Abstract
Hepatitis C virus (HCV) is a leading cause of chronic hepatitis in the world. The study of viral entry and infection has been hampered by the inability to efficiently propagate the virus in cultured cells and the lack of a small-animal model. Recent studies have shown that in insect cells, the HCV structural proteins assemble into HCV-like particles (HCV-LPs) with morphological, biophysical, and antigenic properties similar to those of putative virions isolated from HCV-infected humans. In this study, we used HCV-LPs derived from infectious clone H77C as a tool to examine virus-cell interactions. The binding of partially purified particles to human cell lines was analyzed by fluorescence-activated cell sorting with defined monoclonal antibodies to envelope glycoprotein E2. HCV-LPs demonstrated dose-dependent and saturable binding to defined human lymphoma and hepatoma cell lines but not to mouse cell lines. Binding could be inhibited by monoclonal anti-E2 antibodies, indicating that the HCV-LP-cell interaction was mediated by envelope glycoprotein E2. Binding appeared to be CD81 independent and did not correlate with low-density lipoprotein receptor expression. Heat denaturation of HCV-LPs drastically reduced binding, indicating that the interaction of HCV-LPs with target cells was dependent on the proper conformation of the particles. In conclusion, our data demonstrate that insect cell-derived HCV-LPs bind specifically to defined human cell lines. Since the envelope proteins of HCV-LPs are presumably presented in a virion-like conformation, the binding of HCV-LPs to target cells may allow the study of virus-host cell interactions, including the isolation of HCV receptor candidates and antibody-mediated neutralization of binding.
Hepatitis C virus (HCV) is a major cause of posttransfusion and community-acquired hepatitis (2, 3, 13, 34). The majority of HCV-infected individuals develop chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (34, 46). Treatment options for chronic HCV infection are limited, and a vaccine to prevent HCV infection is not available (31, 33, 34).
HCV has been tentatively classified in a separate genus (Hepaticivirus) of the family Flaviviridae (4, 36, 43). The virion contains a positive-stranded RNA genome of approximately 9.6 kb. The genome consists of a highly conserved 5′ noncoding region followed by a long open reading frame of 9,030 to 9,099 nucleotides (nt) that is translated into a single polyprotein of 3,010 to 3030 amino acids (aa) (4, 36). Processing of the polyprotein occurs with a combination of host and viral proteases. The HCV structural proteins comprise the putative nucleocapsid or core protein and the two envelope glycoproteins, E1 and E2 (4, 36, 43). The cleavage of structural proteins from the polyprotein is catalyzed by a host signal peptidase. Envelope proteins E1 and E2 are transmembrane proteins consisting of a large N-terminal ectodomain and a C-terminal hydrophobic anchor. E1 and E2 are posttranslationally modified by extensive N-linked glycosylation (for review see references 16 and 21). The envelope glycoproteins have been shown to assemble into a noncovalent heterodimer which is retained in the endoplasmic reticulum (15). This heterodimer is believed to be the prebudding form of an HCV glycoprotein complex (16). In insect cells, the HCV structural proteins have been shown to assemble into enveloped virus-like particles (HCV-LPs) (5) with morphological, biophysical, and antigenic properties similar to those of putative virions isolated from HCV-infected humans (5, 8, 9). In contrast to individually expressed envelope glycoproteins, the E1-E2 heterodimers of insect cell-derived HCV-LPs are presumably presented in a native, virion-like conformation (8).
Although a detailed analysis of the viral genomic organization has led to the identification of various genetic elements (4) and the establishment of subgenomic replicons (10, 35), the study of viral entry and infection is still hampered by the inability to propagate the virus efficiently in cultured cells and the limited animal tropism of the virus. The chimpanzee is the only nonhuman host serving as a model for HCV infection (18, 29, 52).
Binding of individually expressed recombinant glycoprotein E2 to human cell lines has been used as a surrogate model for binding of virus to host cells, allowing the study of antibody-mediated neutralization of binding (44). Using this surrogate assay, Pileri at al. have demonstrated that envelope glycoprotein E2 interacts with the large extracellular loop of cellular membrane protein CD81 (41), a member of the tetraspanin family (32). CD81 has been suggested as a HCV receptor candidate (20, 21), and an E2-CD81 interaction may play a role in T-cell activation (48).
In this study, we demonstrate that HCV-LPs derived from infectious clone H77C bind efficiently to human cell lines and represent a novel model for the study of virus-host interactions.
(This study was presented in part in abstract form at the 7th and 8th International Meeting of Hepatitis C and Related Viruses, 3 to 7 December 2000, Gold Coast, Australia, and 2 to 5 September 2001, Paris, France.)
MATERIALS AND METHODS
Antibodies and recombinant proteins.
Mouse anti-core (C1 and C2), anti-E1 (A4), and anti-E2 (E2G and E2H) monoclonal antibodies (MAbs) directed against an HCV genotype 1a strain were described previously (5, 25) and were generously provided by H. B. Greenberg (Division of Gastroenterology, Department of Medicine, Stanford University School of Medicine, Palo Alto, Calif.). Mouse anti-E1 (MAb 159) and anti-E2 (MAb 917) MAbs directed against an HCV genotype 1b strain were provided by J. Y. N. Lau (Schering-Plough Corporation, Kenilworth, N.J.). Mouse anti-E2 MAb 3E5 was generously provided by M. Houghton (Chiron Corp., Emeryville, Calif.). Mouse MAbs (H33, H50, H53, and H60) directed against conformation-dependent epitopes of E2 were described previously (15, 39). An additional panel of anti-E2 antibodies (4F7, 4E5, and 7B7) directed against conformation-dependent epitopes was a generous gift from M. Da Silva Cardoso (14). Mouse anti-human CD81 MAbs 5A6 and 1D6 were described previously (20, 27) and were provided by S. Levy (Divison of Oncology, Department of Medicine, Stanford University School of Medicine). A mouse anti-human low-density lipoprotein (LDL) receptor (LDLr) MAb (C7 clone) (1, 51) was obtained from Progen Biotechnik Corp., Heidelberg, Germany. A mouse antigalactosidase (anti-LacZ) MAb was obtained from Boehringer Mannheim, Mannheim, Germany. Human sera containing antibodies against HCV were obtained from patients with chronic hepatitis C and high-titer anti-HCV antibodies as decribed previously (5). Purified recombinant E2 (aa 384 to 715) protein (44) was generously provided by M. Houghton (Chiron). Recombinant soluble glutathione S-transferase-CD81 large extracellular loop (LEL) fusion proteins (CD81-LEL) were generous gifts from S. Levy and were described in detail elsewhere (27).
Cell lines.
