Abstract
The Cdc24 protein plays an essential role in chromosomal DNA replication in the fission yeast Schizosaccharomyces pombe, most likely via its direct interaction with Dna2, a conserved endonuclease–helicase protein required for Okazaki fragment processing. To gain insights into Cdc24 function, we isolated cold-sensitive chromosomal suppressors of the temperature-sensitive cdc24-M38 allele. One of the complementation groups of such suppressors defined a novel gene, pfh1+, encoding an 805 amino acid nuclear protein highly homologous to the Saccharomyces cerevisiae Pif1p and Rrm3p DNA helicase family proteins. The purified Pfh1 protein displayed single-stranded DNA-dependent ATPase activity as well as 5′ to 3′ DNA helicase activity in vitro. Reverse genetic analysis in S.pombe showed that helicase activity was essential for the function of the Pfh1 protein in vivo. Schizosaccharomyces pombe cells carrying the cold-sensitive pfh1-R20 allele underwent cell cycle arrest in late S/G2-phase of the cell cycle when shifted to the restrictive temperature. This arrest was dependent upon the presence of a functional late S/G2 DNA damage checkpoint, suggesting that Pfh1 is required for the comple tion of DNA replication. Furthermore, at their permissive temperature pfh1-R20 cells were highly sensitive to the DNA-alkylating agent methyl methanesulphonate, implying a further role for Pfh1 in the repair of DNA damage.
INTRODUCTION
The study of chromosomal DNA replication in eukaryotic cells has greatly benefited from the amenability of the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe to genetic analysis. Collections of conditional-lethal cell division cycle (cdc) mutants generated by random mutagenesis of both yeasts have played an important role in facilitating the identification of a large number of protein factors essential for different stages of the replication process. In S.pombe, for example, genes encoding all three essential replicative DNA polymerases have been identified in this way (reviewed in 1).
In S.pombe the cdc24-M38 mutant was initially isolated as one of the cdc mutants (2). Cells carrying this temperature-sensitive mutation halt cell cycle progression when shifted to the restrictive temperature, becoming arrested in late S-phase with incompletely replicated DNA (2–4). The cdc24+ gene encodes an essential 501 amino acid protein with no apparent homologs in other species (3,4).
Two lines of evidence indicate possible interactions between Cdc24 and DNA polymerase δ (Pol δ). First, Cdc24 physically and genetically interacts with two accessory proteins for DNA polymerase δ: the large subunit of replication factor C (RF-C) and proliferating cell nuclear antigen (PCNA), encoded by rfc1+ and pcn1+, respectively (4–6). Secondly, overexpression of the cdm1+ gene, encoding a non-essential subunit of Pol δ, suppresses the temperature-sensitive lethality of the cdc24-M38 mutation (7).
The dna2+ gene was also identified as a multicopy suppressor of a temperature-sensitive cdc24 mutant (3). dna2+ interacts genetically, not only with cdc24+, but also with cdc1+ and cdc27+ (encoding the essential small subunits of Pol δ), cdc17+ (encoding S.pombe DNA ligase I), and rad2+ (encoding the S.pombe homolog of the nuclease Fen-1) (8). Fen-1 is required for the Okazaki fragment processing in the mammalian SV40 in vitro DNA replication system (9). Dna2 binds to Cdc24 strongly in the yeast two-hybrid assay system (8). This helicase–endonuclease is required for DNA replication, but not for the bulk DNA synthesis (8). This phenotype is the same as those of cdc24 and cdc17.
In addition to having weak DNA helicase activity (10,11), budding yeast Dna2p also has single-stranded DNA-specific endonuclease activity (12). Indeed, in vitro studies have suggested that Dna2 acts as a nuclease in Okazaki fragment maturation during lagging strand DNA synthesis (13,14, reviewed in 15). Taken together, these genetic and biochemical results suggest the most likely function of Cdc24 is in the process of Okazaki fragment maturation, perhaps as a regulator of Dna2 activity, or as an adaptor molecule, linking Dna2 to Pol δ.
DNA helicase molecules play important roles in various DNA transactions including DNA replication, recombination and repair (16–18). In budding yeast, in addition to Dna2p, at least three other DNA helicases appear to be involved in DNA replication. The MCM helicase complex has DNA helicase activity and is proposed to act by unwinding duplex DNA at replication origins (19). In addition, this complex moves with replication fork after initiation of DNA synthesis (20), and is also required for the elongation phase of DNA synthesis (21). The MCM protein complex is therefore a good candidate for the replicative helicase that unwinds DNA duplex ahead of the moving replication fork. Two additional S.cerevisiae DNA helicases, Pif1p and Rrm3p, are also likely to have functions in the DNA replication (reviewed in 22). Pif1p is 5′ to 3′ DNA helicase (23,24) that functions both in the nucleus and in mitochondria. In the nucleus, Pif1p inhibits both de novo telomere formation and telomere elongation (25,26) and in this way protects cells against gross chromosome rearrangements (27). In the mitochondria, Pif1p is involved both in mtDNA repair and recombination (23,28). Rrm3p, a putative DNA helicase protein highly homologous to Pif1p, is suggested to be a replicative DNA helicase specific for ribosomal DNA (29). Interestingly, Pif1p and Rrm3p have opposite functions in replication fork movement at rDNA (29). As a result Pif1p and Rrm3p are thought to have similar substrate specificities but to exert different effects on them. The Pif1p/Rrm3p subfamily of DNA helicases is conserved from yeast to human (26). However there are no reports of detailed analysis of Pif1p subfamily helicases other than in S.cerevisiae.
Here we report the isolation of the S.pombe pfh1+ gene, which encodes a DNA helicase homologous to Pif1p subfamily DNA helicases, as a suppressor mutation of the cdc24-M38 mutant. We present evidence that Pfh1 DNA helicase is essential for cell viability (in contrast to the case of S.cerevisiae homologs Pif1p and Rrm3p) and that Pfh1 is required for cell cycle progression in late S-phase and for the proper response to DNA-damaging agents. These results lead us to propose that Pfh1 performs essential functions in both chromosomal DNA replication and DNA repair in S.pombe.
MATERIALS AND METHODS
Fission yeast strains and media
The strains of S.pombe used in this study are listed in Table 1. Media were prepared as described previously (30–33).
Table 1. Strains used in this study.
| Strain name | Genotype |
|---|---|
| EV3A | h– leu1-32 |
| M131 | h– cdc24-M38 leu1-32 |
| K164-9 | h– cdc25-22 |
| H303-12 | h– pfh1-R20 leu1-32 |
| H276-9 | h– pfh1-R23 leu1-32 |
| E29 | h– pfh1 << pfh1-GFP-ura4+ ura4-D18 leu1-32 |
| E39 | h– rad1-1 leu1-32 |
| H329 | h+/h– pfh1+/pfh1::ura4+ ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 |
| U1-2 | h+/h– ade6-M210/ade6-M216 ura4+/ura4-D18 leu1-32/leu1-32 |
Vectors and libraries
The pALSK+, pcL and pREP vectors were described previously (34–36). The S.pombe cDNA library used was described previously (4). Schizosaccharomyces pombe genomic DNA libraries were constructed by inserting restriction enzyme-digested wild-type DNA into pALSK+ vector.
Isolation of cdc24-M38 suppressor mutants
Approximately 2.9 × 109 exponentially growing h– cdc24-M38 leu1-32 cells at 23°C in YE (32) medium were plated onto YE plates (0.5–2.0 × 107 cells per plate) and incubated at 36°C for 3 days. Spontaneous revertant colonies were then replica plated and incubated at 18°C for 3–5 days to test for cold sensitivity. Colonies that showed cold sensitivity (cs) were selected and the suppression and cold sensitivity were confirmed by streaking onto YE plates and incubating at 36 and 18°C. To test whether both the cdc24-suppressor and cs phenotypes were caused by a single gene mutation, mutants were back-crossed with cdc24-M38 mutant and examined for the temperature and cold sensitivities. To test whether the suppressing mutations were intra or extragenic, they were crossed with h+S leu1 and analyzed. To classify into complementation groups, we generated h+S cdc24-M38 pfh1-R23 mutant, crossed with other cdc24 suppressor mutants and analyzed by random spore analysis.
