Abstract
Evidence is provided that plant transcription factors can be efficiently reprogrammed to dominant- negative functions by the use of a repressor domain of the engrailed (en) gene from Drosophila. Ectopic expression of translational fusions between the en298 N-terminus and the complete coding regions of the SHOOTMERISTEMLESS, APETALA3, PISTILLATA and KNAT1 transcription factors results in trans-dominant functions which phenocopy loss-of-function mutants. The combination of the dominant-negative en298-STM function with the hormone-binding domain of the glucocorticoid receptor provides strong evidence that phenocopies rely on the incorporation of the chimeric protein into the nuclear compartment. By this dominant-negative approach KNAT1 was rapidly identified to be encoded by the BREVIPEDICELLUS locus. Dominant-negative chimeric proteins may be of wide use to elucidate biological functions of plant transcriptional activators and may be suitable to study protein–protein interactions in planta.
INTRODUCTION
Plant transcription factors mostly belong to gene families containing numerous members. An example of how large such families can grow is the myb family comprised of at least 136 genes in Arabidopsis (1). Homeobox and MADS box genes are also found in multiple copies comprising several subfamilies in all plant species analysed so far (2,3). In genetic screens for loss-of-function alleles, only a few of these genes have been associated with a mutant phenotype. One explanation for numerous silent family members may be genetic redundancy; each gene-specific contribution may be subtle or dependent on special conditions. This fraction of silent genes in plants and in other genomes provides a major challenge in molecular biology because it is foreseen to oppose a functional analysis also in reverse genetic approaches.
To identify biological functions of transcription factors in Drosophila and amphibian development, an alternative method has been successfully used. A pioneering experiment was performed in Drosophila where the DNA-binding homeodomain of the Engrailed (En) gene product was replaced by its Fushi tarazu (Ftz) counterpart and expression of the chimeric gene controlled by a heat-shock promoter. After induction, the sequence-specificity of the Ftz homeodomain directed the strong transcriptional en repressor function to ftz target genes resulting in phenocopies of ftz loss-of-function alleles in transgenic progeny (4). In addition to the En298 repressor domain, the Krüppel associated box (5) is now used in animal systems where these repressor domains are combined frequently only with the DNA-binding domain of the transcription factor in question (6). Functional studies are subsequently performed either in transient gene expression experiments, e.g. tissue culture cell lines, where the c-MYB DNA-binding domain combined with the En298 domain causes G1 cell cycle arrest or apoptosis (7). In contrast, and more informative in respect to developmental questions, have been injections of chimeric RNA transcripts into oocytes, zygotes or developing embryos, a method which identified the Xenopus Brain Factor 2 (XBF-2) as a transcriptional repressor converting ectoderm into neural tissue (8).
Similar experiments in plants are restricted by the lack of functional repressor domains. However, plant transcription factors share a significant degree of similarity with their animal counterparts. Essential subunits of the transcription initiation complex are so similar that X-ray data originally obtained for the Arabidopsis TATA box binding protein (9) have been generalised to animal species (10). Also conserved are DNA-binding motifs like the homeodomain (2), the myb-motif (11) and the MADS box (3) or dimerisation motifs such as leucine zippers (12) or helix–loop–helix domains (13). In addition, transcriptional activation domains of the VP16 protein from Herpes simplex virus (14) and the GAL4 gene product from yeast (15) have been shown to be functional in plants. For the VP16 activation domain, direct interactions with TAFII31 (16) in the transcription initiation complex are known, and similar interactions can thus be implicated in plants. This obvious conservation of transcription factors and basic transcriptional machinery between plant and animal species made it tempting to test whether animal repressor domains might be functional in plants.
One criterion of functionality could be dominant-negative functions provided by chimeric gene products in analogy to the pioneering En-Ftz experiment in Drosophila (4). For an initial study we decided to test three genes: SHOOT MERISTEMLESS (STM) (17) an essential function in the shoot apical meristem (SAM), APETALA3 (AP3) (18) and PISTILLATA (19) contributing to flower development. While STM belongs to the class of homeodomain proteins, the floral organ identity genes AP3 and PI encode MADS-box proteins, which depend on heterodimerisation to achieve transcriptional control function (19). The three genes are not only members of different classes of plant transcription factors, but their mutant phenotypes affect different developmental stages, the early vegetative phase or rather late floral development.
Here, we describe ectopic expression experiments performed with en298-STM and en298-AP3 or en298-PI translational fusions, which result in transdominant phenocopies of stm or ap3/pi phenotypes in >75% of the primary transformants. The performance of the CHimeric Repressor Interference System (CHRIS) with unknown gene functions was tested with the KNAT1 gene (20), a rather close relative to STM, which is also expressed in the SAM. Although at the time of this study KNAT1 was not associated with any known mutant, the dominant-negative en298-KNAT1 fusion phenotype rapidly revealed the relationship to the BREVIPEDICELLUS locus.
MATERIALS AND METHODS
Chimeric gene constructions
As a precursor to all translational fusions a 929 bp AflII–BamHI fragment of the cDNA clone D2B (21) was inserted between the CaMV 35S promoter and the poly(A) signal into NotI/BamHI cleaved pRTΩNot-Asc vector (22). These cDNA sequences cover the natural translation start and encode the first 298 amino acid residues of the Drosophila Engrailed protein. Unique NcoI and BamH1 sites were created behind the en298 N-terminus, resulting in the pCHRIS vector, and used to insert the complete protein coding regions which were amplified by PCR with appropriate restriction site additions. The STM coding sequences were kindly provided by Dr R. Simon, the AP3 cDNA clone was a gift from Dr Zsuzsanna Schwarz Sommer (MPI für Züchtungsforschung) and the PI and KNAT1 reading frames were isolated via RT–PCR. The entire open reading frames (ORFs) were fused to the en298 N-terminus, the ATG start being inherent in the NcoI site. The initial C-terminal deletions of STM made use of unique restriction sites in the STM coding sequence; all N-terminal deletions and subdomains were created via PCR. The engineering of a BamHI site upstream from the STM TGA stop codon allowed the C-terminal fusion to the hormone-binding domain of the glucocorticoid receptor. The complete chimeric en298-STM reading frame was transferred to the binary vector pBI-ΔGR (23). Generally, translational fusions and PCR products were verified by DNA sequencing. For complementation of the bp-1 mutant the KNAT1 coding region was inserted into the pRTΩNot-Asc vector (22).