The human hepatoma cell lines HuH-7 and HepG2 were generous gifts from T. Jake Liang, National Institutes of Health, Bethesda, Md. (6, 7), and Peter Hafkemeyer, Department of Medicine II, University of Freiburg, Freiburg, Germany. Human MOLT-4 and Daudi lymphoma cells were obtained from Paul Fisch, Tumorbiology Center Freiburg, Freiburg, Germany. Mouse hepatoma Hepa 1-6 and lymphoma SP2-0 cell lines and COS-7 cells from African green monkeys were generously provided by Leonhard Mohr and Ursula Schultz, Department of Medicine II, University of Freiburg, respectively. HuH-7, HepG2, COS-7, and Hepa 1-6 cells were maintained in Dulbecco modified Eagle medium (Gibco Life Technologies, Gaithersburg, Md.) containing 10% fetal calf serum (FCS) (PAA Laboratories, Linz, Austria). MOLT-4, Daudi, and SP2-0 cells were maintained in RPMI medium (Gibco Life Technologies) containing 10% FCS (PAA Laboratories) or 10% lipoprotein-deficient human serum (LPDS; obtained from M. Nauck, Division of Clinical Chemistry, Department of Medicine, University of Freiburg). HepG2, COS-7, Hepa 1-6, and SP2-0 cells were originally obtained from the American Type Culture Collection. Maintenance of Spodoptera frugiperda Sf9 insect cells (obtained from Gibco Life Technologies) was described in detail elsewhere (5). LDL concentrations in FCS were determined by K. Winkler (Division of Clinical Chemistry, Department of Medicine, University of Freiburg) as described elsewhere (50a).
Recombinant plasmids and baculoviruses.
pCVH77C, containing the cDNA needed to generate an infectious full-length HCV genome (52), was a generous gift from J. Bukh and R. Purcell, Hepatitis Viruses Section, NIAID, National Institutes of Health. It was used to generate recombinant baculovirus BVHCV.S1a. Shuttle plasmid pFastBacHCV.S1a was generated by subcloning an NruI-Tth111I fragment (nt 269 to 2828) of pCVH77C (52) into the EcoRI and SpeI sites of pFastBac (Gibco Life Technologies; partial digestion with NruI). The EcoRI, Tth111I, and SpeI sites were blunt ended with the Klenow fragment before ligation. The correct sequence of pFastBacHCV.S1a was confirmed by multiple restriction enzyme digestions and sequencing. Plasmid pCDHCV.S1a, containing the cDNA for the HCV structural proteins of pCVH77C under the control of a cytomegalovirus promoter, was generated by ligating the EcoRI-XbaI fragment of pFastbacHCV.S1a into pCDNA3 (Invitrogen, Carlsbad, Calif.).
Synthesis and purification of HCV-LPs in insect cells.
The procedures used for the expression and purification of HCV-LPs were described previously (5). For binding assays, sucrose gradient-purified particles were dialyzed overnight at 4°C against phosphate-buffered saline (PBS) by using Spectrapor DispoDialyzer CE10 (Spectrum Labs Inc., Rancho Dominguez, Calif.). The HCV-LP E2 concentration was determined by using an E2-specific enzyme-linked immunosorbent assay (ELISA) as described recently (39). Highly purified recombinant E2 (aa 384 to 715) protein and anti-E2 antibody 3E5 served as calibration reagents for E2 quantitation (39). For electron microscopy, partially purified HCV-LPs were fixed and processed as described previously (5, 8). For comparison of HCV protein expression in insect and mammalian cells, HuH-7 cells were transfected with pCDHCV.S1a or pCDNA3 by using liposome-mediated gene transfer (Lipofectamine; Gibco Life Technologies). At 72 h posttransfection, cells were lysed and HCV protein expression was analyzed by immunoblotting as described previously (5).
HCV-LP ELISA.
HCV-LPs, produced and partially purified as described above, were diluted in PBS and used to coat (1 μg of total protein/well) 96-well ELISA plates (ProBindIII; Becton Dickinson) by overnight incubation at 4°C. The plates were then blocked with PBS containing 5% low-fat dry milk (Carnation; Nestle Food Corp., Glendale, Calif.) and 4% goat serum (KPL Laboratories, Gaithersburg, Md.) for 4 h and incubated with monoclonal antienvelope antibodies diluted in PBS containing 5% dry milk for 1 h. The plates were then incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Amersham, Arlington Heights, Ill.) diluted 1:1,000 in PBS-5% dry milk for 1 h and washed six times. The particle-bound antibodies were quantitated by colorimetric reaction by measuring the optical density (OD) at 490 nm with an Abbott Laboratories (Chicago, Ill.) OPD detection kit. All samples and dilutions were tested in duplicate. The HCV-LP ELISA cutoff value was determined to be 2.5 times the mean value for the control (OD of insect cell control preparation). The anti-HCV-LP titer was determined by the end-point dilution method to be the highest dilution factor that gave an OD below the cutoff value of the ELISA.
Analysis of binding of HCV-LPs and E2 to cell lines.
The binding of HCV-LPs and recombinant E2 to cell lines was accomplished as described recently (44). Human or mouse cells (1.5 × 105 or 1 × 106 cells per assay) were incubated with HCV-LPs or recombinant E2 protein at various concentrations in PBS (final volume, 100 or 200 μl) for 1 h at 4°C. The concentration of the HCV-LP E2 protein or the individually expressed E2 protein was determined by the E2-specific ELISA (described above). Nonbound HCV-LPs or E2 protein was removed by three centrifugations in PBS at 200 × g for 5 min at 4°C. Cells were subsequently incubated for 40 min at 4°C with anti-E2 or mouse isotype control MAb. The cells were washed four times in PBS at 4°C, subsequently resuspended in 200 μl of PBS, and incubated with phycoerythrin (PE)-conjugated anti-mouse IgG antibody. Cell-bound fluorescence was analyzed with a FACScan or FACScalibur flow cytometer (Becton Dickinson) by using Lysis II or Cellquest 3.11 software. These programs produce histograms of each sample and calculate the mean fluorescence intensity (MFI) of the cell population, which directly relates to the surface density of PE-labeled E2 bound to hepatocytes (44). The MFI values of cells with or without HCV-LPs or E2 protein and with isotype control antibody or anti-E2 antibody were compared. The threshold for positivity was set for each experiment by flow cytometric analysis of hepatocytes incubated with a control insect cell preparation or PBS or incubated with anti-E2 and PE-conjugated secondary antibodies (44).
