy of its gene (lanes 5 and 7) and from that carrying nine copies (lane 8). A 2.5 µg aliquot of cell extract was subjected to SDS–PAGE in lane 1, while a 4-fold greater quantity of the cell extract was analyzed in lanes 2–8. The protein detected in all of the cell extracts (lanes 1–8) is an endogenous protein in CHO cells. (B) The epidermal growth factor receptor (EGFR) gene containing an amber codon in position 1068 (lanes 1 and 3) and the wild-type EGFR gene (lane 2), each with a FLAG tag, were introduced into CHO cells. The B.stearothermophilus suppressor tRNATyr and the E.coli TyrRS pair were expressed in the cells (lane 3). The faint band migrating at the level of the E.coli TyrRS (lanes 1 and 2) was due to endogenous proteins that respond to the anti-FLAG antibody.
It has been reported that B.stearothermophilus tRNATyr can be aminoacylated by E.coli TyrRS (22), although this Bacillus tRNATyr has a different set of nucleotides from that in E.coli tRNATyr, in the positions involved in the tertiary interactions, and thus contains both boxes A and B (Fig. 2). We found that co-expression of the B.stearothermophilus suppressor tRNA and the E.coli TyrRS suppressed the amber mutation in ras(Am) with an efficiency of 15% (Fig. 4A, lane 5). This suppression did not occur in the absence of either this tRNA (lane 6) or the enzyme (lane 7). The requirement for E.coli TyrRS for suppression indicates that the Bacillus suppressor tRNATyr is not aminoacylated by the endogenous aaRSs in the cell. On the other hand, the expression of E.coli TyrRS, from the transiently transfected plasmid, by itself hardly changed the growth rate of the CHO cells.
The suppression efficiency for this heterologous pair of tRNATyr·TyrRS was significantly lower than the efficiency for the human suppressor tRNATyr or the efficiencies for other suppressor tRNAs working in mammalian cells (20–40%) (28,29). Since the suppression efficiency can be improved by increasing the dose of the suppressor tRNA gene (30), a plasmid carrying nine copies of the Bacillus suppressor tRNATyr gene was constructed and was introduced into the CHO cells together with the plasmid carrying the E.coli TyrRS gene. The suppression efficiency was thus improved up to 24% (Fig. 4A, lane 8), a value comparable to that for the human suppressor tRNATyr. Hereafter, we used this plasmid for expression of the Bacillus suppressor tRNATyr.
The pair of the Bacillus suppressor tRNATyr and the E.coli TyrRS also suppressed the amber mutation in human embryonic kidney 293 cells, with an efficiency similar to that in the CHO cells (data not shown). Furthermore, an amber mutation in the epidermal growth factor receptor gene was suppressed with an efficiency of 20% (Fig. 4B), while an amber mutation in the Aequorea victoria green fluorescent protein gene was also suppressed with similar efficiency (data not shown). These genes and the ras(Am) gene have different codon contexts around the amber codon.
3-Iodo-l-tyrosine incorporation in response to an amber codon
For 3-iodo-l-tyrosine incorporation into proteins, we considered the following two points. First, mammalian cells have the transport machinery to assimilate canonical amino acids, as well as a variety of their analogs, from the environment (31). Second, the wild-type TyrRSs from eukaryotes and prokaryotes do not recognize 3-iodo-l-tyrosine as a substrate (32–34). Therefore, 3-iodo-l-tyrosine probably has no toxicity caused by its misincorporation at tyrosine positions, while a variant TyrRS that can recognize 3-iodo-l-tyrosine is necessary for its attachment to the suppressor tRNA.