Plant transformation, dexamethasone induction and analysis of transgenic plants
For transfer into the Arabidopsis genome the translational en298 fusions in pCHRIS were inserted into pGPTV-BAR AscI (22) by use of the AscI sites flanking the CaMV 35S promoter and poly-A signal. The complete binary vectors were transferred into Agrobacterium tumefaciens GV310 and the resulting strains were used for infiltration of Arabidopsis immature inflorescences (24). Transgenic plants were selected for BASTA resistence except en298-STM/GR transgenics which carried the kanamycin-resistance marker. Hormone induction was performed by spraying wild-type control plants or transgenic en298-STM/GR seedlings at the four- to six-leaf stage with 1 µM dexamethasone. Plants were photographed every day in the same orientation and magnification. Pictures were processed with the Adobe Photoshop 7.0 software on a Macintosh G4 computer.
Total RNA of Arabidopsis seedlings or inflorescences was extracted as described (25) and poly(A)+ RNA enriched by oligo-dT chromatography. RNA samples were separated on formaldehyde agarose gels (1.2%) and transferred to Hybond N+ membranes (Amersham Buchler). Filters were hybridised overnight either at 42°C in 50% formamide, 6× SSPE, 5× Denhardt’s, 0.5% SDS and 250 mg/ml CT-DNA or at 68°C in 0.7 M Na-phosphate pH 7.2, 7% SDS. Washing was performed at 68°C in 0.2× SSPE, 0.1% SDS. Probes labelled by random oligonucleotide priming were sufficient to visualise the transgene transcript in total or poly(A)+ RNA. Single-stranded RNA probes were needed to confirm the endogenous STM transcript in poly(A)+ RNA. First strand cDNA synthesis for RT–PCR was performed with the Superscript system (Gibco-BRL) primed with oligo-dT on total RNA (5 µg). PCRs generally contained 1 µM primer, 0.2 mM NTPs and 0.2 U Taq polymerase and were routinely run with 30 cycles. A unique forward primer CTCCCTAAAGAAGCTCGTCAAC located in the STM coding region was combined with the STM-specific (CGCATAACAATAGAACACCAAAAGG) or the pCHRIS-specific (CCTTATCTGGGAACTACTCAC) reverse primer to discriminate between the native or chimeric transcript, respectively.
RESULTS
Chimeric gene constructions and plant transformation
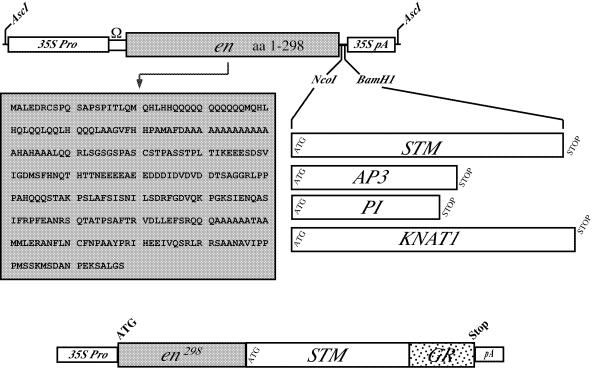
For the construction of translational fusions between the Drosophila Engrailed (En) repressor domain and plant transcription factors a 929 bp cDNA fragment encoding 298 amino acids of the en298 N-terminus (21) was cloned behind the CaMV 35S promoter into the vector pRTΩNot (22). Two restriction sites, NcoI and BamHI, inserted behind the repressor domain allowed the directional cloning of protein coding regions in the resulting pCHRIS vector. The ATG inherent in the NcoI site (CCATGG) allows in-frame fusions between the en298 coding sequences and the native methionine start codon. In this study, the STM, AP3, PI and KNAT1 (17–20) coding regions were inserted. Translation of all chimeric proteins always starts from the translation start codon of the Drosophila en ORF.
To enable subsequent fusion with the hormone-binding domain of the rat glucocorticoid receptor in the vector pBI-ΔGR (23) a unique BamHI site was introduced before the stop codon into the STM ORF. The C-terminal addition of the hormone-binding domain in the chimeric En298-STM/GR protein should interfere with nuclear uptake until application of the dexamethasone hormone. All the different constructs schematically represented in Figure 1 were transferred into the Arabidopsis genome via A.tumefaciens mediated T-DNA transfer.
Figure 1.
CHRIS expression cassette. The TMV Ω untranslated leader sequence and the engrailed N-terminal coding sequences (1–298 amino acids; grey box) were inserted between the CaMV 35S promoter and polyadenylation signal. The En298 protein sequence is indicated below the CHRIS cassette. The STM, AP3, PI and KNAT1 coding regions (drawn to scale) were inserted directionally between the unique NcoI and BamHI cloning sites of the pCHRIS vector. For comparison the en298-STM/GR cassette in pBI-ΔGR is indicated below.
Constitutive expression of chimeric en298-STM, en298-AP3 or en298-PI constructs mimics loss-of-function alleles
Abnormalities in the development of transgenic en298-STM plants became evident during the earliest stages of seedling development. Compared to wild-type (Fig. 2A), where the SAM is always enclosed by several leaf primordia, the single leaf in the transgenic en298-STM plant (Fig. 2B) originates between the petioles of both cotyledons. The close-up (Fig. 2C) shows that an apical dome is missing. Sometimes SAM activity in en298-STM plants is completely terminated after the initiation of a single leaf. Most transgenic seedlings, however, cease development after four to seven leaf primordia, which all emerge in the absence of a detectable apical dome. The photograph in Figure 2D shows leaf 3 originating between the petioles of leaves 1 and 2. These trans-dominant alterations phenocopy weak stm alleles and have been reproduced in various independent large-scale experiments (>1000 transgenic T1 progeny) and are generally found in >75%, and sometimes up to 90%, of the primary transformants.
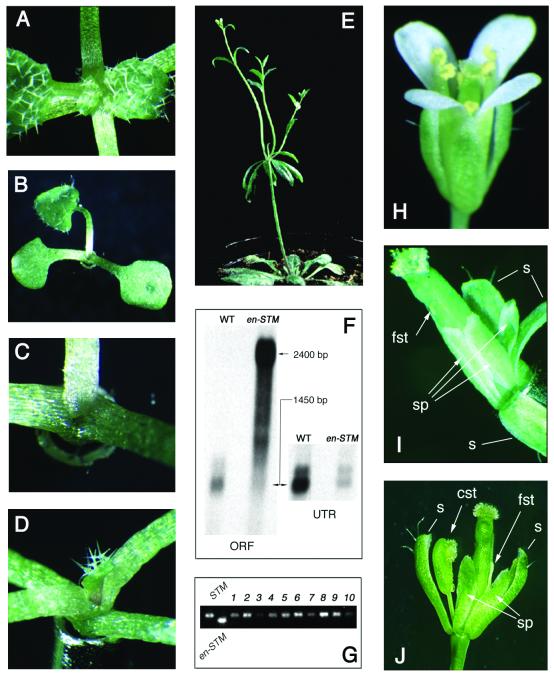
Figure 2.