To study whether the binding of HCV-LPs can be blocked by recombinant CD81-LEL, HCV-LPs were preincubated with soluble CD81-LEL (50 μg/ml for 1 h at 25°C; human or African green monkey CD81), and the binding of HCV-LPs to cell lines was assessed by fluorescence-activated cell sorting (FACS) as described above. To assess whether binding could be blocked by anti-CD81 or anti-LDLr antibody, HuH-7 cells or MOLT-4 cells were incubated with anti-CD81 or anti-LDLr antibody concentrations previously shown to saturate CD81 (MAb 1D6/5A6; 10 μg/ml) (20) or LDLr (MAb C7; 20 μg/ml) (1) prior to the addition of HCV-LPs. Binding of HCV-LPs in the presence of anti-CD81 or anti-LDLr antibody was detected by using a biotinylated anti-E2 antibody and streptavidin-PE. For the assessment of antibody-mediated neutralization of binding, sucrose gradient-purified HCV-LPs (corresponding to 0.25 μg of HCV-LP E2) were incubated with antienvelope MAbs (1:50 dilution, corresponding to an antibody concentration of 20 μg/ml) overnight at 4°C (final volume, 200 μl in PBS). HCV-LP-antibody complexes were added to MOLT-4 cells, and the binding of HCV-LPs was detected as described above. To examine whether trypsin pretreatment of cells can alter HCV-LP binding, MOLT-4 cells were exposed to trypsin (0.05 to 0.5%; Gibco Life Technologies) for 5 min, and trypsin activity was neutralized by the addition of medium containing 10% FCS and repetitive washing of the cells with PBS.
RESULTS
Synthesis of HCV-LPs derived from infectious clone H77C.
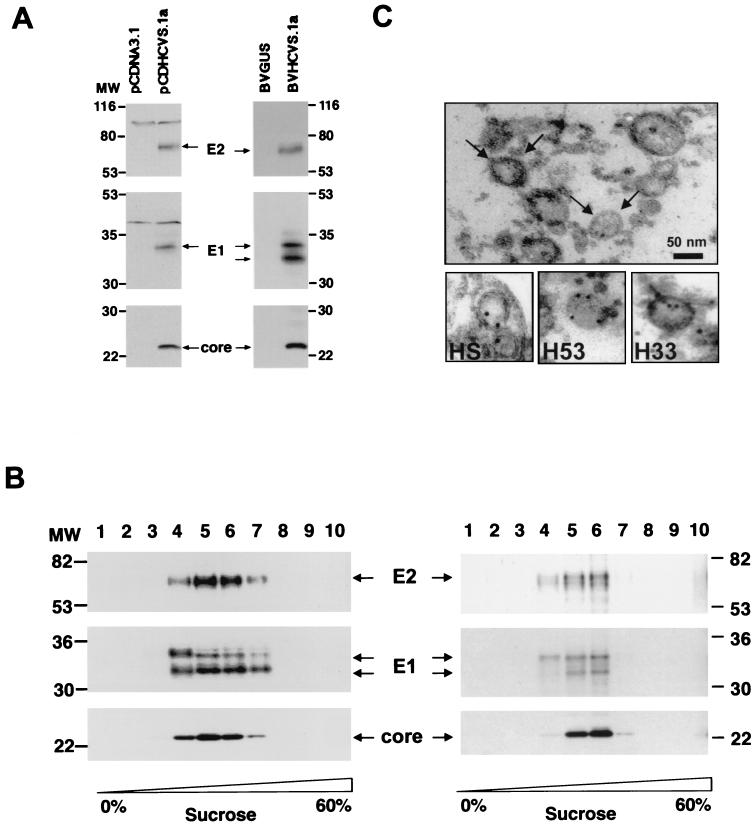
Clone H77C is an HCV prototype genome isolated from a well-chararacterized individual with chronic hepatitis. RNA transcribed from this clone is infectious in the chimpanzee model (52). To analyze whether H77C structural proteins assemble into HCV-LPs in insect cells, a recombinant baculovirus (BVHCV.S1a) was generated containing the coding sequences for the core, E1, E2, and p7 proteins and 21 aa of the NS2 protein as well a short stretch of the 5′ untranslated region (HCV nt 269 to 2828) of H77C (Fig. 1A) . BVHCV.S1a directed the efficient production of HCV structural proteins in insect cells, as demonstrated by immunoblotting of insect cell lysates (Fig. 1A). Analysis of insect cell lysates by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting with MAbs against the core, E1, and E2 proteins revealed appropriate posttranslational processing of HCV structural proteins. The size of the core protein was identical to that of the core protein expressed in a mammalian tissue culture system (Fig. 1A). The sizes of the E1 and E2 proteins were similar (E1, 33 and 34 kDa; E2, 68 kDa) (Fig. 1A) but not identical to the sizes of envelope proteins expressed in mammalian cells. The different pattern of envelope protein expression was due to differences in glycosylation between the two protein expression systems, as demonstrated by endoglycosidase H digestion (data not shown). In mammalian cells, E1 was expressed as a single glycosylated protein (34 kDa), whereas E1 expression in insect cells resulted in the formation of two glycosylated forms of E1 (33 and 34 kDa) (Fig. 1A).
FIG. 1.
Synthesis and purification of HCV-LPs derived from infectious clone H77C. (A) Expression of HCV structural proteins. Sf9 insect cells were infected with control baculovirus BVGUS (containing the cDNA for β-glucuronidase [GUS]) or baculovirus BVHCV.S1a. To compare the expression of HCV structural proteins between insect and human cells, human hepatoma HuH-7 cells were transiently transfected with control plasmid pCDNA3.1 or plasmid pCDHCV.S1a, containing the same cDNA for the HCV structural proteins as baculovirus BVHCV.S1a. At 96 h postinfection or at 72 h posttransfection, Sf9 and HuH-7 cells were lysed and subjected to SDS-PAGE and immunoblotting with anti-core (C1 and C2; bottom panel), anti-E1 (E1A; middle panel), and anti-E2 (E2H; top panel) MAbs. Sizes (in thousands) of protein molecular weight (MW) markers are indicated on the left and right. HCV proteins are indicated in the center. (B) Sucrose velocity (left) and equilibrium (right) centrifugation of HCV-LPs. At 96 h postinfection, BVHCVS.1a-infected insect cells were lysed and subjected to low-speed centrifugation. For velocity centrifugation, the supernatant was layered onto a 10 to 60% sucrose gradient and centrifuged at 200,000 × g for 2.5 h at 4°C. Ten fractions were collected from the top and analyzed by SDS-PAGE and immunoblotting as described for panel A. For equilibrium centrifugation, lysates of insect cells infected with BVHCVS.1a were subjected to low-speed centrifugation, and the supernatant was pelleted over a 30% sucrose cushion. The pellet containing HCV-LPs was resuspended and subjected to equilibrium centrifugation (22 h; 4°C; 150,000 × g) by using a 20 to 60% sucrose gradient. Ten fractions were collected and analyzed as described for panel A. Molecular weights (MW) (in thousands) of protein markers and HCV proteins are indicated. (C) Electron microscopy of HCV-LPs. (Top panel) HCV-LPs purified by sucrose equilibrium centrifugation were fixed and visualized by electron microscopy (arrows) as described in Materials and Methods. (Bottom panels) Immunogold labeling of HCV-LPs with serum from an HCV-infected human (human serum [HS]) and two MAbs (H33 and H53) directed against conformation-dependent epitopes of the E2 protein. Bar, 50 nm.