We employed E.coli TyrRS(V37C195), which has been reported to recognize 3-iodo-l-tyrosine efficiently (the kcat/Km value for amino acid activation is 3.3 × 103/M/s) and l-tyrosine 10-fold less efficiently (the kcat/Km is 3.2 × 102/M/s) (19). Furthermore, TyrRS(V37C195) activates l-tyrosine 10 000-fold less efficiently, as compared with the kcat/Km value (2.3 × 106/M/s) for the wild-type enzyme. The wild-type TyrRS and TyrRS(V37C195) were each expressed in CHO cells, together with the Bacillus suppressor tRNATyr and the ras(Am) gene. These enzymes were expressed at similar levels (Fig. 5A and B). In the absence of 3-iodo-l-tyrosine (Fig. 5A), amber suppression was observed for both enzymes, although the yield of the ras(Am) products for TyrRS(V37C195) was only 40% of that for the wild-type enzyme. This indicates that, in the absence of competing 3-iodo-l-tyrosine, TyrRS(V37C195) still recognizes l-tyrosine and incorporates it into the amber position. 3-Iodo-l-tyrosine was then added, to a final concentration of 0.3 mM, in the growth medium, which contained l-tyrosine at a 2-fold higher concentration (Fig. 5B). This concentration of 3-iodo-l-tyrosine had no apparent effect on cell growth. In the presence of 3-iodo-l-tyrosine, the suppression efficiency for TyrRS(V37C195) was improved to a level comparable to that for the wild-type enzyme, suggesting that 3-iodo-l-tyrosine was efficiently taken up by the cell and was incorporated into the protein through recognition by TyrRS(V37C195).
Figure 5.
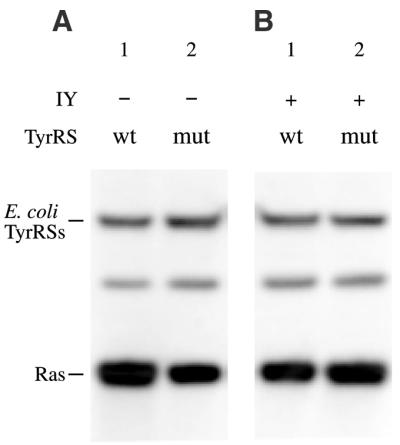
Western blots for the detection of amber suppression in the absence (A) and presence (B) of 3-iodo-l-tyrosine. The ras(Am) gene was introduced into CHO cells (all lanes). The wild-type E.coli TyrRS (lanes 1 in A and B) or TyrRS(V37C195) (lanes 2 in A and B) was expressed in the cells, along with the B.stearothermophilus suppressor tRNATyr. The presence or absence of 3-iodo-l-tyrosine (IY) and the type of expressed TyrRS, wt for wild-type TyrRS and mut for TyrRS(V37C195), are indicated above the lanes.
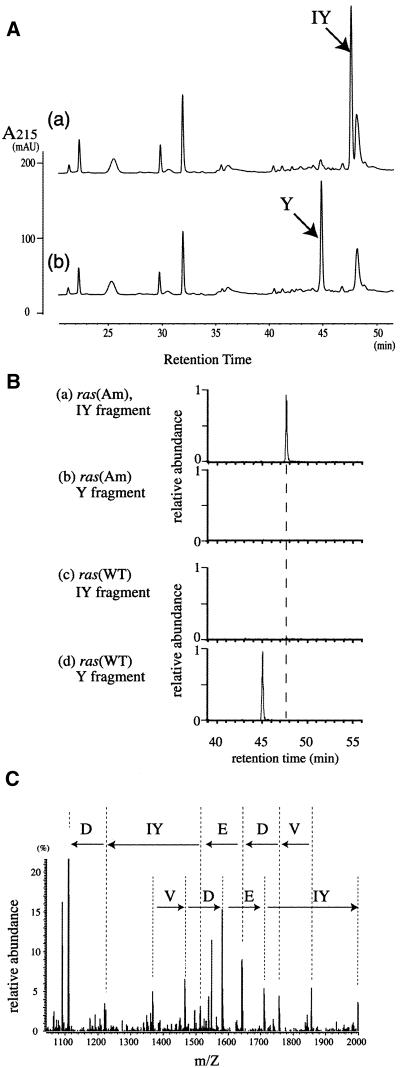
To confirm 3-iodo-l-tyrosine incorporation and to determine its occupancy at the amber position (position 32), the ras(Am) and ras(WT) products were analyzed by LC-MS. These ras products were each synthesized in CHO cells expressing the Bacillus suppressor tRNATyr and TyrRS (V37C195) in the presence of 3-iodo-l-tyrosine. The purified Ras proteins were digested with Achromobacter protease I (Lys-C) and the produced fragments were separated from each other by liquid chromatography (Fig. 6A). The amino acids incorporated at position 32 were analyzed in the mass chromatograms, each for a specific average mass, along with the liquid chromatography retention time. Two fragments, each corresponding to the region from Ser17 to Lys42, contained iodotyrosine and tyrosine at position 32 (designated the IY and Y fragments, respectively).
Figure 6.