Flow cytometry
Flow cytometry was performed as described previously (37) by using the FACScan system, the CellFIT cell cycle analysis program, and LYSIS II software (Becton Dickinson).
Pulsed-field gel electrophoresis
Agarose plugs were prepared as described previously (4). Pulsed-field gel electrophoresis was carried out in a 0.7% agarose gel with a Gene Navigator (Pharmacia) apparatus for 80 h in 0.5× TAE buffer at 50 V with a switching time of 30 min.
Isolation of pfh1+ genes
The pfh1-R20 and pfh1-R23 mutant cells were transformed with S.pombe cDNA and genomic DNA libraries (33). The cells were incubated on MMA (33) plates at 34°C for 16 h and 18°C for 10–14 days. Plasmids were recovered from transformants and confirmed for their suppression activity. Both pfh1+ cDNA and genomic DNA were isolated.
Gene disruption
A null mutant of pfh1+ was constructed as follows. The internal 1.5 kb EcoRV fragment containing 60% of the pfh1+ coding region was replaced with the 1.8 kb HindIII-excised ura4+ gene. The 6.2 kb MscI fragment containing disrupted pfh1 gene was transfected into a diploid strain. Stable ura+ transformants were obtained, and successful disruptants were identified by Southern blot analysis.
Analysis of germinating spores
Spores were isolated by treatment with helicase (32,38). Diploid strains were sporulated on MEA (32) plates for 7 days at 30°C. Spores formed were washed with water and incubated at 30°C for 18 h in 40 ml of water containing 0.1 ml of helicase (IBF). After extensive washing with water, the spores were cultured in minimal medium containing adenine and leucine at 30°C to induce preferential germination of pfh1Δ (ura+) cells. Germinating cells were analyzed by 4′,6-diamidino-2-phenylindole (DAPI) staining and flow cytometric analysis.
Cell cycle northern analysis
The cell cycle was synchronized by the cdc25-22 block and release method as described previously (30). Northern blot analysis was performed as described previously (39).
Plasmids for overexpression of wild-type and mutant Pfh1 proteins
An EcoRI restriction site was introduced immediately upstream of the pfh1+ open reading frame (ORF) by polymerase chain reaction (PCR). After subcloning into pGBT9 (Clontech), the amplified DNA sequences were confirmed by sequencing. Next, plasmid pGBT9-Pfh1 containing the intact pfh1+ ORF and 3′-UTR between EcoRI–BamHI sites was first digested with PstI, and the linearized plasmid was then partially digested with EcoRI. The PstI and EcoRI fragment containing the full-length pfh1+ was isolated and ligated into PstI/EcoRI-cleaved pET43.1a (Novagen) to obtain pET43-Pfh1. This procedure resulted in the fusion of sequences encoding the NusA protein to the 5′ end of the pfh1+ ORF. In this construct, six histidine residues are also present between NusA and Phf1 protein sequences, which facilitates the detection and purification of the recombinant protein. A single amino acid change (lysine into glutamate at amino acid position 338) was introduced into the pfh1+ ORF in pET43-Pfh1 using the Quick-Change™ Site Directed Mutagenesis Kit (Stratagene). The site-directed mutagenesis for the single amino acid change was performed according to the manufacturer’s (Stratagene) recommendation using two oligonucleotides: 5′-GC TCT GCT GGA ACA GGT GAA TCT GTT CTC CTC CG-3′ and 5′-CG GAG GAG AAC AGA TTC ACC TGT TCC AGC AGA GC-3′. The underlined nucleotides denote the mutagenic change of nucleotides. The resulting plasmid was named pET-Pfh1K338E.
Purification of the recombinant Pfh1 protein
Escherichia coli BL21 harboring either pET43-Pfh1 or pET-Pfh1K338E was grown to saturation in 5 ml of LB medium containing 50 µg/ml ampicillin. The preparative medium (500 ml of LB, 50 µg/ml ampicillin) was inoculated with 1 ml of the seed culture. The culture was then incubated at 18°C with vigorous shaking (250 r.p.m.) to an OD600nm = 0.5–0.6, followed by the addition of 1 mM IPTG and incubation for an additional 18 h. Cells were harvested by centrifugation (8600 g, 7 min) and the cell pellet was resuspended in 30 ml of lysis buffer (25 mM Tris–HCl pH 7.5, 10% glycerol, 500 mM NaCl, 10 mM imidazole, 1 mM PMSF, 1 mM benzamidine, 1.25 µg/ml leupeptin and 0.625 µg/ml pepstatin A). Cells were disrupted by sonication (seven cycles of a 1 min pulse and a 2 min cooling interval). The extracts were cleared by centrifuging at 37 000 r.p.m. for 1 h in a Beckman 45 Ti rotor and the supernatant (4.2 mg/ml, 30 ml) was loaded onto a Ni2+-charged HiTrap-chelating column (1 ml; Pharmacia Biotech). The column was washed with 10 column volumes of lysis buffer, and eluted with 10 ml of elution buffer (80 mM imidazole, 25 mM Tris–HCl pH 7.5, 10% glycerol, 500 mM NaCl, 1 mM PMSF, 1 mM benzamidine, 1.25 µg/ml leupeptin, 0.625 µg/ml pepstatin A and 0.02% NP-40). The eluted fractions (1.4 mg/ml, 10 ml) were pooled and dialyzed against T buffer (25 mM Tris–HCl pH 7.5, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine, 1.25 µg/ml leupeptin, 0.625 µg/ml pepstatin A and 0.02% NP-40) containing 300 mM NaCl (buffer T300; hereafter, the number indicates the concentration of NaCl added to the buffer T). The dialysate was then loaded onto a HiTrap heparin column (1 ml; Pharmacia Biotech). The column was washed with 10 ml of buffer T300, and eluted with a linear gradient (300–700 mM) of NaCl in buffer T (15 ml). Fractions containing the DNA-dependent ATPase activity were pooled and concentrated 10-fold. Aliquots (0.4 ml) were loaded onto a Superdex 200 HR 10/30 column (24 ml; Pharmacia Biotech). The column was eluted with buffer T500, and the fractions were assayed for DNA helicase and single-stranded DNA dependent ATPase activities (see below). The active fractions (>95% in purity) were stored at –80°C. The mutant recombinant Pfh1 protein was purified with the same procedure without enzymatic assays.
Preparation of helicase substrates
DNA substrates used to measure helicase activity of Pfh1 were constructed by hybridizing an oligonucleotide (5′-CGG ACG CTC GAC GCC ATT AAT AAT GTT TTC-3′) to ΦX174 single-stranded circular (ssc) DNA and purified as described (40). The oligonucleotide-based partial duplex substrates to determine the unwinding direction of Pfh1 were prepared as described previously (12). Briefly, oligonucleotides H90 (5′-TGG GCT CAC GTG GTC GAC GCT GGA GGT GAT CAC CAG ATG ATT GCT AGG CAT GCT TTC CGC AAG AGA ACG GGC GTC TGC GTA CCC GTG CAG-3′) and H3 (5′-CTG CAC GGG TAC GCA GAC GCC-3′) were annealed to construct a 5′-overhang partial duplex DNA substrate; oligonucleotides H90 and H5 (5′-CAG CGT CGA CCA CGT GAG CCC-3′) were used to assemble a 3′-overhang substrate. Both substrates contain a 21-bp duplex DNA region. Labeling of the 5′ ends of oligonucleotides was performed by incorporating [γ-32P]ATP with T4 polynucleotide kinase. The substrates were gel-purified prior to use: their specific activities were within 1000–2000 c.p.m./fmol.
ATPase and helicase assays
DNA-dependent ATPase activity was measured in a reaction mixture (20 µl) containing 50 mM Tris–HCl pH 8.5, 2 mM DTT, 2 mM MgCl2, 0.25 mg/ml bovine serum albumin (BSA), 150 µM cold ATP, 8.25 nM [γ-32P]ATP (>3000 Ci/mmol), and 50 ng of M13 sscDNA when necessary. After incubation at 37°C for 10 min, an aliquot (1 µl) was spotted onto a polyethyleneimine–cellulose plate (J. T. Baker, USA) and developed in 0.5 M LiCl/1.0 M formic acid. The products were analyzed using a phosphoimager (Bio-Rad).