Analysis of en298-STM, en298-AP3 or en298-PI transgenic plants. (A) Six-leaf stage wild-type seedling. Note the small leaflets enclosing the wild-type SAM in comparison with the en298-STM transgenic seedling (B), with only a single leaf emerging between the two cotyledons. (C) Close-up of the apex of the same transgenic seedling as in (B) showing the absence of an apical dome. (D) Emerging third leaf in a weaker stm phenocopy. (E) Inflorescence emerging from an axillary meristem. Note the airborne rosette comprised of several cauline leaves and three secondary inflorescences initiated from axillary meristems after the primary inflorescence meristem has lost activity. Most of these secondary inflorescences often develop a few fertile flowers. (F) RNA gel-blot experiment performed with 5 µg of poly(A)+ RNA pooled from 10 individual seedlings. The size of the transcripts is indicated on the margin. The same filter was hybridised with a probe of the STM coding region (ORF, left) or the 3′-UTR (right), which is absent in the chimeric en298-STM transcript. (G) RT–PCR experiment to confirm transcriptional activity of the native STM gene in individual transgenic seedlings (1–10). The native amplicon (STM) was discriminated from the chimeric product (en298-STM) by combining a unique forward primer with reverse primers specific to the 3′-UTR of the native STM transcript or the pCHRIS vector. Single-tube RT–PCR experiments with all three primers failed to detect the endogenous transcripts because the unique forward primer achieved saturation in excess of the chimeric amplicons. (H) Wild-type Arabidopsis flower with four sepals, four petals (white), six stamens and two fused central carpels. (I) Transgenic en298-AP3 flower with sepaloid petals (sp) and filamentous stamen (fst) in whorls 2 and 3. The first and fourth whorl, sepals(s) and carpels, respectively, are unaffected. A single sepal has been removed for better detection of the homeotic transformations in whorls 2 and 3. (J) Slightly more phenotypic variation is observed in transgenic en298-PI flowers in addition to sepaloid petals (sp) and filamentous stamen (fst): carpeloid stamens (cst) frequently replace the filamentous appendages in whorl 3.
Concomitantly, or after a short break, axillary meristems are activated. The resulting secondary shoot axes may develop from normal rosettes, but functional deficiencies in these axillary meristems become evident during the inflorescence phase. A lack of internode elongation in most en298-STM plants results in airborne rosettes of several cauline leaves (Fig. 2E). Characteristic is a cycling back and forth between elongating and non-elongating internodes resulting in multiple airborne rosettes. Exceptionally, such transgenic plants never flower, but most transgenic plants develop a few fertile flowers. The inheritance of phenocopies was analysed in 20 lines originating from primary en298-STM transformants (T1). BASTA resistant T2 progeny were obtained from 18 T1 plants and stm phenocopies were recovered in 16 families, although with variable strength. The absolute numbers of affected progeny plants varied between different families, however, none showed a 3:1 segregation of the BASTA resistance marker. The phenotypic variability therefore is presumably related to multiple T-DNA copies and the expression level of the transgene; low expression levels could also account for the failure to recover phenocopies in two of the 20 families.
To exclude silencing of the endogenous STM gene, transgenic en298-STM phenocopy plants were subjected to RNA gel-blot and RT–PCR analysis. The chimeric transcripts derived from the CaMV 35S promoter were easily detectable in total or poly(A)+ RNA probed with the STM coding regions (Fig. 2F). Although the high abundance of chimeric transcripts interfered with the detection of the shorter native mRNA in these gel-blot experiments, discrimination could be achieved by probing with the natural 3′-untranslated region (UTR) sequences that are lacking the en298-STM transgene (Fig. 2F, right). The reduction in transcript level is not due to the absence of the STM transcripts in individual T1 progeny, because the native transcripts could be detected by RT–PCR experiments in each individual phenocopy plant (Fig. 2G). The level of native STM transcripts is always significantly lower in transgenic seedlings than in wild-type, which coincides with the absence of an apical dome in the phenocopy plants.
In contrast to the phenotypes during vegetative and inflorescence development in en298-STM transgenic plants, the phenocopies caused by ectopic expression of the en298-AP3 or en298-PI construct were exclusively restricted to the flower. In 76% (en298-AP3: 109/143) or 83% (en298-PI: 159/183) of the primary transformants obtained with each chimeric construct, respectively, petals were converted to sepals in the second floral whorl and filamentous or carpeloid structures (en298-PI) or the complete absence of organs replaced stamens in the third whorl (Fig. 2, compare wild-type in H with transgenic flowers depicted in I and J). Both homeotic transformations are characteristic for either ap3 or pi mutant alleles. A higher phenotypic variability was observed in en298-PI transgenic plants, which reflects the range of phenotypes from weak to strong pi alleles and thus presumably indicates quantitative differences in the expression level of the transgene. Ectopic expression of the En298-AP3 or the En298-PI chimeric protein therefore phenocopies the loss of B-function in flower development. Back-crossing of en298-AP3 transgenic plants to wild-type showed inheritance of phenocopies to subsequent generations always strictly co-segregating with the BASTA resistance marker. Therefore, the constitutive expression of three translational fusion proteins comprised of the Drosophila En298 transcriptional repressor domain and the entire STM, AP3 or PI polypeptides results in very specific phenocopies of loss-of-function alleles.
Phenocopies rely on the incorporation of the chimeric En298-STM protein into the nuclear compartment
To address the molecular mechanism the chimeric En298-STM polypeptide was expressed in a C-terminal fusion with the hormone-binding domain of the glucocorticoid receptor in transgenic Arabidopsis plants. Due to this addition, the resulting En298-STM/GR protein should accumulate in the cytoplasm. A linkage between the dexamethasone treatment and alterations in SAM activity thus should depend on nuclear import of the protein and strongly argues against homology-based post-transcriptional gene silencing (26,27).