LC-MS analyses of the ras and ras(Am) products. (A) The liquid chromatographic profiles, detected with a UV spectrometer, for the fragments from the ras(Am) products (chart a) and the ras products (chart b). (B) The mass chromatograms for the IY fragment (charts a and c) and the Y fragment (charts b and d) from the ras(Am) products (charts a and b) and the ras products (charts c and d). The Y fragment consists of residues 17–42, SALTIQLIQNHFVDEYDPTIEDSYRK, of the Ras protein. Tyr32, underlined in the sequence, was replaced by iodotyrosine in the IY fragment. (C) The tandem mass spectrum of the IY fragment. The partial sequence in the direction from the N-terminus to the C-terminus reads as Val, Asp, Glu, iodotyrosine and Asp in that order.
In the analysis of the ras(Am) products, the IY fragment was strongly observed (Fig. 6B, chart a), indicating efficient incorporation of 3-iodo-l-tyrosine. The partial sequence of this IY fragment, determined by tandem mass spectrometry, confirmed that iodotyrosine was actually incorporated at position 32 (Fig. 6C). On the other hand, no Y fragment was detected (Fig. 6B, chart b), indicating that 3-iodo-l-tyrosine inhibited the incorporation of l-tyrosine at position 32. Further analysis of the LC-MS data showed that no other canonical amino acids were incorporated at this position (data not shown). The occupancy of iodotyrosine at position 32 was thus estimated to be >95% by the present LC-MS analysis. In contrast, for the ras(WT) products synthesized in the presence of 3-iodo-l-tyrosine, the IY fragment was not observed (Fig. 6, chart c), with the expected detection of the Y fragment (chart d). This demonstrates that the endogenous TyrRS in the CHO cell does not recognize 3-iodo-l-tyrosine and TyrRS(V37C195) does not recognize the endogenous tRNATyr, because either misrecognition would incorporate 3-iodo-l-tyrosine in place of tyrosine.
Conditional incorporation of 3-iodo-l-tyrosine regulated by inducible expression of the variant TyrRS
Escherichia coli GlnRS has been expressed in mammalian cells, from a tetracycline-regulated promoter, for induced suppression (35). We created a CHO cell line that stably maintains the TyrRS(V37C195) gene expressed from another type of tetracycline-regulated promoter (designated CHO-YS cells). The ras(Am) gene and the Bacillus suppressor tRNATyr gene were then transiently introduced into the CHO-YS cells. TyrRS(V37C195) was expressed when tetracycline was present in the growth medium (Fig. 7, lanes 1 and 2), but was not expressed without the inducer (lane 3). The expression level was twice the level for TyrRS(V37C195) that was expressed from the plasmid transiently introduced into the cells. The ras(Am) products were detected in the presence of both 3-iodo-l-tyrosine and tetracycline, with a suppression efficiency of 30% (lane 1). The quality of the ras(Am) products, as well as that of ras(WT) produced similarly in CHO-Y cells, was analyzed by LC-MS and was thus shown to be identical to their quality under TyrRS(V37C195) expression from the plasmid; >95% of the ras(Am) products contained 3-iodo-l-tyrosine at the amber position, while 3-iodo-l-tyrosine was not detected in the ras(WT) products (data not shown).
Figure 7.
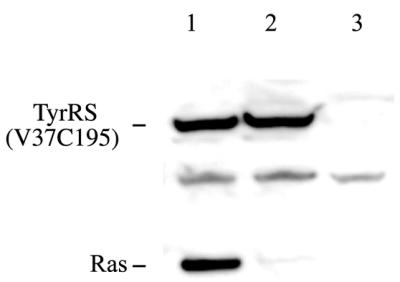
Western blots for detection of inducible amber suppression for 3-iodo-l-tyrosine incorporation into Ras protein. The ras(Am) gene was introduced into CHO-Y cells, together with the Bacillus suppressor tRNATyr gene (lanes 1–3). Tetracycline was added to the growth medium, in the presence (lane 1) or the absence (lane 2) of 3-iodo-l-tyrosine, or neither was added to the medium (lane 3).