Helicase activity was measured in a reaction mixture (20 µl) containing 50 mM Tris–HCl pH 8.5, 2 mM DTT, 2 mM MgCl2, 2 mM ATP, 0.25 mg/ml BSA, and the 5′-32P labeled partial duplex DNA substrate (15 fmol). Reactions were incubated at 37°C for 10 min. Reactions were stopped with 4 µl of 6× stop solution [60 mM EDTA pH 8.0, 40% (w/v) sucrose, 0.6% SDS, 0.25% bromophenol blue and 0.25% xylene cyanol]. The reaction products were subjected to electrophoresis for 0.5 h at 150 V through a 12% polyacrylamide gel containing 0.1% SDS in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). The gel was dried on DEAE–cellulose paper and autoradiographed. Labeled DNA products were quantified with the use of a phosphoimager.
Construction of pREP81-pfh1 K338R
To construct pREP81-pfh1, an NdeI restriction enzyme recognition site was introduced immediately upstream of the pfh1+ ORF by PCR and subcloned into pREP81 expression vector. pREP81-pfh1 K338R, lysine to arginine substitution (AAA to AGA) at amino acid position 338, was generated by PCR-mediated site-directed mutagenesis. This mutation generated a BglII restriction site. The DNA sequences amplified by PCR were confirmed by DNA sequencing.
Construction of pfh1-GFP strain
Appropriate restriction enzyme recognition sites were introduced to flanking sequences of pfh1 and the EGFP from pEGFP-N1 (Clontech) by PCR. Sequences encoding the C-terminal half of Pfh1 (1.1 kb EcoRI–NotI fragment), EGFP and the nmt1 downstream sequences (transcriptional terminator) from pREP41 were subcloned into pBS-ura4+ plasmid. The DNA sequences amplified by PCR were confirmed by DNA sequencing. The plasmid obtained by this method was then linearized by XbaI digestion, and transformed into h– ura4-D18 leu1-32 cells. Stable ura+ tranformants were selected and homologous recombination at the pfh1+ locus was confirmed by PCR.
RESULTS
Isolation of pfh1 mutants as cdc24 suppressor mutants
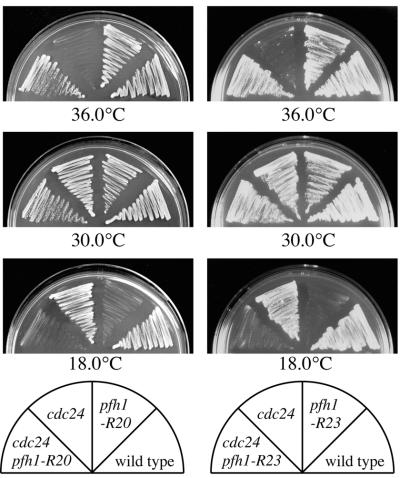
To isolate new factors interacting genetically with cdc24+, we set out to isolate cdc24 suppressor mutants using the temperature-sensitive allele cdc24-M38. The nonsense mutation in cdc24-M38 produces a C-terminally truncated 369 amino acid protein instead of the full-length 501 amino acid protein (3). Exponentially growing cdc24-M38 cells were spread onto YE plates and incubated at 36°C to select for spontaneous revertant colonies. To allow the cloning of mutated genes by complementation and to facilitate further genetic analysis, we selected colonies that possessed the additional property of cold sensitivity together with the suppression of temperature sensitivity of cdc24-M38 by microscopic examination following replica plating. In this way we isolated 37 clones that can grow at 36°C but not at 18°C (see Materials and Methods). For example, Figure 1 shows two of these mutations, pfh1-R20 and pfh1-R23, that suppressed the temperature sensitivity of the cdc24-M38 mutation and gained cold sensitivity (Fig. 1).
Figure 1.
Suppression of the temperature sensitivity of the cdc24-M38 by the pfh1-R20 and pfh1-R23 mutations. The indicated cells were streaked on YE plates and incubated for 3 days at 30°C, 3 days at 36°C or 5 days at 18°C, respectively.
In order to identify the genes responsible for these suppressor mutants, we screened the S.pombe DNA libraries that complemented the cold sensitivity of these mutants. Since we isolated a novel DNA helicase gene, pfh1+ (for PIF1 helicase homolog) among the isolated genes (see below), we selected this gene and corresponding mutants for further analysis.
pfh1 mutant cells arrest their cell cycle with 2C DNA content
By classification into complementation groups, we found that five independent pfh1 mutants including pfh1-R20 and pfh1-R23 were isolated as cdc24 suppressor mutants. The analysis of the other 32 cdc24 suppressor mutants will be described elsewhere.
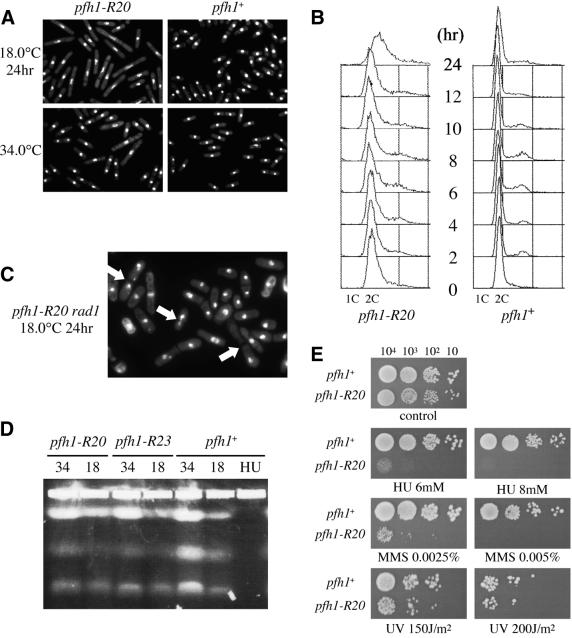
To analyze Pfh1 function by using pfh1 mutant cells, we first generated pfh1-R20 and pfh1-R23 single mutants by genetic crossing and tetrad dissection. These two single pfh1 mutants were still cold sensitive for growth (Fig. 1). To analyze the phenotype of these mutants, exponentially growing cells at 34°C were shifted down to 18°C and analyzed by DAPI staining. At 34°C, pfh1-R20 cells were slightly elongated compared with pfh1+ cells (Fig. 2A), suggesting that the Pfh1-R20 mutant protein has reduced activity even at the permissive temperature of 34°C. After the temperature shift to 18°C, pfh1-R20 cells ceased cell proliferation and arrested with elongated morphology and a single nucleus (Fig. 2A). This phenotype suggested that the pfh1+ gene was required for cell cycle progression. To analyze the pfh1-R20 arrest point in the cell cycle, DNA content was analyzed by flow cytometry. After the temperature shift to 18°C, pfh1-R20 cells arrested with a 2C DNA content. No obvious delay in S-phase was apparent (Fig. 2B). Thus, pfh1-R20 mutant cells appear to arrest at late S- or G2-phase of the cell cycle. Similar results were obtained from the analysis of the second pfh1 allele, pfh1-R23 (data not shown).
Figure 2.
Properties of pfh1-R20 cells. (A) Morphology of pfh1-R20 cells. pfh1-R20 and pfh1+ cells were grown to mid-log phase in minimal medium at 34°C and shifted down to 18°C. Cells were collected at 24 h after temperature shift, fixed with 70% ethanol and stained with DAPI. (B) Flow cytometric analysis of pfh1-R20 cells. Exponentially growing cells at 34°C were shifted down to 18°C as indicated in (A). Samples were taken at indicated times after temperature shift and analyzed by flow cytometry. (C) DAPI staining of pfh1-R20 rad1-1 cells. pfh1-R20 rad1-1 cells from microcolonies formed at 34°C were inoculated on YE plates and incubated at 18°C for 24 h. The cells were then fixed with 70% ethanol and stained with DAPI. Arrows indicate cells that underwent aberrant mitosis. (D) Pulsed-field gel electrophoresis of the chromosomes. Indicated strains growing exponentially at 34°C in minimal medium supplemented with leucine were shifted down to 18°C and incubated for 16 h. Samples were prepared from these cells and exponentially growing cells at 34°C. HU indicates wild-type cells treated with 12 mM HU at 34°C for 4 h. Chromosomes were separated with pulsed-field gel electrophoresis. (E) pfh1-R20 cells are sensitive to MMS and HU. Approximately 104, 103, 102 and 10 cells (from left to right) of exponentially growing pfh1-R20 and wild-type cells in YEL (32) at 34°C were spotted in YE plates or YE plates containing MMS (0.0025, 0.005%), HU (6, 8 mM) or irradiated by UV (150, 200 J/m2). Plates were incubated at 34°C for 2 days (without drug) or 3 days (with drug or UV irradiated).