In order to partially purify virus-like particles, lysates of baculovirus-infected insect cells were subjected to sucrose equilibrium and velocity gradient centrifugation. HCV-LPs banded in defined fractions of sucrose velocity and equilibrium gradients, as indicated by the colocalization of HCV structural proteins in these fractions (Fig. 1B). The sucrose gradient sedimentation pattern of HCV-LPs derived from H77C was similar to that obtained for HCV-LPs derived from strain HCV-J cDNA (5), indicating similar biophysical properties of the particles (data not shown). The density of the fractions demonstrating immunoreactivity for HCV-LPs was 1.14 to 1.20 g/cm3 in sucrose equilibrium gradients (fractions 5 and 6 in Fig. 1B). To confirm that the colocalization of HCV structural proteins in the gradient fractions did not represent randomly assembled protein aggregates, sucrose gradient fractions showing immunoreactivity for HCV structural proteins were examined by transmission electron microscopy. Electron microscopy demonstrated the presence of abundant enveloped virus-like particles in defined sucrose gradient fractions (Fig. 1C), confirming the formation of HCV-LPs from H77C cDNA. Particles consisted of an electron-dense core structure surrounded by a bilayer lipid membrane (Fig. 1C). The majority of the particles had a diameter of 40 to 60 nm (Fig. 1C). Immunogold affinity electron microscopy demonstrated labeling of HCV-LPs with human serum-derived anti-HCV antibodies (HS) and anti-E2 MAbs (H33 and H53) directed against conformation-dependent epitopes (Fig. 1C, bottom panels). The particle morphology of H77C-derived HCV-LPs was similar to that of HCV-LPs derived from strain HCV-J (5).
Epitope mapping of H77C-derived HCV-LP envelope proteins.
To analyze the surface topology of HCV-LPs derived from H77C and screen for antienvelope antibodies useful for the detection of HCV-LPs binding to cell lines, we studied the interaction of HCV-LPs with well-defined antienvelope MAbs. Using HCV-LPs as a capture antigen in an ELISA, we demonstrated that HCV-LPs interacted specifically with a large panel of antienvelope MAbs (Table 1). The results indicate that a variety of epitopes (Table 1) are presented on the E1 and E2 ectodomains of the outer surface of the particles. The binding of anti-E2 MAb 2F10, directed against a defined epitope of the E2 hypervariable region, demonstrated that the E2 hypervariable region is accessible on the HCV-LP envelope. HCV-LPs interacted with two different panels (14, 15) of anti-E2 MAbs directed against conformation-dependent epitopes (Table 1), confirming that HCV-LPs contain properly folded E2 protein.
TABLE 1.
Immunoreactivities of HCV-LP (H77C) with anti-E1 and anti-E2 MAbsa
| Antibody | Anti-HCV-LP (H77C) titer | Sequence (HCV genotype) | Source or reference |
|---|---|---|---|
| Anti-E1 | |||
| 14H11B2 | 1:500 | ITGHRMAWDMMMNW (aa 313–326; 1b) | E. Depla |
| ITGHRMAWDMMMNW (H77C) | |||
| 11B708 | 1:16,000 | SIVYEAADMIMHT (aa 212–224; 1b) | E. Depla |
| SIVYEAADAILHT (H77C) | |||
| 159 | 1:1,000 | YEVRNVSGIYHVTNDCSNS (aa 192–210; 1b) | J. Y. N. Lau |
| YQVRNSSGLYHVTNDCPNS (H77C) | |||
| E1A | 1:256,000 | ND (1a) | 25 |
| Anti-E2 | |||
| E2G | 1:16,000 | ND (1a) | 25 |
| E2H | 1:64,000 | GSWHINRTALNCND (aa 437–450; 1a) | 25 |
| GSWHINSTALNCNE (H77C) | |||
| 3E5 | 1:512,000 | SGAPTYSW (aa 522–529; 1a) | M. Houghton |
| SGAPTYSW (H77C) | |||
| 16A6 | 1:32,000 | GVPTYNWG (aa 523–530; 1b) | E. Depla |
| GAPTYSWG (H77C) | |||
| 17H10 | <1:50 | TRGLVSLFSPGSAQKIQLVN (aa 396–415; 1b) | E. Depla |
| TAGLVGLLTPGAKQNIQLIN (H77C) | |||
| 2F10 | 1:2,000 | GLVSLF (aa 398–403; 1b) | E. Depla |
| GLVGLL (H77C) | |||
| 4H6 | <1:50 | VGTTDRFGVPTYNWG (aa 516–530; 1b) | E. Depla |
| VGTTDRSAPTYSWGA (H77C) | |||
| H33 | 1:4,000 | Conformation dependent (1a) | 15 |
| H50 | 1:4,000 | Conformation dependent (1a) | 15 |
| H53 | 1:4,000 | Conformation dependent (1a) | 15 |
| H60 | 1:2,000 | Conformation dependent (1a) | 15 |
| 4F7 | 1:100 | Conformation dependent (1a) | 14 |
| 4E5 | 1:50 | Conformation dependent (1a) | 14 |
| 7B7 | 1:100 | Conformation dependent (1a) | 14 |
Titers of antienvelope MAbs against HCV-LPs (1 μg of total protein per well) were determined by end-point dilution as described in Materials and Methods. All antibodies were diluted to a concentration of approximately 0.5 mg/ml prior to titration. The mapped epitopes of the MAbs are shown in comparison with the H77C amino acid sequence (NCBI accession number AAB67036). ND, not determined.
Dose-dependent and saturable binding of HCV-LPs to defined human cell lines.