On the other hand, the ras(Am) products were hardly detected in the absence of 3-iodo-l-tyrosine (lane 2) and were not detected in the absence of the inducer (lane 3). These observations showed that tetracycline effectively conditioned 3-iodo-l-tyrosine incorporation into the ras(Am) product through induction of TyrRS(V37C195) expression. A comparison of Figure 7 with Figure 5A shows that l-tyrosine incorporation in the absence of 3-iodo-l-tyrosine was significantly lower than that for the case that TyrRS(V37C195) was expressed from the plasmid. This had not been expected, and was observed in more than three independent experiments. The molecular mechanism underlying this phenomenon is a subject of further study.
DISCUSSION
It has been shown that an amber suppressor derived from E.coli tRNAGln can function in mammalian cell, in the presence of its cognate enzyme (20,35). The difficulty of extending this approach to other tRNA·aaRS systems is that many prokaryotic tRNAs do not contain the complete promoter for their expression in eukaryotes. Although some of the residues in the promoter sequence exist in most of the tRNA species, the remaining residues must be generated by base substitutions, which might impair the suppressor activity of the tRNA. In fact, the suppressors derived from E.coli tRNATyr failed to exhibit suppressor activity in the presence of E.coli TyrRS. This difficulty was overcome by utilizing the natural diversity in the tRNA sequences that can be recognized by an aaRS species; B.stearothermophilus suppressor tRNATyr, together with E.coli TyrRS, can cause amber suppression in mammalian cells.
It has been proposed that suppressor tRNAs that can work in mammalian cells are potential gene therapy agents for various human diseases associated with nonsense mutations (36,37). The possible toxicity due to a constitutive suppressor activity may be avoided by the use of an inducible suppression system depending on a tRNA·aaRS pair orthogonal to the mammalian translation system (35). The present study developed a second orthogonal tRNA·aaRS pair functional in mammalian cells. The inducible suppression systems may also be useful for the conditional gene expression required in cell biology studies (38).
Orthogonal tRNA·aaRS pairs are also useful for alloprotein synthesis in living cells, when the aaRS is engineered to be specific for an unnatural amino acid. The genetic code of E.coli has thus been engineered so that tyrosine analogs, assimilated from the growth medium, can be incorporated at the amber positions of proteins (17,18). The present study showed that this approach can be extended to a living eukaryotic system; 3-iodo-l-tyrosine in the growth medium, which also contained l-tyrosine at a 2-fold higher concentration, was actually taken up by the cell and almost exclusively occupied the amber position in the proteins. Although TyrRS(V37C195) expressed from the plasmid incorporates l-tyrosine in the absence of 3-iodo-l-tyrosine, we found that inducible expression of the variant enzyme significantly decreases the level of tyrosine incorporation. Although the feasibility of aaRS engineering has been demonstrated with M.jannaschii and E.coli TyrRSs (17–19), its scope remains to be expanded to include unnatural amino acids that are largely different from the canonical amino acids in their chemical or structural characteristics.
The mammalian system developed in the present study is advantageous for the incorporation of 3-iodo-l-tyrosine into eukaryotic proteins, because some of them are not properly folded into their native conformations in prokaryotic systems. 3-Iodo-l-tyrosine incorporation allows 125I-labeling of proteins for tracing them in cell fractions and may be useful for the X-ray crystallographic analysis of proteins. A phosphorylated 3-iodo-l-tyrosine residue in a peptide is selectively accepted by an engineered Src homology 2 (SH2) domain, but not by the wild-type SH2 domain (manuscript in preparation). Thus, 3-iodo-l-tyrosines inserted within cellular signaling proteins may be utilized to modulate communication between the proteins. In thyroglobulins, the precursors of thyroid hormones, 3-iodo-l-tyosine residues exist naturally, due to post-translational modification. Thus, the genetic assignment of unnatural amino acids will provide a useful tool for regulating the post-translational modification of mammalian proteins.
Acknowledgments
ACKNOWLEDGEMENT
We thank Dr T. Masaki (Ibaraki University) for providing the Achromobacter protease I (Lys-C).