The absence of pfh1+ triggers the late S/G2 DNA damage checkpoint
To investigate whether pfh1+ is required for the completion of S-phase, we attempted to analyze the phenotype of a pfh1 rad1 double mutant. Rad1 is required for DNA replication/damage checkpoint function (41,42): the rad1-1 mutant fails to arrest the cell cycle when DNA is unreplicated or damaged. If pfh1+ is required for the completion of DNA replication, pfh1 mutant cells would be expected to enter mitosis when combined with the rad1-1 mutation. To test this, we generated pfh1-R20 rad1-1 double mutants by tetrad analysis. However, we found that pfh1-R20 rad1-1 cells grew very slowly, forming only microcolonies at 34°C. These cells were streaked on YE plates and incubated at 18°C for 24 h and examined by DAPI staining. Cells displaying nuclear abnormalities suggestive of inappropriate entry into mitosis were frequently observed (Fig. 2C). These phenotypes were also observed in cells grown even at a permissive temperature of 34°C. Similar results were obtained from the analysis of pfh1-R23 rad1-1 cells (data not shown). These results strongly suggest that pfh1+ is required for S-phase completion. To investigate this further, we constructed pfh1 double mutants with Δchk1 and Δcds1 mutants (43–45). Both Chk1 and Cds1 are downstream kinases of the DNA replication/damage checkpoint pathway. By the analysis of these mutant cells at 18°C, we found that checkpoint arrest required Chk1 but not Cds1: elongated cells were not observed in pfh1-R20 Δchk1 or in pfh1-R23 Δchk1, whereas pfh1-R20 Δcds1 and pfh1-R23 Δcds1 cells were elongated at 18°C (data not shown). This result suggested that loss of Pfh1 function triggers a chk1+-dependent late S/G2 DNA damage checkpoint pathway. Similar checkpoint dependencies were reported with other S-phase cell division cycle mutants, such as pol3 (46) and cdc17 (43–45).
The chromosomes of pfh1 mutant cells were analyzed by pulse-field gel electrophoresis in which incompletely replicated chromosomes fail to enter the gel. In contrast to hydroxyurea (HU)-arrested cells, the chromosomes of pfh1-R20 and pfh1-R23 cells cultured at a restrictive temperature of 18°C entered the gel (Fig. 2D), suggesting that the chromosomal defects that accumulate in pfh1 mutant cells at the restrictive temperature, while sufficient to activate the S/G2 checkpoint, are insufficient to prevent the chromosomal DNA from entering the gel.
The pfh1 mutant is sensitive to methyl methanesulphonate (MMS) and HU
To analyze further the function of pfh1+, we tested the sensitivity of pfh1-R20 and pfh1-R23 mutants to the DNA replication inhibitor HU and the DNA-damaging agents MMS and UV. This was achieved by spotting cells onto YE plates containing HU or MMS, or by irradiating cells spotted onto YE plates with UV. As shown in Figure 2E, pfh1-R20 mutant cells showed only a slightly increased sensitivity to UV compared with pfh1+ cells but were highly sensitive to both HU and MMS. Similar results were obtained with pfh1-R23 cells (data not shown). Therefore, the pfh1+ gene was required for the proper response to nucleotide pool depletion (the consequence of HU addition) and for repairing DNA damage caused by MMS.
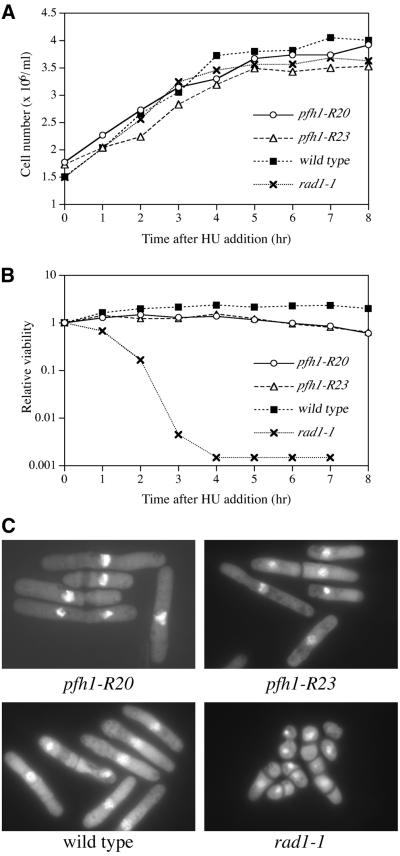
There are two possible explanations for the sensitivity to HU. The mutant cells may be inherently checkpoint defective, i.e. entry into mitosis may result in the presence of HU, with lethal consequences. Alternatively, the cells may arrest normally in HU, but be unable to recover from the arrest to resume normal growth. In order to distinguish between these possibilities, we exposed pfh1 mutant cells to HU transiently and investigated their viability and morphology over a period of time (Fig. 3A–C). As shown in Figure 3A, cell number increase in the pfh1-R20 and pfh1-R23 cultures ceased following HU exposure: the elongated morphology of pfh1-R20 and pfh1-R23 cells at 6 h incubation in HU indicated that checkpoint function was intact in these cells, in contrast to rad1-1 cells which showed cut phenotype without cell elongation (Fig. 3C). These results indicated that HU sensitivity of pfh1 mutant cells arose not from a checkpoint defect but from a problem in the recovery process.
Figure 3.
HU sensitivity of pfh1 mutant cells. (A) Growth curve after HU addition. HU (12 mM, final concentration) was added to asynchronous culture of the indicated strains at 34°C in minimal medium supplemented with leucine. Cell aliquots were taken every hour. (B) pfh1-R20 and pfh1-R23 cells are sensitive to HU. The viability was assayed by colony formation and expressed as relative viability compared with the viability at 0 h. (C) Cell morphology of indicated strains. Cells cultured in the presence of 12 mM HU for 6 h were fixed with 70% ethanol and stained with DAPI.
Isolation of the pfh1+ gene
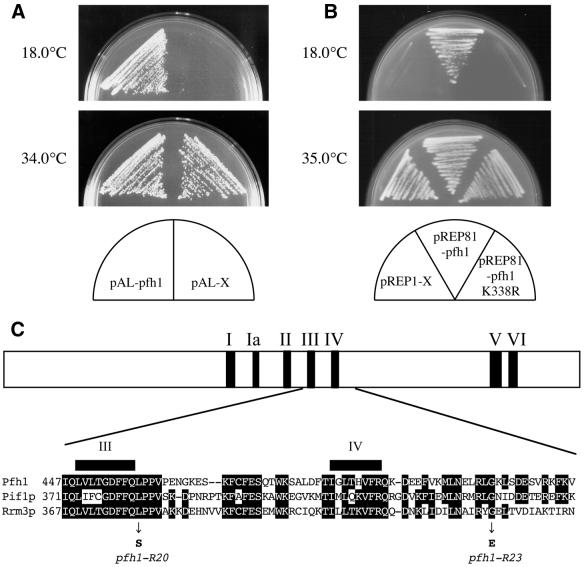
To isolate the pfh1+ gene, both S.pombe genomic DNA and SV40 promoter-driven cDNA libraries were screened for clones that could suppress the cold sensitivity of pfh1-R20 and pfh1-R23 cells. We isolated three cDNA clones and one genomic clone that rescued the mutants (Fig. 4A). All three cDNA clones contained the same ORF, and this ORF was also present in the genomic DNA clone. The ORF was identical to a gene located on the S.pombe chromosome II cosmid clone SPBC887 (EMBL accession no. AL033388) at nucleotides 28298–30756. (Note that we confirmed the presence of the predicted 41 bp intron in the ORF by comparison between cDNA and genomic DNA sequences.) To examine the relationship between this ORF and pfh1+, we constructed a strain in which the ura4+ gene was integrated immediately downstream of the newly isolated ORF. By crossing cells carrying this allele with the pfh1-R20 mutant, we were able to demonstrate tight linkage between the ura4+ marker and the cold sensitivity caused by the pfh1-R20 mutation. Based on these results, we concluded that the ORF is indeed pfh1+.