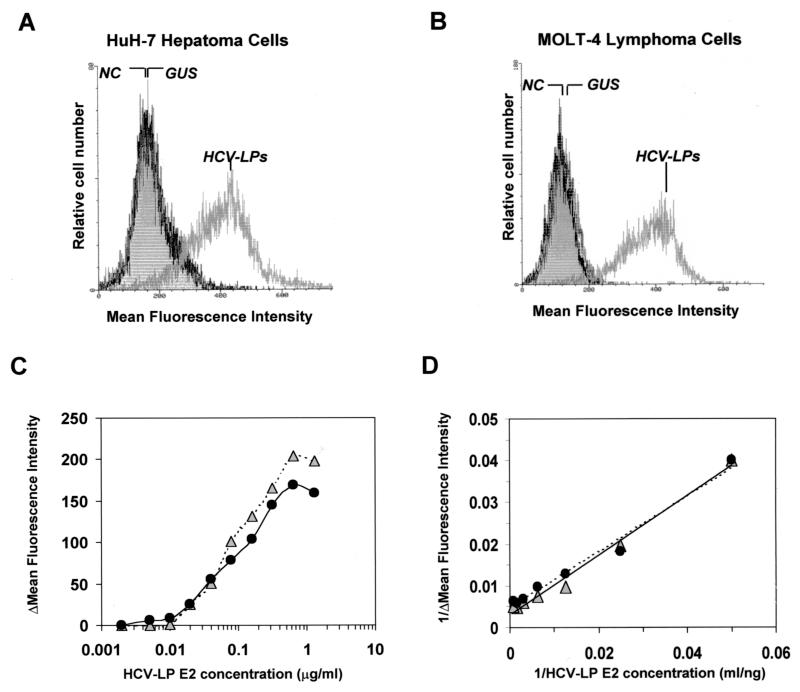
We next assessed whether insect cell-derived HCV-LPs of H77C can be used to examine virus-cell interactions. Dialyzed, sucrose gradient-purified HCV-LPs were incubated with human and mouse cell lines, and the cellular binding of HCV-LPs was studied by FACS analysis with anti-E2 antibodies. Anti-E2 antibodies 16A6E7, 3E5, and E2H were suitable for the detection of HCV-LP binding. HCV-LP binding could also be detected with anti-E1 antibody E1A. These antibodies were characterized by strong interactions with HCV-LPs and low or no cross-reactivity (background staining) with the target cell lines. HCV-LPs displayed dose-dependent and saturable binding to human hepatoma and lymphoma cell lines HuH-7 and MOLT-4, respectively (Fig. 2). In contrast, no measurable binding to these cell lines was observed when control insect cell preparations were used (Fig. 2A and B). Binding reached a plateau at an HCV-LP E2 concentration of approximately 0.5 to 1 μg/ml. Half-maximal saturation of binding was present at an HCV-LP E2 concentration of approximately 50 to 200 ng/ml (Fig. 2C). Although saturation of HCV-LP binding of HuH-7 and MOLT-4 cells was reached at different HCV-LP E2 concentrations, binding analysis with the double-reciprocal-plot method (11, 44) revealed similar HCV-LP binding profiles for MOLT-4 and HuH-7 cells (Fig. 2D). HCV-LPs also exhibited strong binding to human hepatoma and B-cell lymphoma lines HepG2 and Daudi (Table 2). No significant binding was observed to mouse hepatoma or lymphoma cell lines at HCV-LP E2 concentrations saturating the binding of HuH-7 or MOLT-4 cells (Table 2). Binding to MOLT-4 cells appeared to be partially dependent on bivalent cations, since binding could be enhanced in the presence of calcium and magnesium (data not shown).
FIG. 2.
Dose-dependent and saturable binding of HCV-LPs to human hepatoma and lymphoma cells. (A and B) Flow cytometry of HCV-LPs bound to HuH-7 and MOLT-4 cells. HuH-7 or MOLT-4 cells were incubated with HCV-LPs (corresponding to approximately 0.5 μg of HCV-LP E2/ml) or similarly prepared lysates derived from BVGUS-infected insect cells as a negative control. After extensive washing with PBS, the binding of HCV-LPs or control preparations (from insect cells infected with a recombinant baculovirus containing the cDNA for β-glucuronidase [GUS]) was analyzed by FACS with an anti-E2 MAb (16A6) or an isotype control antibody (negative control [NC]) and PE-conjugated anti-mouse IgG. The MFI (shown on the x axis) of the cell population relates to the surface density of HCV-LPs bound to the cells. (C and D) Dose-dependent and saturable binding of HCV-LPs to target cells. HuH-7 cells (triangles) and MOLT-4 cells (circles) were incubated with increasing concentrations of HCV-LPs, and particle binding was analyzed as described above. (C) The net MFI for each HCV-LP concentration (y axis) was calculated by subtracting the MFI for the negative control (GUS) with anti-E2 and PE-conjugated anti-mouse IgG antibodies from that obtained with the respective HCV-LP concentration (x axis). (D) MFI and HCV-LP concentration are plotted as reciprocal values (double-reciprocal plot) (11). The data show the results of a representative experiment performed in duplicate.
TABLE 2.
Binding of HCV-LPs to cell linesa
| Cell line | Source | Tissue | Change in MFI forb:
|
tE2 bindingc (reference) | |
|---|---|---|---|---|---|
| HCV-LP binding | CD81 expression | ||||
| HuH-7 | Human | Hepatoma | 204 | 237 | + (20) |
| HepG2 | Human | Hepatoma | 404 | 4 | − (20) |
| Daudi | Human | B-cell lymphoma | 113 | 439 | + (20, 44) |
| MOLT-4 | Human | T-cell lymphoma | 170 | 494 | + (20, 44) |
| COS-7 | African green monkey | Transformed kidney | 138 | 392 | − (20) |
| SP2-0 | Mouse | B-cell myeloma | 4 | 8 | NR |
| Hepa 1–6 | Mouse | Hepatoma | 9 | 3 | NR |
The binding of HCV-LPs to defined human and nonhuman cell lines was carried out as described in Materials and Methods. Human CD81 expression was analyzed by FACS with anti-human CD81 antibody 1D6 (10 μg/ml).
Data are shown as the change in the MFI relative to the data for the control (insect cell control preparation and anti-E2 antibody for HCV-LPs or isotype control antibody for CD81) for a representative experiment performed in duplicate. A value of >20 indicates a positive signal for binding.
Presence (+) or absence (−) of binding of recombinant C terminally truncated E2 protein (tE2) to cell lines. NR, not reported.