REFERENCES
- 1.Brunner J. (1993) New photolabeling and crosslinking methods. Annu. Rev. Biochem., 62, 483–514. [DOI] [PubMed] [Google Scholar]
- 2.Cotton G.J. and Muir,T.W. (1999) Peptide ligation and its application to protein engineering. Chem. Biol., 6, R247–R256. [DOI] [PubMed] [Google Scholar]
- 3.Koide H., Yokoyama,S., Kawai,G., Ha,J., Oka,T., Kawai,S., Miyake,T., Fuwa,T. and Miyazawa,T. (1988) Biosynthesis of a protein containing a nonprotein amino acid by Escherichia coli: l-2-aminohexanoic acid at position 21 in human epidermal growth factor. Proc. Natl Acad. Sci. USA, 85, 6237–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noren C.J., Anthony-Cahill,S.J., Griffith,M.C. and Schultz,P.G. (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science, 244, 182–188. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson W.A., Horton,J.R. and LeMaster,D.M. (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J., 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak M.W., Kearney,P.C., Sampson,J.R., Saks,M.E., Labarca,C.G., Silverman,S.K., Zhong,W., Thorson,J., Abelson,J.N., Davidson,N. et al. (1995) Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science, 268, 439–442. [DOI] [PubMed] [Google Scholar]
- 7.Dougherty D.A. (2000) Unnatural amino acids as probes of protein structure and function. Curr. Opin. Chem. Biol., 4, 645–652. [DOI] [PubMed] [Google Scholar]
- 8.Yabuki T., Kigawa,T., Dohmae,N., Takio,K., Terada,T., Ito,Y., Laue,E.D., Cooper,J.A., Kainosho,M. and Yokoyama,S. (1998) Dual amino acid-selective and site-directed stable-isotope labeling of the human c-Ha-Ras protein by cell-free synthesis. J. Biomol. NMR, 11, 295–306. [DOI] [PubMed] [Google Scholar]
- 9.Furter R. (1998) Expansion of the genetic code: site-directed p-fluoro-phenylalanine incorporation in Escherichia coli. Protein Sci., 7, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bain J.D., Diala,E.S., Glabe,C.G., Wacker,D.A., Lyttle,M.H., Dix,T.A. and Chamberlin,A.R. (1991) Site-specific incorporation of nonnatural residues during in vitro protein biosynthesis with semisynthetic aminoacyl-tRNAs. Biochemistry, 30, 5411–5421. [DOI] [PubMed] [Google Scholar]
- 11.Short G.F. III, Laikhter,A.L., Lodder,M., Shayo,Y., Arslan,T. and Hecht,S.M. (2000) Probing the S1/S1′ substrate binding pocket geometry of HIV-1 protease with modified aspartic acid analogues. Biochemistry, 39, 8768–8781. [DOI] [PubMed] [Google Scholar]
- 12.Pollitt S.K. and Schultz,P.G. (1998) A photochemical switch for controlling protein-protein interactions. Angew. Chem. Int. Ed., 37, 2104–2107. [DOI] [PubMed] [Google Scholar]
- 13.Murakami H., Hohsaka,T., Ashizuka,Y. and Sisido,M. (1998) Site-directed incorporation of p-nitrophenylalanine into streptavidin and site-to-site photoinduced electron transfer from a pyrenyl group to a nitrophenyl group on the protein framework. J. Am. Chem. Soc., 120, 7520–7529. [Google Scholar]
- 14.Bain J.D., Switzer,C., Chamberlin,A.R. and Benner,S.A. (1992) Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature, 356, 537–539. [DOI] [PubMed] [Google Scholar]
- 15.Hirao I., Ohtsuki,T., Fujiwara,T., Mitsui,T., Yokogawa,T., Okuni,T., Nakayama,H., Takio,K., Yabuki,T., Kigawa,T. et al. (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nature Biotechnol., 20, 177–182. [DOI] [PubMed] [Google Scholar]
- 16.Hecht S.M., Alford,B.L., Kuroda,Y. and Kitano,S. (1978) “Chemical aminoacylation” of tRNA’s. J. Biol. Chem., 253, 4517–4520. [PubMed] [Google Scholar]
- 17.Wang L., Brock,A., Herberich,B. and Schultz,P.G. (2001) Expanding the genetic code of Escherichia coli. Science, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 18.Wang L., Brock,A. and Schultz,P.G. (2002) Adding l-3-(2-naphthyl)alanine to the genetic code of E. coli.J. Am. Chem. Soc., 124, 1836–1837. [DOI] [PubMed] [Google Scholar]
- 19.Kiga D., Sakamoto,K., Kodama,K., Kigawa,T., Matsuda,T., Yabuki,T., Shirouzu,M., Harada,Y., Nakayama,H., Takio,K. et al. (2002) An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA, 99, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drabkin H.J., Park,H. and RajBhandary,U.L. (1996) Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Mol. Cell. Biol., 16, 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhrer C., Xie,L., Kellerer,S., Varshney,U. and RajBhandary,U.L. (2001) Import of amber and ochre suppressor tRNAs into mammalian cells: a general approach to site-specific insertion of amino acid analogues into proteins. Proc. Natl Acad. Sci. USA, 98, 14310–14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedouelle H. (1990) Recognition of tRNATyr by tyrosyl-tRNA synthetase. Biochimie, 72, 589–598. [DOI] [PubMed] [Google Scholar]
- 23.van Tol H., Stange,N., Groos,H.J. and Beier,H. (1987) A human and a plant intron-containing tRNATyr gene are both transcribed in a HeLa cell extract but spliced along different pathways. EMBO J., 6, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J.-H., Saito,K. and Yokoyama,S. (2002) Chimeric receptor analyses of the interactions of the ectodomains of ErbB-1 with epidermal growth factor and of those of ErbB-4 with neuregulin. Eur. J. Biochem., 269, 2323–2329. [DOI] [PubMed] [Google Scholar]
- 25.Sprague K.U. (1994) Transcription of eukaryotic tRNA genes. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. AMS Press, Washington, DC, pp. 31–50.