Figure 4.
Isolation of the pfh1+ gene, structure of Pfh1 protein and positions of mutations. (A) Isolation of pfh1+. Cold-sensitive pfh1-R20 cells carrying indicated plasmids were streaked on an MM (30) plate and incubated at the indicated temperatures. (B) pREP81-pfh1 K338R was unable to suppress pfh1-R20 cells. pfh1-R20 cells carrying the indicated plasmids were incubated as in (A). (C) Structure of Pfh1 protein is shown by an open bar. Seven conserved helicase motifs are indicated by shaded boxes. The part of predicted amino acid sequence of S.pombe pfh1+, containing motifs III and IV, is shown in single letter code and aligned with Pif1p and Rrm3p of S.cerevisiae. Identical amino acids are boxed. The mutations occurred in pfh1-R20 and pfh1-R23 alleles are indicated.
DNA sequencing revealed that the pfh1+ gene encodes an 805 amino acid protein with a calculated molecular mass of ∼90 kDa. A database search revealed that Pfh1 is related to the Pif1p and Rrm3p proteins of S.cerevisiae, with amino acid identities of 35 and 33%, respectively. Pif1p is a 5′ to 3′ DNA helicase implicated in mitochondrial DNA recombination and repair, telomere length regulation, and replication fork progression in ribosomal DNA (23,25,28,29). The related Rrm3p protein has the opposite effect on replication fork progression in ribosomal DNA. Pfh1 protein contains seven conserved helicase motifs (47). The locations of these helicase motifs are shown in Figure 4C.
Alleles of pfh1-R20 and pfh1-R23 have point mutations in conserved amino acids
To determine the nature of the mutation in pfh1-R20 and pfh1-R23, these alleles were amplified by PCR. The PCR products were then sequenced directly and compared with the wild-type pfh1+ DNA sequence. We found one point mutation in each allele (Table 2), each predicted to bring about a single amino acid change in the mutant proteins. Both mutations affected amino acids conserved among Pfh1, Pif1p and Rrm3p (Fig. 4C). These were a Leu458 to Ser change in pfh1-R20 and a Gly508 to Glu change in pfh1-R23, respectively. These residues (Leu458 and Gly508) are therefore crucial for Pfh1 activity at low temperature.
Table 2. Identification of pfh1 alleles.
| Mutant allele | R20 | R23 |
|---|---|---|
| Wild-type sequence | TTA Leu458 | GGG Gly508 |
| Mutant sequence | TCA Ser458 | GAG Glu508 |
The pfh1+ gene is essential for viability
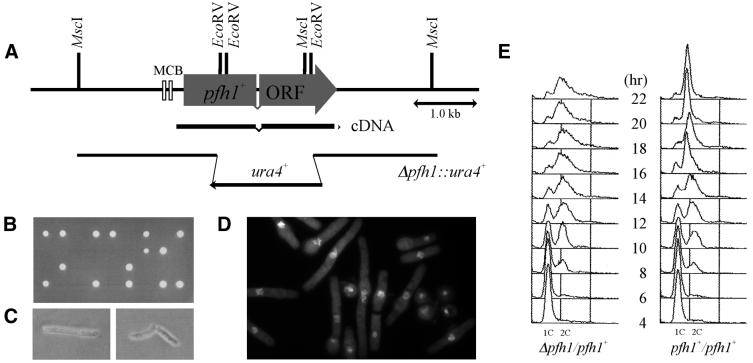
To further investigate the function of pfh1+, diploid cells lacking one copy of the pfh1+ gene were constructed by one-step gene replacement using a ura4+ gene cassette (Fig. 5A). Analysis of tetrads resulting from sporulation of the Δpfh1/pfh1+ cells demonstrated that two viable and two inviable spores were present in each ascus (Fig. 5B) and that all the viable spores were auxotrophic for uracil (i.e. pfh1+). pfh1+ is therefore an essential gene. Microscopic observation revealed that Δpfh1 spores could germinate and undergo up to three divisions before arresting as elongated cells (Fig. 5C). We confirmed that the expression of a pfh1+ cDNA under control of the SV40 promoter suppressed this lethal phenotype (data not shown).
Figure 5.
Analysis of the pfh1+ gene and Δpfh1 cells. (A) Restriction map of the pfh1+ gene. The ORF, pfh1+ cDNA and the positions of two MCB sequences are shown. The gene contains one intron. The EcoRV DNA fragment was replaced with a ura4+ gene cassette for generating Δpfh1 cells. (B) pfh1+ is an essential gene. Tetrads generated from pfh1+/pfh1::ura4+ diploid cells were dissected on YE plates and incubated at 30°C for 4 days. (C) Terminal phenotype of Δpfh1 cells. Cells that germinated from single Δpfh1 spores on a YE plate were photographed. (D) DAPI staining of germinating Δpfh1 cells. Δpfh1 spores derived form pfh1+/pfh1::ura4+ were preferentially germinated in minimal medium lacking uracil. Germinating cells cultured for 18 h were fixed with 70% ethanol and stained with DAPI. (E) Flow cytometric analysis of pfh1 null cells after spore germination. Spores derived from pfh1+/pfh1::ura4+ and ura4+/ura4-D18 control strain were germinated in minimal medium lacking uracil at 30°C. Germinating cells were collected every 2 h, fixed with 70% ethanol and analyzed by flow cytometry. The positions of the 1C and 2C DNA peaks are indicated.
Bulk DNA synthesis occurred in Δpfh1 cells
To monitor the progression of DNA synthesis in Δpfh1 cells, we performed flow cytometric analysis during spore germination. Spores prepared from Δpfh1/pfh1+ diploid cells were inoculated into minimal medium lacking uracil, such that Δpfh1 (ura+) spores would germinate preferentially. At 18 h after inoculation, Δpfh1 cells showed elongated cell morphology suggesting cell cycle delay (Fig. 5D). However, there was no obvious delay in DNA synthesis during germination (Fig. 5E). Thus, pfh1+ is likely to be dispensable for bulk DNA synthesis.
Cell cycle northern analysis of the pfh1+ gene
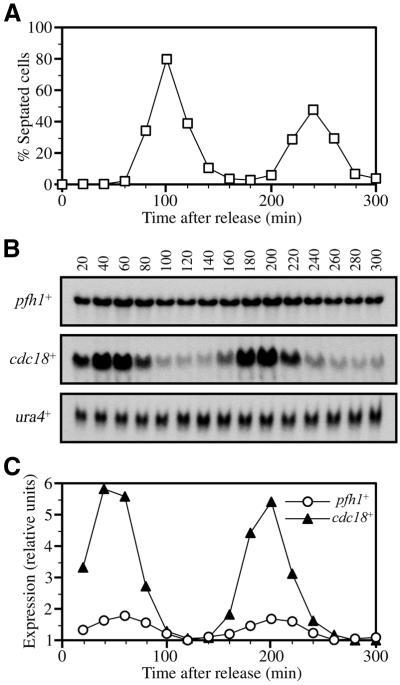
One feature of the pfh1+ gene is the presence of two MluI cell cycle box (MCB) sequences 382 and 637 bp upstream of the start of the pfh1+ ORF. MCB elements are binding sites for the S-phase-specific transcription factor complex Res1/Res2/Cdc10 (reviewed in 48). This suggested to us that pfh1+ mRNA levels might vary through the cell cycle, with maximal expression in S-phase. To examine the expression of pfh1+ during the cell cycle, we synchronized S.pombe cells by the cdc25-22 block and release method (Fig. 6A). RNA samples were prepared every 20 min following return to the permissive temperature and analyzed by northern hybridization using probes specific for pfh1+, cdc18+ (previously shown to peak at the G1/S boundary) and ura4+ (known to remain constant through the cell cycle). As expected, expression of cdc18+ peaked at G1/S while expression of ura4+ was constant through the cell cycle (Fig. 6B and C). The levels of pfh1+ transcripts also showed a degree of cell cycle variation, peaking at G1/S and decreasing in G2-phase (Fig. 6B and C). Unlike cdc18+ mRNA, however, pfh1+ message was still present in G2-phase cells.