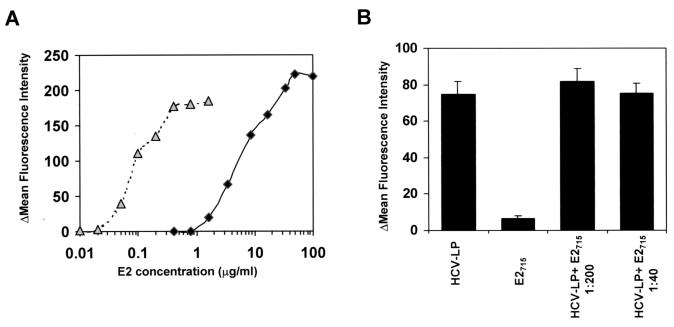
The binding of HCV-LPs was then compared with the binding of E2 protein C terminally truncated at aa 715 (E2715) (44). HCV-LP E2 and E2715 binding was assessed side by side with anti-E2 MAb 3E5. With this antibody, HCV-LPs exhibited a binding profile similar to that shown in Fig. 2, although the saturation of binding appeared to be reached at slighty different HCV-LP E2 concentrations. HCV-LP E2 and E2715 demonstrated markedly different binding profiles for human hepatoma cells. A 50- to 100-fold lower concentration was required for half-maximal saturation of binding for HCV-LP E2 than for C-terminally truncated E2715 (Fig. 3A). Excess E2715 protein did not inhibit the binding of HCV-LPs in direct competition experiments (Fig. 3B). These data are consistent with different conformations of HCV-LP E2 and C-terminally truncated E2715 resulting in different interactions with cellular binding molecules.
FIG. 3.
Differential binding of HCV-LPs and C-terminally truncated E2715 to human hepatoma cells. (A) Comparative analysis of HCV-LP and E2715 binding to HuH-7 cells. HuH-7 cells were incubated with increasing concentrations of HCV-LP E2 (triangles) and E2715 (squares). Binding was analyzed by FACS with anti-E2 MAb 3E5 or an isotype control antibody (negative control) and PE-conjugated anti-mouse IgG. The net MFI for each HCV-LP or E2715 concentration (y axis) was calculated by subtracting the MFI for the negative control (GUS control preparation for HCV-LPs or PBS for E2715) with anti-E2 and PE-conjugated anti-mouse IgG antibodies from that obtained with the respective HCV-LP or E2715 concentration (x axis). The data show the results of a representative experiment performed in duplicate. (B) Binding of HCV-LPs to HuH-7 cells in the presence of E2715. Prior to incubation of HuH-7 cells with HCV-LPs (0.25 μg/ml), HuH-7 cells were incubated with E2715 (50 μg/ml, corresponding to an HCV-LP E2/E2715 ratio of 1:200, or 10 μg/ml, corresponding to an HCV-LP E2/E2715 ratio of 1:40). The binding of HCV-LPs was measured by using an anti-E1 (E1A) antibody (Table 1). Binding was calculated as the mean and standard deviation net MFI for a representative experiment performed in triplicate.
To assess whether the cellular binding of HCV-LPs was dependent on particle conformation, we compared the binding profiles for native and heat-denatured HCV-LPs. Heat denaturation appeared to result in a loss of particle structure, as indicated by heat-induced precipitation of protein aggregates and a loss of cosedimentation of HCV structural proteins in sucrose gradients. The loss of the native envelope conformation in the majority of heat-denatured HCV-LPs was also evident from the failure of heat-denatured HCV-LPs to interact with anti-E2 antibody H53 in an HCV-LP ELISA (titer, <1:100). Compared to native HCV-LPs, denatured HCV-LPs exhibited strongly reduced binding to human cell lines, indicating that binding was dependent on the native conformation of the HCV-LP envelope (Fig. 4). In contrast, heat denaturation of recombinant E2715 resulted only in a minor reduction of cellular binding.
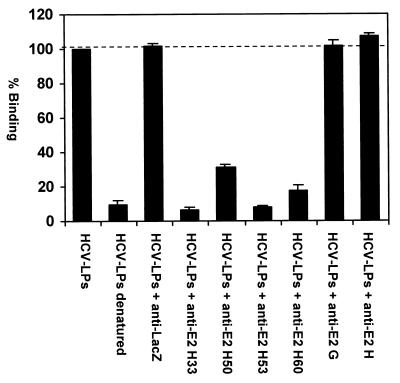
FIG. 4.
Inhibition of HCV-LP binding to MOLT-4 cells by anti-E2 antibodies. For the assessment of antibody-mediated neutralization of binding, sucrose gradient-purified HCV-LPs at a subsaturating concentration (corresponding to approximately 0.25 μg of HCV-LP E2/ml) were incubated with antienvelope MAbs or an isotype control MAb (anti-LacZ) (1:50 dilution, corresponding to an antibody concentration of 20 μg/ml) overnight at 4°C. HCV-LP-antibody complexes were added to MOLT-4 cells for 1 h at 4°C. After removal of nonbound HCV-LP-antibody complexes by washing of cells in PBS, the binding of HCV-LPs was detected by FACS with anti-E2 MAb 16A6 as described in the legend to Fig. 2. Data are shown as percent binding relative to the binding of HCV-LPs without antibody (100%). Binding was calculated as the mean (range) net MFI for a representative experiment performed in duplicate.
A previous study (5) demonstrated that a defined, limited amount of mild detergent in a cell lysis buffer can increase the yield of particles without destroying the HCV-LP envelope. We confirmed this observation by comparing the functional properties of HCV-LPs from insect cells lysed by repeated freezing-thawing or by use of a cell lysis buffer containing 0.1% Nonidet P-40 (5). HCV-LPs from preparations obtained under both lysis conditions demonstrated similar cellular binding.
To study whether HCV-LP binding requires an interaction of the HCV E2 protein with the target cell membrane, we assessed HCV-LP binding to MOLT-4 cells in the presence of anti-E2 MAbs. HCV-LP binding to MOLT-4 cells was inhibited in the presence of anti-E2 antibodies directed against conformation-dependent epitopes (Fig. 4). In contrast, incubation of particles with an irrelevant isotype control MAb (anti-LacZ) did not result in inhibition of binding in parallel experiments (Fig. 4). Inhibition of binding was dependent on the ratio of HCV-LP E2 protein to anti-E2 antibodies, indicating a dose-dependent effect of anti-E2 antibodies on HCV-LP binding. These data suggest that HCV-LP binding to MOLT-4 cells appears to be E2 dependent.
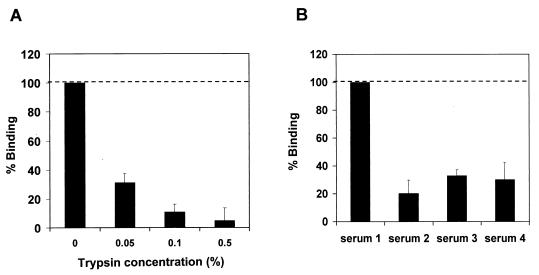
HCV-LP binding was sensitive to pretreatment of cells with trypsin, indicating that attachment of HCV-LPs is mediated by a cellular protein (Fig. 5A). Interestingly, the use of different sera for the cultivation of MOLT-4 cells strongly affected HCV-LP binding (Fig. 5B). These data indicate that the expression of the cell surface protein required for HCV-LP binding may be induced or repressed by a mediator(s) present in defined serum preparations.