- 26.Pallanck L., Pak,M. and Schulman,L. (1994) tRNA discrimination in aminoacylation. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. AMS Press, Washington, DC, pp. 371–394.
- 27.Ha J.-M., Ito,Y., Kawai,G., Miyazawa,T., Miura,K., Ohtsuka,E., Noguchi,S., Nishimura,S. and Yokoyama,S. (1989) Conformation of guanosine 5′-diphosphate as bound to a human c-Ha-ras mutant protein: a nuclear Overhauser effect study. Biochemistry, 28, 8411–8416. [DOI] [PubMed] [Google Scholar]
- 28.Young J.F., Capecchi,M., Laski,F.A., RajBhandary,U.L., Sharp,P.A. and Palese,P. (1983) Measurement of suppressor transfer RNA activity. Science, 221, 873–875. [DOI] [PubMed] [Google Scholar]
- 29.Capone J.P., Sedivy,J.M, Sharp,P.A. and RajBhandary,U.L. (1986) Introduction of UAG, UAA and UGA nonsense mutations at a specific site in Escherichia coli chloramphenicol acetyltransferase gene: use in measurement of amber, ochre and opal suppression in mammalian cells. Mol. Cell. Biol., 6, 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buvoli M. Buvoli,A. and Leinwand,L. (2000) Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol. Cell. Biol., 20, 3116–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen H.N. (1990) Role of amino acid transport and countertransport in nutrition and metabolism. Physiol. Rev., 70, 43–77. [DOI] [PubMed] [Google Scholar]
- 32.Calendar R. and Berg,P. (1966) The catalytic properties of tyrosyl ribonucleic acid synthetases from Escherichia coli and Bacillus subtilis. Biochemistry, 5, 1690–1695. [DOI] [PubMed] [Google Scholar]
- 33.Scherberg N.H., Seo,H. and Hynes,R. (1978) Incorporation of radioiodotyrosines into proteins formed during cell-free translation. J. Biol. Chem., 253, 1773–1779. [PubMed] [Google Scholar]
- 34.Ohno S., Yokogawa,T. and Nishikawa,K. (2001) Changing the amino acid specificity of yeast tyrosyl-tRNA synthetase by genetic engineering. J. Biochem. (Tokyo), 130, 417–423. [DOI] [PubMed] [Google Scholar]
- 35.Park H. and RajBhandary,U.L. (1998) Tetracycline-regulated suppression of amber codons in mammalian cells. Mol. Cell. Biol., 18, 4418–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temple G.F., Dozy,A.M., Roy,K.L. and Kan,Y.W. (1982) Construction of a functional human suppressor tRNA gene: an approach to gene therapy for β-thalassaemia. Nature, 296, 537–540. [DOI] [PubMed] [Google Scholar]
- 37.Atkinson J. and Martin,R. (1994) Mutations to nonsense codons in human genetic disease: implications for gene therapy by nonsense suppressor tRNAs. Nucleic Acids Res., 22, 1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunes S. and Steller,H. (1991) Ablation of Drosophila photoreceptor cells by conditional expression of a toxin gene. Genes Dev., 5, 970–983. [DOI] [PubMed] [Google Scholar]