Figure 6.
Cell cycle northern blot analysis of the pfh1+ gene. h– cdc25-22 cells were arrested in late G2 and then released to the permissive temperature. Cell aliquots were taken every 20 min. The growth of the synchronized cells was followed for two generations. (A) The cell cycle profile was monitored by measuring the percentage of septated cells at each time point. (B) The expression of pfh1+, cdc18+ and ura4+ were examined by northern hybridization. (C) The relative mRNA levels of pfh1+ and cdc18+. The mRNA level of pfh1+, cdc18+ and ura4+ shown in (B) were quantified using BAS2000 (Fuji Film). The relative ratio against ura4+ was calculated and plotted. The lowest signals were adjusted as one relative unit.
Purification of recombinant wild-type and mutant Pfh1 proteins
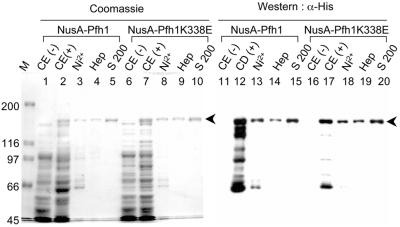
In order to obtain recombinant Pfh1 protein we inserted the intact pfh1+ ORF into the E.coli expression plasmid pET43.1a (see Materials and Methods) which resulted in the fusion of NusA at the N-terminus of Pfh1. The NusA fusion to Pfh1 greatly improved the solubility and stability of the enzyme, which otherwise had extremely poor solubility and was heavily degraded when expressed in E.coli (data not shown). In parallel, we isolated a mutant protein Pfh1K338E, in which an amino acid was changed in the ATP-binding motif (motif 1) critical for ATP hydrolysis. Both wild-type NusA-Pfh1 and mutant NusA-Pfh1K338E were purified as described in Materials and Methods. We analyzed the purified proteins to examine the purity of enzyme preparations as shown in Figure 7. Fractions from each purification step, which included Ni2+-agarose, HiTrap heparin and Superdex 200 chromatography, were subjected to 8% SDS–PAGE along with crude extracts prepared from uninduced and induced E.coli BL21 cells harboring pET43-Pfh1 or pET43-Pfh1K338E. The gel was either Coomassie stained (Fig. 7, lanes 1–10) or analyzed by western blot (Fig. 7, lanes 11–20). Both wild-type NusA-Pfh1 and mutant NusA-Pfh1K338E were efficiently expressed in E.coli in soluble forms (Fig. 7, lanes 2 and 7, respectively), and both proteins were purified to near homogeneity using the purification procedure described in Materials and Methods (Fig. 7, lanes 5 and 10). Some of the smaller polypeptides present in the fractions eluted from the Ni2+-charged HiTrap-chelating column (Fig. 7, lanes 3 and 8) appear to be proteolyzed products since they were detected by monoclonal antibody specific for penta-histidine in western blot analyses (Fig. 7, lanes 13 and 17).
Figure 7.
Purification of recombinant wild-type and mutant Pfh1 proteins. Crude extracts prepared from uninduced (lanes 1, 6, 11 and 16) and induced (lanes 2, 7, 12 and 17) E.coli BL21 cells harboring pET43-Pfh1 (lanes 1–5 and 11–15) and pET43-Pfh1K338E (lanes 6–10 and 16–20) were subjected to 8% SDS–PAGE along with fractions from each purification step as indicated, and the gel was either Coomassie stained (lanes 1–10) or analyzed in western blot analyses (lanes 11–20) using monoclonal antibody specific for penta-histidine (α-His; Qiagen). The numbers on the left indicate the molecular sizes (in kDa) of marker proteins (M; Bio-Rad). M, molecular weight marker; CE(–), crude extracts (20 µg) prepared from uninduced E.coli cells; CE(+), crude extracts (20 µg) prepared from induced E.coli cells; Ni2+, fractions (1 µg for lanes 3 and 8; 400 ng for lanes 13 and 18) eluted from the Ni2+-charged HiTrap-chelating column; Hep, fractions (500 ng for lanes 4 and 9; 200 ng for lanes 14 and 19) from the HiTrap heparin column; S 200, fractions (500 ng for lanes 5 and 10; 200 ng for lanes 15 and 20) obtained from the Superdex 200 column. Arrowheads indicate the position of recombinant wild-type or mutant NusA-Pfh1 proteins.
ATPase and helicase activities of Pfh1
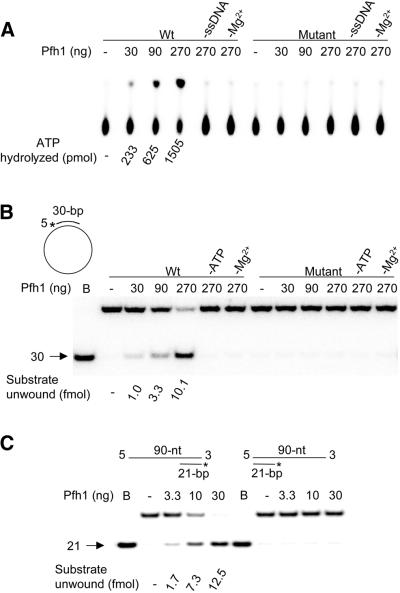
With purified wild-type NusA-Pfh1, we were able to detect ssDNA-dependent ATPase activity as shown in Figure 8A. The amount of ATP hydrolyzed was proportional to the amounts of enzyme added, resulting in 233, 625 and 1505 pmol of ATP hydrolysis with 30, 90 and 270 ng, respectively, of enzyme. ATP hydrolysis by NusA-Pfh1 was dependent on the presence of ssDNA and Mg2+. In contrast, ATP hydrolysis was not detected by NusA-Pfh1K338E when we used the same amounts of enzyme as wild-type protein (Fig. 8A). This result confirms that the Pfh1 has intrinsic ssDNA-dependent ATPase activity. We next examined whether NusA-Pfh1 possesses helicase activity and found that wild-type NusA-Pfh1 was able to unwind the 30 bp partial duplex DNA present in ΦX174 sscDNA, whereas the mutant NusA-Pfh1K338E failed to do so. This is consistent with the result above. The unwinding activity of wild-type Pfh1 was also dependent on the presence of ATP and Mg2+. The removal of NusA tag by thrombin cleavage neither increased nor decreased the ability of the recombinant NusA-Pfh1 enzyme to hydrolyze ATP (data not shown).
Figure 8.
Hydrolysis of ATP and unwinding of duplex DNA by Pfh1 enzymes. (A) DNA-dependent ATPase activities of wild-type Pfh1 and mutant Pfh1K338E were measured as described in Materials and Methods. The amounts of enzymes (Wt, NusA-Pfh1; mutant, NusA-Pfh1K338E) added and omissions of M13 sscDNA (–ssDNA) and Mg2+ (–Mg2+) were as indicated. The amounts of ATP hydrolyzed are presented at the bottom. (B) Helicase activities of NusA-Pfh1 and NusA-Pfh1K338E were measured as described in Materials and Methods. The reactions were incubated at 37°C for 10 min. The controls without ATP (–ATP) or MgCl2 (–Mg2+) were as indicated. The structure of the partial duplex ΦX174 sscDNA substrate used was shown. The asterisks indicate 32P-labeled ends. An arrow indicates the position where the labeled oligonucleotides migrated. The amounts of substrate unwound are presented at the bottom. (C) Schematic structures of substrates used are shown at the top. The asterisks indicate 32P-labeled ends. The indicated amounts of NusA-Pfh1 in a 20 µl reaction mixture were incubated with 15 fmol of either 5′- or 3′-overhang 21-bp duplex DNA substrate at 37°C for 10 min. The products were analyzed on a 12% polyacrylamide gel. Lanes labeled B denote boiled substrate controls. An arrow indicates the position where the labeled oligonucleotides migrated. The amounts of substrate unwound are presented at the bottom.