Although some of the primary en298-STM/GR transformants (T1) exhibited weak stm phenocopies, several primary transformants showed wild-type development. To test for dexamethasone inducibility, we focused on one T2 family with phenotypically normal kanamycin resistant seedlings. After wild-type development to the four- to five-leaf stage, 10 T2 transgenic seedlings were sprayed with dexamethasone and photographed daily to document the developmental progress. As controls, wild-type plants sprayed with dexamethasone or untreated transgenic progeny were analysed concomitantly. In the 6 day interval depicted in Figure 3, five to six new leaves emerged in the control plants, in contrast to transgenic en298-STM/GR plants, where only a single leaf appeared after spraying with dexamethasone. The hormone treatment also affected growth of pre-existing leaves: the petioles hardly elongated and the leaf blades remained closely attached to the shoot axis. The result depicted in Figure 3 is representative for 7 out of 10 transgenic progeny. The remaining three transgenic progeny showed no response to the hormone application. However, RNA gel-blot analysis showed the absence of the en298-STM/GR transcript which was detected at high levels in the responsive progeny (data not shown). The arrest in SAM activity therefore was strictly correlated with transcription of the transgene and the three non-responding escapes are presumably caused by insufficient transgene expression. One line carrying a single T-DNA insertion has since been propagated for five generations and whilst heterozygous progeny develop normally, homozygous seedlings show mild stm phenocopies in the absence of the hormone. This resembles the mild phenotypes observed among T1 en298-STM/GR primary transformants carrying multiple transgene copies (data not shown) and indicates that at higher concentrations the chimeric En298-STM/GR protein may act outside the plant cell nucleus.
Figure 3.
Dexamethasone-dependent arrest of SAM activity. Wild-type (control, left) or transgenic en298-STM/GR seedlings (right) were sprayed with dexamethasone at the four- to five-leaf stage and photographed daily. While five new leaves emerge in the control seedling (numbered +1 to +5), only a single new leaf is detectable in the transgenic en298-STM/GR seedlings in the depicted 6 day interval. Development in untreated transgenic en298-STM/GR siblings was also comparable to wild-type and resulted in four to five leaves.
Squelching versus transcriptional repression
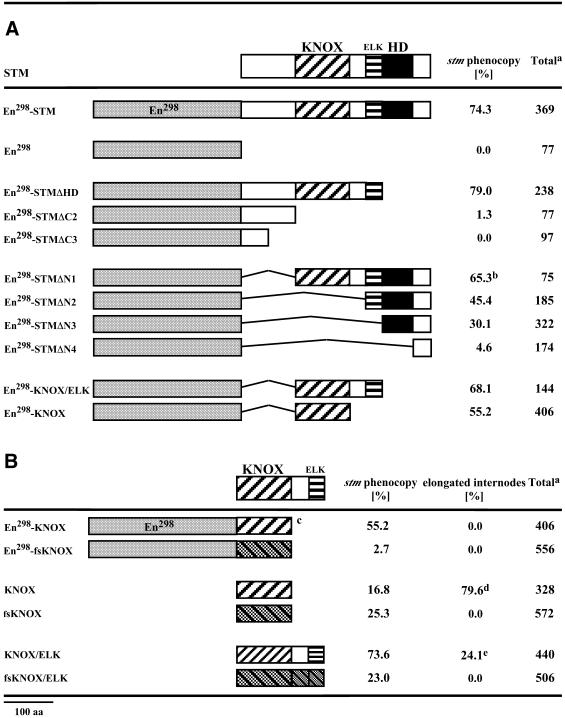
As an alternative to transcriptional repression, excessive chimeric En298-STM/GR protein in the cytoplasm could interact with potential partner proteins and thus deplete the native STM protein from its essential interaction partners—a mechanism frequently referred to as squelching. Consistent with this assumption, the specificity of floral phenocopies in en298-AP3 and en298-PI transgenic plants might be explained by the obligatory dimerisation of AP3 and PI monomers to the active AP3/PI heterodimer if an excess of En298-AP3 titrates the native PI protein or vice versa. To further investigate the molecular mechanism we created C- and N-terminal deletions of the STM protein and expressed these deletion polypeptides either fused or unfused to the En298 repressor domain in transgenic Arabidopsis plants. The whole series of deletion constructs is shown in Table 1A relative to conserved domains in the STM protein. The phenocopy frequency during vegetative development for each construct was determined 3 weeks after sowing (seven-leaf stage in wild-type).
Table 1. Deletional analysis of the STM protein.
aTotal number of analysed transgenic plants.
b48% of transgenic plants show a stm phenocopy and lobed leaves.
cSee also (A).
d23.2% of transgenic plants have elongated internodes and show a stm phenocopy.
e3% of transgenic plants have elongated internodes and show a stm phenocopy.
Results with chimeric En298-STM deletion polypeptides that lack the DNA-binding homeodomain support squelching as a mechanism. For example, the frequency of phenocopies obtained with the C-terminal deletion polypeptide that lacks the homeodomain (En298-STMΔHD) is as high as that obtained with the complete en298-STM coding region. Overall, the phenocopy frequency shows some direct correlation to the length to the STM cDNA fragment, but a major contribution is provided by the KNOX or MEINOX domain (28). Consistent with this assumption, 55% of the primary transformants obtained with the En298-KNOX(STM) fusion showed the characteristic dominant-negative phenotype. A further increase (68%) is observed in the presence of the ELK domain (En298-KNOX/ELK), which might contribute a nuclear localisation signal (29). Smaller KNOX subdomains did not cause any detectable phenotype (data not shown). These phenocopies strictly depend on the translational fusion between the En298 and the KNOX(STM) domains, as a frameshift version (En298-fs-KNOX(STM)), uncoupling translation in the KNOX(STM) domain does not affect plant development, similar to control plants transformed with the empty pCHRIS vector. Phenocopies, therefore, are strictly dependent on the expression of the chimeric En298-KNOX(STM) protein.