FIG. 5.
HCV-LP binding to MOLT-4 cells is sensitive to trypsin pretreatment and is modulated by serum in cell culture medium. (A) Trypsin pretreatment and HCV-LP binding. To examine whether trypsin pretreatment of cells could alter HCV-LP binding, MOLT-4 cells were exposed to trypsin (concentrations indicated on the x axis) for 5 min, and trypsin activity was neutralized by the addition of medium containing 10% FCS and repetitive washing of the cells with PBS. The binding of HCV-LPs was then assessed as described in the legend to Fig. 2 (binding without trypsin pretreatment, 100%). Binding was calculated as the mean and standard deviation net MFI for a representative experiment performed in triplicate. (B) HCV-LP binding and serum in cell culture medium. MOLT-4 cells were cultured as described in Materials and Methods by using four different sera (sera 1 to 4). HCV-LP binding was assessed as described in the legend to Fig. 2. Binding was calculated as the mean (range) net MFI for a representative experiment performed in duplicate (binding to cells maintained in medium containing serum 1, 100%).
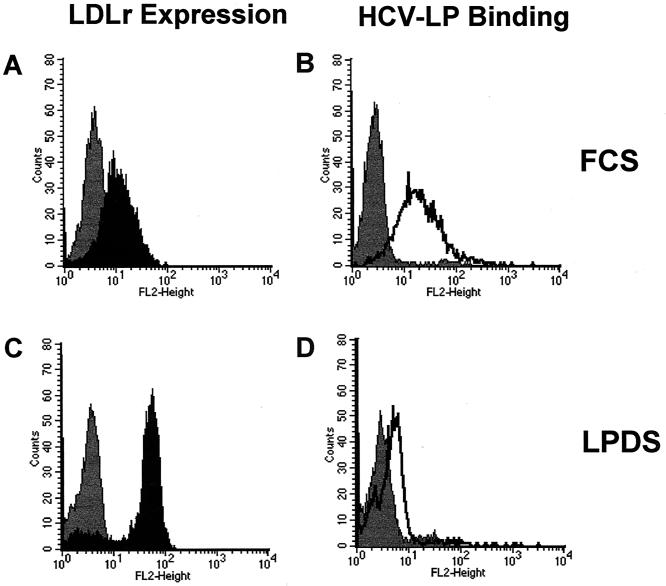
To assess whether different LDL concentrations in sera resulted in variable LDLr expression mediating the observed difference in HCV-LP binding, we determined FCS LDL concentrations and studied HCV-LP binding in cells grown in medium with LPDS. FCS LDL concentrations (range, 15.6 to 25.5 mg/dl) did not correlate with HCV-LP binding. Although culturing of MOLT-4 cells in LPDS resulted in a strong upregulation of LDLr expression (Fig. 6C), HCV-LP binding was markedly decreased compared to that in MOLT-4 cells cultured in medium with lipoprotein-rich FCS (Fig. 6D). Preincubation of MOLT-4 or HuH-7 cells with a recently characterized anti-LDLr antibody (1, 51) did not inhibit HCV-LP binding (data not shown). These data indicate that the observed relationship between cell culture serum and HCV-LP binding is not due to variable levels of MOLT-4 LDLr expression induced by different FCS LDL concentrations.
FIG. 6.
HCV-LP binding and LDLr expression in MOLT-4 cells. To study whether HCV-LP binding correlates with LDLr expression, MOLT-4 cells were incubated for 40 h in medium containing 10% lipoprotein-rich FCS (serum 1 of Fig. 5) (A and B) or 10% LPDS (C and D). (A and C) LDLr expression in MOLT-4 cells grown in FCS (A) or LPDS (C). LDLr expession was measured by flow cytometry of MOLT-4 cells by using anti-LDLr antibody C7 (10 μg/ml) as described in Materials and Methods (black graphs). (B and D) HCV-LP binding to MOLT-4 cells grown in FCS (B) or LPDS (D). Assessment of HCV-LP binding (0.5 μg of HCV-LP E2/ml) was performed as described in the legend to Fig. 2 (unshaded graphs). Background fluorescence (grey graphs) was measured by using an isotype control MAb (A and C) or an insect cell control preparation (GUS) and anti-E2 antibody (B and D). Fluorescence intensity (FL2-Height) and relative cell number (counts) are shown. Data represent an analysis of four independent experiments performed in duplicate or triplicate.
HCV-LP binding and CD81.
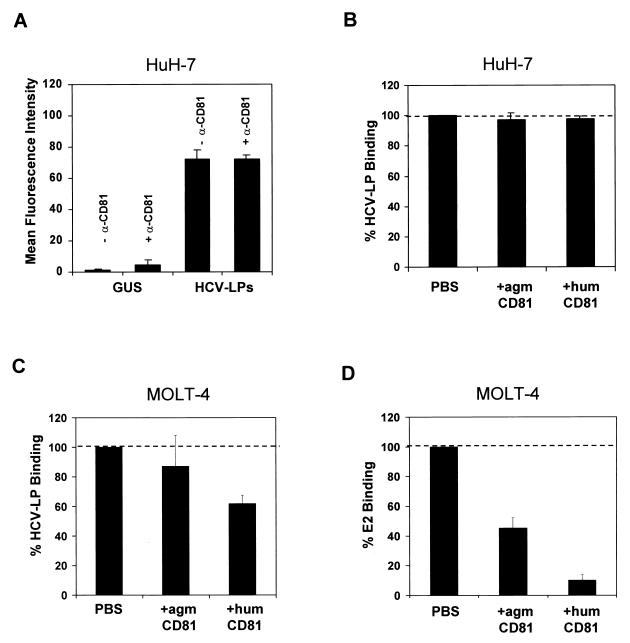
To study whether the binding of HCV-LPs to cell lines is dependent on the interaction with E2 binding protein CD81, we analyzed the binding of HCV-LPs to cell lines expressing no CD81 or CD81 from African green monkeys. Interestingly, HCV-LPs displayed strong binding to the human hepatoma cell line HepG2. Unlike HuH-7, MOLT-4, and Daudi cells, HepG2 cells do not express CD81 on their cell surface (Table 2 and reference 20). Furthermore, HCV-LPs demonstrated measurable binding to COS-7 cells (Table 2), expressing nonhuman CD81 with a low affinity for recombinant E2 (20, 27). These data provided indirect evidence that HCV-LP binding to cell lines does not require the presence of human CD81 on the cell surface.