Since the S.cerevisiae Pif1p helicase unwinds DNA in the 5′ to 3′ direction (23,24), we tested whether S.pombe Pfh1 unwinds duplex DNA in the same direction as Pif1p. As shown in Figure 8C, NusA-Pfh1 utilized a partial duplex substrate with a 5′ tail only, but not the substrate with a 3′ tail, demonstrating that Pfh1 translocates and unwinds duplex DNA in the 5′ to 3′ direction like S.cerevisiae Pif1p. It is noteworthy that unwinding by Pfh1 was more efficient with oligonucleotide-based substrates than with ΦX174-based substrate. For example, >80% of substrate was unwound with 30 ng of Pfh1 (Fig. 8C) with oligonucleotide-based substrate, while >270 ng of Pfh1 was required for comparable unwinding of the ΦX174-based substrate (Fig. 8B). This is likely to be due to the presence of bulk ssDNA in ΦX174 sscDNA that may sequester a large fraction of enzyme.
Catalytic activity is essential for Pfh1 function in vivo
To examine the essentiality of catalytic activity of Pfh1 in vivo, we constructed plasmid pREP81-pfh1K338R expressing Pfh1-K338R, which contains a mutation in the ATP-binding motif. pREP81-pfh1 and pREP81-pfh1K338R were transformed to pfh1-R20 cells and we examined the suppression ability of cold sensitivity. As shown in Figure 4B, pREP81-pfh1 was able to complement the cold sensitivity of pfh1-R20 cells, whereas pREP81-pfh1K338R was not. In addition, Δpfh1/pfh1+ diploid cells transformed with these plasmids were sporulated and examined for the viability. Similarly, pREP81-pfh1 but not pREP81-pfh1K338R could complement the lethality of Δpfh1 cells (data not shown). Thus, catalytic activity of Pfh1 DNA helicase is essential for cell viability.
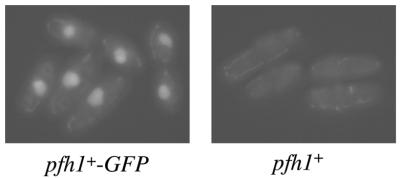
Pfh1 is a nuclear protein
As described above, the budding yeast Pfh1 homolog Pif1p has been shown to function both in the nucleus and in mitochondria in S.cerevisiae. To determine the intracellular localization of S.pombe Pfh1, we constructed a strain that expressed a Pfh1-GFP fusion protein from the pfh1+ genomic locus, driven by native pfh1+ promoter (see Materials and Methods). Exponentially growing Pfh1-GFP expressing cells were then analyzed by fluorescence microscopy. The fluorescent signal of the Pfh1-GFP protein was observed in the nucleus (Fig. 9), consistent with the notion that the Pfh1 protein functions in chromosomal DNA replication. Although some fluorescence could also be detected in the cytoplasm of Pfh1-GFP expressing cells (in ribbon-like patterns reminiscent of previously published images of S.pombe mitochondria), these were also seen, at similar intensity, in wild-type cells lacking the GFP protein (Fig. 9). Unlike Pif1p, therefore, Pfh1 in S.pombe appears to be exclusively nuclear.
Figure 9.
Nuclear localization of Pfh1-GFP. Exponentially growing S.pombe cells expressing Pfh1-GFP under the control of the pfh1+ promoter (left) and wild-type cells (right) in minimal medium were analyzed by fluorescent microscopy.
DISCUSSION
The Cdc24 protein plays an important role in chromosomal DNA replication in fission yeast. Cells carrying temperature-sensitive mutations in the cdc24 gene undergo cell cycle arrest when shifted to the restrictive temperature, becoming arrested with incompletely replicated chromosomes prior to the onset of mitosis (2–4). Previously, cdc24+ has been shown to interact with three genes encoding subunits of the DNA polymerase δ holoenzyme complex (Pol δ–PCNA–RF-C) in S.pombe, as well as with the dna2+ gene which encodes a nuclease-helicase required for Okazaki fragment processing. Overexpression of either pcn1+, rfc1+ or cdm1+ (encoding the fission yeast PCNA protein, the large subunit of replica tion factor C, and the D-subunit of DNA polymerase δ, respectively) or dna2+ is sufficient to rescue the temperature-sensitive phenotype of cdc24 mutant alleles (3,4,7). In the case of Pcn1, Rfc1 and Dna2, these genetic interactions reflect physical interactions between the encoded proteins. Co-immunoprecipitation experiments have shown physical associations, direct or indirect, between Cdc24 and PCNA, and between Cdc24 and Rfc1 (4). Yeast two-hybrid analysis suggests that Cdc24 and Dna2 interact directly (8).
In order to isolate novel factors that interact with cdc24+, we identified cold-sensitive chromosomal suppressors of the temperature-sensitive cdc24-M38 mutant allele (2). Amongst 37 such mutants, we mapped five to a single complementation group. We cloned the corresponding gene, which we named pfh1+, and here describe the results of in vivo and in vitro analysis of the function of Pfh1, an 805 amino acid protein homologous to Pif1p and Rrm3p DNA helicase family members in S.cerevisiae (reviewed in 22).
To elucidate the in vivo function of pfh1+, we analyzed the cold-sensitive pfh1-R20 and pfh1-R23 cells as well as the pfh1-disrupted cells. All the data, including genetic interactions (Fig. 1), mutant phenotype (Figs 2, 3 and 5), mRNA expression (Fig. 6), 5′ to 3′ DNA helicase activity (Fig. 8), essentiality of this activity in vivo (Fig. 4B), and cellular localization (Fig. 9) suggest that Pfh1 has an essential function in chromosomal DNA replication. However, like Cdc24 and Dna2, Pfh1 is not required for bulk DNA synthesis (Figs 2B and 5E).
To understand the enzymatic function of Pfh1, we purified recombinant protein expressed in E.coli to near homogeneity and demonstrated that Pfh1 protein has ssDNA-dependent ATPase and 5′ to 3′ DNA helicase activities (Fig. 8). In addition, we introduced a point mutation at a lysine residue in the ATP-binding pocket predicted to be essential for ATPase activity, demonstrated that the recombinant mutant protein did indeed lack such activity, and introduced the mutant allele into pfh1-R20 and Δpfh1 cells. Since this mutant gene could not complement the cold sensitivity of pfh1-R20 cells nor lethality of Δpfh1 cells, we conclude that the essentiality of pfh1+ is based on its catalytic activity.
What is the cellular function of Pfh1? One fascinating possibility is that Pfh1 functions in Okazaki fragment processing together with the Dna2–Cdc24 complex. Conceivably, the Pfh1 helicase could cooperate with Dna2. Recently, we found that DNA helicase and endonuclease activities of budding yeast Dna2p are tightly coupled to ensure efficient removal of the flap during the Okazaki fragment maturation step (49). In budding yeast, Dna2p helicase activity assists this process by removing secondary structures present in the flap (49). In this context, since purified S.pombe Dna2 lacks any detectable ATPase (helicase) activity (H. Y. Kang and Y. S. Seo, unpublished results), fission yeast Dna2 may require a coupled DNA helicase to execute complete removal of the flap in the Okazaki fragment maturation step. Pfh1 may therefore unwind DNA secondary structures present in the flaps formed by displacement synthesis by DNA polymerase δ (reviewed in 15). Further characterization of the biochemical properties of the Pfh1 helicase in vitro will doubtless prove highly informative.
A further issue to be addressed concerns the extent of Pfh1 function throughout the genome. In budding yeast, the Pfh1 homologs Pif1p and Rrm3p have been termed region-specific helicases (22): Pif1p is believed to function in mtDNA, telomere DNA and rDNA whereas Rrm3p is involved in rDNA replication (where its function opposes that of Pif1p). This raises the possibility that, in S.pombe, the function of Pfh1 (and by implication, perhaps that of Cdc24 also) might be confined to certain regions of the genome only, such as the rDNA repeats on chromosome III. However, it is worth noting that, in contrast to pfh1+, neither PIF1 nor RRM3 is an essential gene in S.cerevisiae. Indeed, budding yeast cells mutated for both genes are viable. Thus, there are clear differences between the two yeast systems.
In addition to its role in chromosomal DNA replication, Pfh1 also appears to have an essential role to play in the repair of DNA damage: pfh1 mutant cells are highly sensitive to exposure to the DNA-alkylating agent MMS. Whether Pfh1 is required to cope with the effects of MMS-induced damage specifically in S-phase remains to be seen.