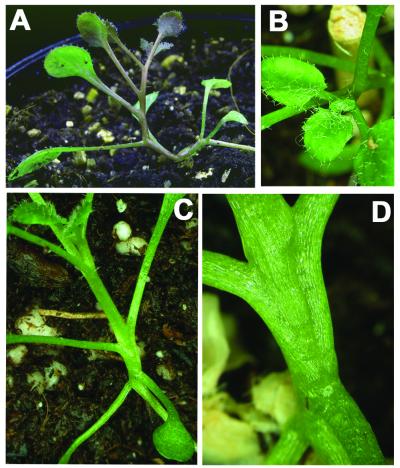
To test for the contribution of the En298 repressor domain, transgenic plants were raised expressing only the KNOX(STM) or KNOX/ELK(STM) polypeptides (Table 1B). Surprisingly, in the absence of the En298 repressor domain, all except 2–3% of the transgenic plants obtained with both constructs exhibited pronounced phenotypes. Severe phenocopies with only one to three leaves were observed in 74% of the 35S::KNOX/ELK transgenic plants and an additional 24% of plants showed internode elongation of rosette phytomers (Fig. 4A) but mostly maintained SAM activity (Fig. 4A and B). The ratio between phenocopies and elongated internodes is reversed in transgenic plants expressing the KNOX domain, where 16% of plants phenocopied a loss of stm function and 79% exhibited elongated internodes, with approximately 1/4 of these additionally showing a late block in SAM activity. This elongation of internodes is a novel phenotype not seen in En298 fusions and associated with alterations in phyllotaxy, e.g. changing from a spiral to a distichous pattern (Fig. 4C and D). This high frequency of phenotypes (97 or 98%) depends on the translation of the KNOX(STM) domain, as only 25 or 23% of stm phenocopies are observed in transgenic plants expressing frameshift versions of the KNOX(STM) or KNOX/ELK(STM) coding regions (Table 1B). The low, but similar, percentage of phenotypic plants observed with both frameshift versions contrasts with results with the en298-fsKNOX(STM) construct, where we have not seen any evidence for co-suppression. However, it indicates that homology-dependent silencing of the endogenous STM gene may contribute a minor fraction of the total frequency dependent on the construct. Although expression of the KNOX(STM) or KNOX/ELK(STM) domain provides dominant-negative functions, the associated novel phenotypes, e.g. internode elongation, indicate that the translational fusion with the En298 repressor domain largely contributes to the specificity of phenocopies.
Figure 4.
Phenotypes in transgenic 35S::KNOXSTM and 35S::KNOX/ELKSTM plants. (A) Elongated internodes in a 35S::KNOXSTM plant, small leaflets enclose the active SAM (B). Internodal elongation is often associated with alterations in phyllotaxy (C), a distichous pattern with two leaves emerging from a single node is depicted in (D).
Expression of the En298-KNAT1 polypeptide phenocopies the brevipedicellus mutant
To further investigate the specificity, we exchanged the STM coding region against the KNAT1 ORF. The KNAT1 expression pattern in the SAM is very similar to that of STM, but, in addition, the KNAT1 gene is transcribed in the cortical layers of the inflorescence and floral pedicel. Transgenic plants expressing en298-KNAT1 exhibited normal vegetative development (compare Fig. 5A and B), but in 75.6% of the primary transformants (508/672) a phenotype was observed in the inflorescence: the length of the internodes and the floral pedicel was reduced and the pedicels bent downwards, so that the orientation of flowers or siliques was opposite to that of the wild-type (compare Fig. 4C and D). This phenotype is reminiscent of the brevipedicellus (bp) mutant (30) which specifically affects inflorescence development. Although the bp-1 allele is in the Landsberg erecta (Ler) ecotype, the expression of en298-KNAT1 in the Columbia background leads to very similar alterations in the angle of side shoots or floral pedicels and in pedicel length relative to the corresponding wild-type background (Table 2). The increased number of secondary inflorescences (Fig. 5E) indicates that apical dominance is reduced in en298-KNAT1 transgenic and bp mutant plants. The elongation of internodes, which is reduced by 68% in bp-1/Ler is only reduced to 30% in en298-KNAT1/Col transgenic plants, and this is reflected in the total height of en298-KNAT1 plants. The number of flowers is unaffected in en298-KNAT1 transgenic and bp mutant plants. We also raised transgenic plants expressing the KNOX(KNAT1) or KNOX/ELK(KNAT1) domains fused to the En298 repressor domain. Strong bp phenocopies with flowers and siliques oriented downwards were observed only with the en298-KNOX/ELK(KNAT1) construct. In the absence of the ELK domain, flowers or siliques emerged, at most, perpendicular to the inflorescence axis. We did not observe an alteration in the SAM function with any of the chimeric KNAT1 genes.
Figure 5.
Transgenic en298-KNAT1 plants phenocopy bp mutants. Vegetative development in transgenic en298-KNAT1 plants is normal [compare wild-type (A) with the transgenic plant (B)]. The wild-type inflorescence (C) is compared with the inflorescence of a transgenic en298-KNAT1 plant (D). (E) Secondary inflorescences (arrows) emerging in a en298-KNAT1 plant. (F) Close-up of the bp inflorescence. (G) Partial complementation of the bp mutant phenotype by 35S::KNAT1 expression. Homozygous bp-1 mutant plants were transformed with the 35S::KNAT1 transgene and selected for BASTA resistance.
Table 2. Comparison between en298-KNAT1/Col and bp-1/Ler plants.
| Genotype | Height (cm) | Number of flowers | Average internode length (cm) | No. of secondary inflorescences | Angle (°) | Pedicel length (cm) | |
|---|---|---|---|---|---|---|---|
| Side shoot | Pedicel | ||||||
| Ler | 17.7 ± 0.2 | 28.3 ± 0.2 | 0.56 ± 0.01 | 0.14 ± 0.03 | 53 ± 2 | 56 ± 2 | 0.52 ± 0.05 |
| bp-1/Ler | 5.9 ± 0.2 | 29.1 ± 0.2 | 0.18 ± 0.01 | 1.8 ± 0.1 | 92 ± 3 | 132 ± 3 | 0.09 ± 0.01 |
| Δ | 11.8/67% | 0.38/68% | 0.43/83% | ||||
| Col | 35.5 ± 0.3 | 43.9 ± 0.3 | 0.66 ± 0.03 | 0.04 ± 0.01 | 62 ± 2 | 77 ± 1 | 0.69 ± 0.1 |
| en298-KNAT1/Col | 23.6 ± 0.4 | 45.2 ± 0.5 | 0.46 ± 0.04 | 2.2 ± 0.2 | 92 ± 1 | 127 ± 1 | 0.20 ± 0.1 |
| Δ | 11.9/34% | 0.2/30% | 0.49/71% |
Plants (n ≥ 20; en298-KNAT1: n = 60) were measured 50 days after sowing. The average internode length is determined for the first 3 cm (bp-1/Ler) or 10 cm (en298-KNAT1/Col) of the inflorescence starting at the point where the first flower arises. Values given are means ± SE.
KNAT1 and bp both map to chromosome 4 and the bp phenocopies in en298-KNAT1 transgenic plants suggested that bp may in fact represent a loss of KNAT1 function. To substantiate this assumption the KNAT1 ORF was expressed behind the constitutive CaMV 35S promoter in the bp-1 mutant background. A resulting 35S::KNAT1 transgenic plant in the bp-1/Ler background is shown in Figure 5G and demonstrates a rescue of the mutant phenotype (compare Fig. 5G and F). The internode and the pedicel are elongated in the 35S::KNAT1 transgenic bp-1/Ler plant compared with the bp-1 mutant and the flowers/siliques point upwards or are oriented at least perpendicular to the stem axis. The strong lobing of rosette leaves is typical of KNAT1 overexpression from the 35S promoter and is due to ectopic activity in the leaves. Both the bp phenocopy in en298-KNAT1 transgenic plants and the complementation of the bp-1 mutant in 35S::KNAT1 plants thus argue that KNAT1 is encoded by the BP locus. A similar conclusion was recently achieved by two other groups (31,32), together with the suggestion that the bp phenotype may largely depend on the erecta mutant background. However, this assumption is not supported by our data for trans-dominant en298-KNAT1 phenocopies in the Columbia background comprising a wild-type ERECTA function.
DISCUSSION
The experimental data demonstrate that ectopic expression of N-terminal fusion proteins between the en298 N-terminus and the complete STM, KNAT1, AP3 or PI coding regions results in dominant-negative functions which mimic loss-of-function alleles. With all four test genes the efficiency is high and the frequency of characteristic phenocopies in primary transformants always exceeds 75%. The trans-dominant phenocopies are specific and distinct from phenotypes observed in overexpression experiments with AP3, PI, STM or KNAT1 (33,34) The homogeneity of the resulting phenotypes and their high frequency in primary transformants provide rapid and reliable information about the contribution of each gene during plant development. Although STM, AP3 and PI were chosen as test genes because of their predictable loss-of-function phenotypes, the association between KNAT1 and the BREVIPEDICELLUS locus became apparent during these studies and is consistent with parallel efforts recently published by other groups (31,32).
The question of the molecular mechanism was addressed in various ways. In combination with the hormone-binding domain of the glucocorticoid receptor, the activity of the En298-STM/GR fusion polypeptide is under control of the dexamethasone hormone, which allows the incorporation of the chimeric protein into the nuclear compartment (23). The inducibility of the En298-STM/GR fusion protein provides evidence that the main mechanism is protein-based, although post-transcriptional gene silencing (26,27) may contribute to the total frequency at a low level. Based on frameshift non-sense versions, e.g. fsKNOX or fsKNOX/ELK, the co-suppression frequency is two to four times lower than that observed with the corresponding sense constructs (see Table 1B). When a frameshift (+1) is introduced C-terminally to the En298 domain and in front of the KNOX domain (en298-fsKNOX), thus uncoupling translation of the repressor domain from the STM sequences, no phenotypic alterations are detectable. The fraction of phenocopy plants attributable to co-suppression therefore depends on the construct: there may be a low contribution of co-suppression, but this cannot account for the high phenocopy frequency.
Besides hormone control in en298-STM/GR transgenic plants, the most compelling argument for a protein-based mechanism is provided by results for KNOX(STM) or KNOX/ELK(STM) transgenic plants. More than 97% of the primary transformants expressing the sense constructs exhibit phenotypic alterations compared with <25% with the nonsense frameshift versions. Most of the frequency therefore depends on the translation of the KNOX domain, which may interact with protein partners and deplete native endogenous proteins including STM from essential interaction partners. Squelching could thus account for the dominant-negative function due to KNOX(STM) overexpression, and resembles results for the removal of the bZIP domain in the tobacco transcription factor TGA1a which interferes with the formation of the ASF-1 complex (35,36). All effects on the SAM are specific to the KNOX(STM) or KNOX/ELK(STM) domains, since the expression of the corresponding KNAT1 subdomains has no detectable consequence for SAM activity.
Remarkably, the internode elongation in transgenic plants expressing KNOX(STM) or KNOX/ELK(STM) polypeptides is not observed in fusions with the En298 repressor domain. All the En298 fusions result in a single phenotype, which mimics a loss of stm function, with only the frequency of phenocopies being variable. The same specificity is observed with the En298-AP3, the En298-PI or the En298-KNAT1 fusions. In each case, the N-terminal fusion of the En298 repressor domain to the full-length protein results in specific dominant-negative functions, which rapidly uncover the biological contribution of the corresponding gene product. Neomorphic phenotypes, e.g. internode elongation as obtained with the KNOX(STM) or the KNOX/ELK(STM) domain, are missing in the presence of the En298 repressor domain which therefore highly increases the specificity of phenocopies.
Contribution of the En298 repressor domain
The 298 amino acid residues of the en gene product fused with the plant sequences here span the eh1 domain conserved in numerous homeodomain repressor proteins (34). The eh1 motif is known to interact with Groucho (37), a transcriptional co-repressor, which is recruited to DNA by a variety of DNA-binding proteins. Groucho homologues have been cloned from several animal species and the corresponding protein in yeast Tup1 is thought either to position nucleosomes over the core promoter (38) or to directly inhibit the RNA–polymerase initiation complex (39). Recalling the similarity in the basal transcriptional machinery between plants and animals (15,40,41) it cannot be excluded that Groucho homologous co-repressor functions exists in plants, although solely based on sequence homology these are not rapidly identified in the Arabidopsis genome. One possibility, therefore, seems that the chimeric En298 fusion proteins act as transcriptional repressors. Expressed from the strong CaMV 35S promoter, their vast excess may displace the native gene products from target gene promoters thus explaining trans-dominance over the functional endogenous gene.
Although the dexamethasone response in en298-STM/GR transgenic plants implicates the cell nucleus as the target compartment, this does not imply that the chimeric protein exhibits an active repressor function. Trans-dominant phenocopies with STM deletion polypeptides are obtained in the absence of the DNA-binding homeodomain. However, a homeodomain-deleted Fushi tarazu (Ftz) polypeptide incapable of binding DNA can control segmentation in Drosophila, through interactions with other proteins (42). In analogy, En298-STM deletion derivatives might still be recruited to target gene promoters with the help of interacting proteins.