Finally, during the preparation of this manuscript, Zakian and co-workers reported the isolation and characterization of the pfh1+ gene on the basis of the protein sequence similarity between Pfh1, Pif1p and Rrm3p (50). For the most part, their results mirror ours. However, in an echo of the situation in S.cerevisiae, these authors also observed telomere shortening in Δpfh1 S.pombe cells, consistent with a role for Pfh1 in telomere length regulation. Still, these authors also demonstrated that telomere length regulation was not the sole essential function of Pfh1 protein by showing that pfh1+ was essential for cell viability even in a strain with circular chromosomes lacking telomeric sequences. Clearly, further work is required to unambiguously assign a function to Pfh1.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K. Okazaki for S.pombe DNA libraries and P. Fantes for the rad1-1 strain. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Wellcome Trust. H.T. is supported by a JSPS postdoctoral fellowship for research abroad. S.M. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science. This work was also supported by a grant from the Creative Research Initiatives Program of the Korean Ministry of Science and Technology (to Y.-S.S.).
REFERENCES
- 1.MacNeill S.A. and Burgers,P.M.J. (2000) Chromosomal DNA replication in yeast: enzymes and mechanisms. In Fantes,P. and Beggs,J. (eds), The Yeast Nucleus. Oxford University Press, Oxford, UK, pp. 19–57.
- 2.Nasmyth K. and Nurse,P. (1981) Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. [DOI] [PubMed] [Google Scholar]
- 3.Gould K.L., Burns,C.G., Feoktistova,A., Hu,C.P., Pasion,S.G. and Forsburg,S.L. (1998) Fission yeast cdc24+ encodes a novel replication factor required for chromosome integrity. Genetics, 149, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka H., Tanaka,K., Murakami,H. and Okayama,H. (1999) Fission yeast Cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol., 19, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kai M., Tanaka,H. and Wang,T.S. (2001) Fission yeast Rad17 associates with chromatin in response to aberrant genomic structures. Mol. Cell. Biol., 21, 3289–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waseem N.H., Labib,K., Nurse,P. and Lane,D.P. (1992) Isolation and analysis of the fission yeast gene encoding polymerase δ. EMBO J., 11, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds N., Watt,A., Fantes,P.A. and MacNeill,S.A. (1998) Cdm1, the smallest subunit of DNA polymerase δ in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr. Genet., 34, 250–258. [DOI] [PubMed] [Google Scholar]
- 8.Kang H.Y., Choi,E., Bae,S.H., Lee,K.H., Gim,B.S., Kim,H.D., Park,C., MacNeill,S.A. and Seo,Y.S. (2000) Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics, 155, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waga S. and Stillman,B. (1994) Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature, 369, 207–212. [DOI] [PubMed] [Google Scholar]
- 10.Budd M.E. and Campbell,J.L. (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl Acad. Sci. USA, 92, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budd M.E., Choe,W.C. and Campbell,J.L. (1995) DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem., 270, 26766–26769. [DOI] [PubMed] [Google Scholar]
- 12.Bae S.H., Choi,E., Lee,K.H., Park,J.S., Lee,S.H. and Seo,Y.S. (1998) Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem., 273, 26880–26890. [DOI] [PubMed] [Google Scholar]
- 13.Bae S.H., Bae,K.H., Kim,J.A. and Seo,Y.S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- 14.Bae S.H. and Seo,Y.S. (2000) Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- 15.MacNeill S.A. (2001) DNA replication: partners in the Okazaki two-step. Curr. Biol., 11, R842–R844. [DOI] [PubMed] [Google Scholar]
- 16.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 17.Matson S.W., Bean,D.W. and George,J.W. (1994) DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays, 16, 13–22. [DOI] [PubMed] [Google Scholar]
- 18.Shiratori A., Shibata,T., Arisawa,M., Hanaoka,F., Murakami,Y. and Eki,T. (1999) Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast, 15, 219–253. [DOI] [PubMed] [Google Scholar]
- 19.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, –6 and –7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 20.Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 21.Labib K., Tercero,J.A. and Diffley,J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 22.Bessler J.B., Torredagger,J.Z. and Zakian,V.A. (2001) The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol., 11, 60–65. [DOI] [PubMed] [Google Scholar]
- 23.Lahaye A., Stahl,H., Thines-Sempoux,D. and Foury,F. (1991) PIF1: a DNA helicase in yeast mitochondria. EMBO J., 10, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahaye A., Leterme,S. and Foury,F. (1993) PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem., 268, 26155–26161. [PubMed] [Google Scholar]
- 25.Schulz V.P. and Zakian,V.A. (1994) The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell, 76, 145–155. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J., Monson,E.K., Teng,S., Schulz,V.P. and Zakian,V.A. (2000) Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science, 289, 771–774. [DOI] [PubMed] [Google Scholar]
- 27.Myung K., Chen,C. and Kolodner,R.D. (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature, 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 28.Foury F. and Kolodynski,J. (1983) pif mutation blocks recombination between mitochondrial rho+ and rho– genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 80, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivessa A.S., Zhou,J.Q. and Zakian,V.A. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell, 100, 479–489. [DOI] [PubMed] [Google Scholar]
- 30.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments with Fission Yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Gutz H., Heslot,V., Leupold,V. and Leprieno,N. (1974) Schizosaccharomyces pombe. In King,R.C. (ed.), Handbook of Genetics. Plenum Press, New York, Vol. 1, pp. 395–446.
- 32.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 33.Okazaki K., Okazaki,N., Kume,K., Jinno,S., Tanaka,K. and Okayama,H. (1990) High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res., 18, 6485–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- 35.Basi G., Schmid,E. and Maundrell,K. (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene, 123, 131–136. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka K., Yonekawa,T., Kawasaki,Y., Kai,M., Furuya,K., Iwasaki,M., Murakami,H., Yanagida,M. and Okayama,H. (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol., 20, 3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costello G., Rodgers,L. and Beach,D. (1986) Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet., 11, 119–125. [Google Scholar]
- 38.MacNeill S.A., Moreno,S., Reynolds,N., Nurse,P. and Fantes,P.A. (1996) The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase δ, binds to Pol3 and Cdc27. EMBO J., 15, 4613–4628. [PMC free article] [PubMed] [Google Scholar]
- 39.Nakashima N., Tanaka,K., Sturm,S. and Okayama,H. (1995) Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J., 14, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J.S., Choi,E., Lee,S.H., Lee,C. and Seo,Y.S. (1997) A DNA helicase from Schizosaccharomyces pombe stimulated by single-stranded DNA-binding protein at low ATP concentration. J. Biol. Chem., 272, 18910–18919. [DOI] [PubMed] [Google Scholar]
- 41.Al-Khodairy F. and Carr,A.M. (1992) DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J., 11, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowley R., Subramani,S. and Young,P.G. (1992) Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J., 11, 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walworth N., Davey,S. and Beach,D. (1993) Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature, 363, 368–371. [DOI] [PubMed] [Google Scholar]
- 44.Al-Khodairy F., Fotou,E., Sheldrick,K.S., Griffiths,D.J., Lehmann,A.R. and Carr,A.M. (1994) Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell, 5, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami H. and Okayama,H. (1995) A kinase from fission yeast responsible for blocking mitosis in S phase. Nature, 374, 817–819. [DOI] [PubMed] [Google Scholar]
- 46.Francesconi S., Grenon,M., Bouvier,D. and Baldacci,G. (1997) p56chk1 protein kinase is required for the DNA replication checkpoint at 37°C in fission yeast. EMBO J., 16, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 48.Okayama H., Nagata,A., Jinno,S., Murakami,H., Tanaka,K. and Nakashima,N. (1996) Cell cycle control in fission yeast and mammals: identification of new regulatory mechanisms. Adv. Cancer Res., 69, 17–62. [DOI] [PubMed] [Google Scholar]
- 49.Bae S.H., Kim,D.W., Kim,J., Kim,J.H., Kim,D.H., Kim,H.D., Kang,H.Y. and Seo,Y.S. (2002) Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem., 277, 26632–26641. [DOI] [PubMed] [Google Scholar]
- 50.Zhou J.Q., Qi,H., Schulz,V.P., Mateyak,M.K., Monson,E.K. and Zakian,V.A. (2002) Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 subfamily of DNA helicases. Mol. Biol. Cell, 13, 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]