Whether the En298 N-terminal sequences actively repress transcription of target genes in the plant cell nucleus remains an open question, because all test proteins presumably rely on partner proteins. In contrast to active repression, competitive modes can be envisioned. Non-functional En298 fusion proteins included into heterodimeric or multimeric protein complexes may displace functional native complexes from target genes, or excessive En298 fusion proteins may interfere with the assembly of functional complexes or their nuclear uptake. Independent of whether the dominant-negative functions rely on transcriptional repression of target gene promoters or are based on competition, the underlying mechanism has to explain how the ubiquitous expression of en298-STM, en298-AP3, en298-PI or en298-KNAT1 constructs can result in precise phenocopies of loss-of-function alleles. Transcribed from the constitutive CaMV 35S promoter, organ specificity associated with the dominant-negative functions cannot reside in transcriptional activity. An attractive assumption is obligatory and cell- or organ-specific protein–protein interactions, which have been shown for the AP3 and PI gene products (43). Although the En298-AP3 or En298-PI fusion proteins are expressed ubiquitously from the 35S promoter, the essential interaction partner is still confined to the second and third floral whorl, thus confining activity to the flower. We cannot discriminate whether excess of the En298-PI or En298-AP3 fusion protein binds the essential AP3 or PI partner protein in non-functional complexes, or whether the En298 fusion is recruited to target genes as heterodimers. In both scenarios, target promoters dependent on the activation via AP3/PI heterodimers would be negatively affected, resulting in a specific B-function phenotype. As there is no evidence for STM or KNAT1 homodimers (M. Cole and W. Werr, unpublished results) the specificity of phenocopies in en298-STM or en298-KNAT1 transgenic plants indicates that protein partners may be confined to the SAM or the inflorescence, respectively. This assumption gains further support since the isolated KNOX domains of STM and KNAT1 cause phenocopies either exclusive to the SAM or the inflorescence. Therefore, the phenocopy specificity most likely relies on compatible protein interactions which are confined to individual cell types or plant organs.
Applications
Dominant-negative functions may provide a rapid method to elucidate the biological function of plant transcription factors. The opposite approach, use of the strong VP16 activation domain fused to the meristem identity gene LEAFY, uncovered transcriptional activation of the organ-identity gene AGAMOUS (44). After completion of the genome-sequencing project the sequence of the whole set of transcription factors in Arabidopsis is known, but for most of these genes biological functions and contributions to regulatory networks remain to be elucidated. As efficient gene disruption systems based on homologous recombination are lacking in plants, this deficiency is circumvented by laborious PCR-based screens for insertion alleles in large transposon or T-DNA populations, so-called gene machines. However, any success in reverse or true genetic screens depends on a unique gene function. Dominant-negative functions as demonstrated here with AP3, PI, KNAT1 and STM may not only be more rapid, but also superior in the case of redundancy. At the protein level, excess of the chimeric gene product might compete not only with its native counterpart, but simultaneously with other family members that possibly mask a desired gene function.
In contrast to gene machines, which will remain restricted to a few model plants only, dominant-negative functions are applicable to all species susceptible to transformation. As the AP3 function in Arabidopsis for example can be partially complemented by the homologous DEFICIENS gene from Antirrhinum (45), it is likely that dominant-negative functions would also be informative in comparisons between species. Since the evolution of regulatory networks probably accounts for the majority of variation between different plant species, the central position of transcription factors in gene regulation makes them an interesting subject for such functional comparisons, both from academic and applied perspectives. On the applied side, dominant-negative functions may be suitable to transfer knowledge elaborated in model systems like Arabidopsis to crop species.
In conclusion, the data described here provide evidence that plant transcription factors can be reprogrammed to dominant-negative functions by fusion to the en298 N-terminus. Although we cannot distinguish between active transcriptional repression and a competitive mode of action, dominant-negative functions may be of wide use in elucidating biological functions of plant transcriptional factors. Reproducibly, with four transcription factors, we observed ∼75% phenocopies of loss-of-function alleles in primary transformants. The N-terminal addition of the En298 repressor domain to the complete coding region of plant transcription factors therefore provides a rapid and efficient method to visualise a biological contribution of a gene of interest.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor Michael Hoch (Universität Bonn) for the gift of the engrailed cDNA clone D2B, and Petra Comelli and Melanie Cole for excellent technical assistance. The STM cDNA clone was kindly provided by Dr Rüdiger Simon (Universität zu Köln), whom we also thank for experimental suggestions and critical reading of the manuscript. The AP3 cDNA clone was kindly provided by Dr Zsuzsanna Schwarz-Sommer (MPI für Züchtungsforschung, Köln). Part of this work was supported through GENOPLANTE (project number NO1999013) and the Deutsche Forschungsgemeinschaft WE-1262/3-1. H.M. was funded by the Graduiertenkolleg ‘Molekulare Analyse von Entwicklungsprozessen’ at the University of Cologne.
REFERENCES
- 1.Riechmann J.L., Heard,J., Martin,G., Reuber,L., Jiang,C., Keddie,J., Adam,L., Pineda,O., Ratcliffe,O.J., Samaha,R.R., Creelman,R., Pilgrim,M., Broun,P., Zhang,J.Z., Ghandehari,D., Sherman,B.K. and Yu,G. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science, 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- 2.Kerstetter R., Vollbrecht,E., Lowe,B., Veit,B., Yamaguchi,J. and Hake,S. (1994) Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell, 6, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riechmann J.L. and Meyerowitz,E.M. (1997) MADS domain proteins in plant development. Biol. Chem., 378, 1079–1101. [PubMed] [Google Scholar]
- 4.John A., Smith,S.T. and Jaynes,J.B. (1995) Inserting the Ftz homeodomain into engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo. Development, 121, 1801–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margolin J.F., Friedman,J.R., Meyer,W.K., Vissing,H., Thiesen,H.J. and Rauscher,F.J.,III (1994) Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl Acad. Sci. USA, 91, 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerli R.R., Segal,D.J., Dreier,B. and Barbas,C.F.,III (1998) Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA, 95, 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyon J.J. and Watson,R.J. (1996) Interference of Myb transactivation activity by a conditional dominant negative protein: functional interference in a cytotoxic T-cell line results in G1 arrest. Gene, 182, 123–128. [DOI] [PubMed] [Google Scholar]
- 8.Conlon F.L., Sedgwick,S.G., Weston,K.M. and Smith,J.C. (1996) Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development, 122, 2427–2435. [DOI] [PubMed] [Google Scholar]
- 9.Nikolov D.B., Hu,S.H., Lin,J., Gasch,A., Hoffmann,A., Horikoshi,M., Chua,N.H., Roeder,R.G. and Burley,S.K. (1992) Crystal structure of TFIID TATA-box binding protein. Nature, 360, 40–46. [DOI] [PubMed] [Google Scholar]
- 10.Juo Z.S., Chiu,T.K., Leiberman,P.M., Baikalov,I., Berk,A.J. and Dickerson,R.E. (1996) How proteins recognize the TATA box. J. Mol. Biol., 261, 239–254. [DOI] [PubMed] [Google Scholar]
- 11.Kranz H.D., Denekamp,M., Greco,R., Jin,H., Leyva,A., Meissner,R.C., Petroni,K., Urzainqui,A., Bevan,M., Martin,C., Smeekens,S., Tonelli,C., Paz-Ares,J. and Weisshaar,B. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J., 16, 263–276. [DOI] [PubMed] [Google Scholar]
- 12.Schena M. and Davis,R.W. (1992) HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl Acad. Sci. USA, 89, 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni M., Tepperman,J.M. and Quail,P.H. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell, 95, 657–667. [DOI] [PubMed] [Google Scholar]
- 14.Wilde R.J., Cooke,S.E., Brammar,W.J. and Schuch,W. (1994) Control of gene expression in plant cells using a 434:VP16 chimeric protein. Plant Mol. Biol., 24, 381–388. [DOI] [PubMed] [Google Scholar]
- 15.Moore I., Galweiler,L., Grosskopf,D., Schell,J. and Palme,K. (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl Acad. Sci. USA, 95, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uesugi M., Nyanguile,O., Lu,H., Levine,A.J. and Verdine,G.L. (1997) Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science, 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 17.Long J.A., Moan,E.I., Medford,J.I. and Barton,M.K. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature, 379, 66–69. [DOI] [PubMed] [Google Scholar]
- 18.Jack T., Brockman,L.L. and Meyerowitz,E.M. (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell, 68, 683–697. [DOI] [PubMed] [Google Scholar]
- 19.Goto K. and Meyerowitz,E.M. (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev., 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- 20.Lincoln C., Long,J., Yamaguchi,J., Serikawa,K. and Hake,S. (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell, 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole S.J., Kauvar,L.M., Drees,B. and Kornberg,T. (1985) The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell, 40, 37–43. [DOI] [PubMed] [Google Scholar]
- 22.Überlacker B. and Werr,W. (1996) Optimized vectors for expression and transfer of large open reading frames in transgenic plants. Mol. Breeding, 2, 293–295. [Google Scholar]
- 23.Lloyd A.M., Schena,M., Walbot,V. and Davis,R.W. (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science, 266, 436–439. [DOI] [PubMed] [Google Scholar]
- 24.Bechtold N. and Pelletier,G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol., 82, 259–266. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmann,J.G., Smith,J.A. and Struhl,K. (1987) Preparation and analysis of RNA. In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley and Sons, New York.
- 26.Cogoni C. and Macino,G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. [DOI] [PubMed] [Google Scholar]
- 27.Maine E.M. (2000) A conserved mechanism for post-transcriptional gene silencing? Genome Biol., 1, REVIEWS1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bürglin T.R., Finney,M., Coulson,A. and Ruvkun,G. (1989) Caenorhabditis elegans has scores of homoeobox-containing genes. Nature, 341, 239–243. [DOI] [PubMed] [Google Scholar]
- 29.Meisel L. and Lam,E. (1996) The conserved ELK-homeodomain of KNOTTED-1 contains two regions that signal nuclear localization. Plant Mol. Biol., 30, 1–14. [DOI] [PubMed] [Google Scholar]
- 30.Koornneef M., Eden,J.v., Hanhart,C.J., Stam,P., Braaksma,F.J. and Feenstra,W.J. (1983) Linkage map of Arabidopsis thaliana. J. Hered., 74, 265–272. [Google Scholar]
- 31.Douglas S.J., Chuck,G., Dengler,R.E., Pelecanda,L. and Riggs,C.D. (2002) KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell, 14, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venglat S.P., Dumonceaux,T., Rozwadowski,K., Parnell,L., Babic,V., Keller,W., Martienssen,R., Selvaraj,G. and Datla,R. (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl Acad. Sci. USA, 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krizek B.A. and Meyerowitz,E.M. (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development, 122, 11–22. [DOI] [PubMed] [Google Scholar]
- 34.Williams R.W. (1998) Plant homeobox genes: many functions stem from a common motif. Bioessays, 20, 280–282. [DOI] [PubMed] [Google Scholar]
- 35.Rieping M., Fritz,M., Prat,S. and Gatz,C. (1994) A dominant negative mutant of PG13 suppresses transcription from a cauliflower mosaic virus 35S truncated promoter in transgenic tobacco plants. Plant Cell, 6, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao Z.H. and Lam,E. (1995) Construction of a trans-dominant inhibitor for members of the TGA family of transcription factors conserved in higher plants. Plant J., 6, 887–896. [DOI] [PubMed] [Google Scholar]
- 37.Tolkunova E.N., Fujioka,M., Kobayashi,M., Deka,D. and Jaynes,J.B. (1998) Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol., 18, 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- 39.Herschbach B.M., Arnaud,M.B. and Johnson,A.D. (1994) Transcriptional repression directed by the yeast alpha 2 protein in vitro. Nature, 370, 309–311. [DOI] [PubMed] [Google Scholar]
- 40.Wilde R.J., Cooke,S.E., Brammar,W.J. and Schuch,W. (1994) Control of gene expression in plant cells using a 434:VP16 chimeric protein. Plant Mol. Biol., 24, 381–388. [DOI] [PubMed] [Google Scholar]
- 41.Nikolov D.B., Hu,S.H., Lin,J., Gasch,A., Hoffmann,A., Horikoshi,M., Chua,N.H., Roeder,R.G. and Burley,S.K. (1992) Crystal structure of TFIID TATA-box binding protein. Nature, 360, 40–46. [DOI] [PubMed] [Google Scholar]
- 42.Copeland J.W., Nasiadka,A., Dietrich,B.H. and Krause,H.M. (1996) Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature, 379, 162–165. [DOI] [PubMed] [Google Scholar]
- 43.Samach A., Kohalmi,S.E., Motte,P., Datla,R. and Haughn,G.W. (1997) Divergence of function and regulation of class B floral organ identity genes. Plant Cell, 9, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busch M.A., Bomblies,K. and Weigel,D. (1999) Activation of a floral homeotic gene in Arabidopsis. Science, 285, 585–587. [DOI] [PubMed] [Google Scholar]
- 45.Irish V.F. and Yamamoto,Y.T. (1995) Conservation of floral homeotic gene function between Arabidopsis and Antirrhinum. Plant Cell, 7, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]