To confirm this hypothesis, we measured the binding of HCV-LPs in the presence of recombinant CD81-LEL and anti-CD81 antibody. To assess whether HCV-LP binding could be blocked by anti-CD81 antibody, HuH-7 and MOLT-4 cells were incubated with anti-CD81 antibody at a concentration previously shown to saturate CD81 (MAb 1D6, 10 μg/ml) prior to the addition of HCV-LPs. The binding of HCV-LPs in the presence of anti-CD81 antibody was detected by using a biotinylated anti-E2 antibody and streptavidin-PE. The binding of HCV-LPs to HuH-7 cells (Fig. 7A) or MOLT-4 cells (data not shown) was not inhibited by anti-CD81 antibody. To study whether the binding of HCV-LPs could be blocked by CD81-LEL, HCV-LPs were preincubated with excess soluble CD81-LEL, and their binding to human hepatoma cells was assessed by FACS analysis. HCV-LP binding to HuH-7 cells was not inhibited by human CD81-LEL or African green monkey CD81-LEL (Fig. 7B). HCV-LP binding to MOLT-4 cells was only partially inhibited (less than 50%) by soluble excess human CD81-LEL (Fig. 7C). To confirm that soluble fusion proteins were functionally intact, we assessed their ability to block the binding of recombinant E2 to MOLT-4 cells as described previously (20, 27). Human CD81-LEL (50 μg/ml) was able to block the binding of recombinant E2 (10 μg/ml) to MOLT-4 cells by 90% (Fig. 7D). These data suggest that HCV-LP-cell interactions require additional cellular surface proteins besides CD81.
FIG. 7.
Binding of HCV-LPs to human HuH-7 hepatoma and MOLT-4 lymphoma cells in the presence of soluble CD81 and anti-CD81 antibody. (A) To assess whether binding could be blocked by anti-CD81 antibody, HuH-7 cells were incubated with anti-CD81 antibody at a concentration previously shown to saturate CD81 (MAb 1D6, 10 μg/ml) for 1 h at 25°C prior to the addition of HCV-LPs. The binding of HCV-LPs (indicated as MFI) in the presence of anti-CD81 antibody was detected by using a biotinylated anti-E2 antibody and streptavidin-PE. GUS, control insect cell preparation. (B and C) To study whether the binding of HCV-LP could be blocked by recombinant CD81-LEL, HCV-LPs (0.25 μg/mg) were preincubated with an excess of soluble CD81-LEL (50 μg/ml; human [hum] or African green monkey [agm] CD81) for 1 h at 25°C, and the binding of HCV-LPs to human HuH-7 cells (B) or MOLT-4 cells (C) was assessed by FACS as described in the legend to Fig. 2. Data are shown as percent binding relative to the binding of HCV-LPs as a positive control (100%). Binding was calculated as net mean MFI for a representative experiment performed in triplicate. (D) Inhibition of E2 binding to MOLT-4 cells by soluble CD81-LEL. E2 (10 μg/ml) was preincubated with CD81-LEL (50 μg/ml) as described for panel C. E2 binding was measured by FACS as described in the legend to Fig. 3.
DISCUSSION
In this study, we demonstrate that the HCV structural proteins derived from infectious clone H77C assemble into HCV-LPs that bind efficiently to defined human cell lines. H77C has been shown to be a functional clone resulting in the successful infection of chimpanzees inoculated with H77C transcripts (52). This observation implies that the H77C genome must contain all the information required for the assembly of functionally intact, infectious virus particles. Using a recombinant baculovirus containing the cDNA for the HCV structural proteins of H77C, we could demonstrate the assembly of HCV-LPs in insect cells. HCV-LP assembly was shown by criteria (electron microscopy with immunogold affinity labeling and sedimentation and purification of HCV-LPs by sucrose gradient centrifugation) similar to those described recently for HCV-LPs derived from strainHCV-J. Extending our previous results, we could demonstrate that the envelope proteins of HCV-LPs strongly interact with defined anti-E2 antibodies directed against conformation-dependent epitopes. This interaction was shown by electron microscopy with immunogold affinity labeling of HCV-LP envelopes with anti-E2 antibodies H53 and H33 (Fig. 1) and reactivity of HCV-LPs with the same anti-E2 antibodies in an HCV-LP-based ELISA (Table 1). Since anti-E2 antibody H53 does not interact with aggregated E2 (15), our results confirm that HCV-LPs indeed contain envelope proteins in a native conformation. This finding is in line with results from a previous study, suggesting that the envelope glycoproteins of HCV-LPs are predominantly noncovalently associated as E1-E2 heterodimers (8). Interactions of HCV-LPs with defined MAbs against the envelope proteins revealed that envelope ectodomains comprising aa 192 to 224 (E1), 313 to 326 (E1), 398 to 403 (E2), 437 to 450 (E2), 523 to 530 (E2), and 644 to 651 (E2) are accessible on the surface of the particles. These domains comprise E2 hypervariable region 1 (aa 398 to 403), including a well-characterized B-cell epitope important for viral escape (49).
HCV-LPs of H77C and HCV-J showed similar biophysical and morphological properties in gradient centrifugation and electron microscopy analyses, indicating that the basic mechanisms of HCV-LP formation may be similar in HCV genotypes 1a and 1b. The synthesis of HCV-LPs of genotypes 1a (strain HCV-H) and 1b (strain HCV-J) will allow evaluation of the genotype-dependent features of HCV-LP antigenicity (HCV-LP-based ELISAs [9, 47]), immunogenicity (HCV-LP vaccine studies [8, 30]), and virus-host interactions (this study). The formation of HCV-LPs derived from infectious clones H77C (52) and H77 (29) was recently reported independently by two other laboratories (M. Triyatni, J. Vergalla, M. Lechmann, and T. J. Liang, 7th International Meeting on Hepatitis C and Related Viruses, Gold Coast, Australia, 3 to 7 December 2000; A. Patel, A. Owsianka, J. Aitken, S. Graham, L. D. Loomis-Price, J. McKeating, and D. Bhella, 7th International Meeting on Hepatitis C and Related Viruses, Gold Coast, Australia, 3 to 7 December 2000).
Although a detailed analysis of the viral genetic organization has led to the identification of various elements required for assembly and replication, the study of viral attachment and entry has been hampered by the inability to efficiently propagate the virus in vitro. Our study demonstrates for the first time that HCV-LPs can be used as a sensitive and specific tool to study virus-host interactions. HCV-LP binding to human hepatoma and lymphoma cell lines was dose dependent and saturable. Binding was specific for human or primate cell lines, as indicated by the lack of HCV-LP binding to mouse cell lines. Binding appeared to be dependent on the proper conformation of the particles: heat denaturation of HCV-LPs resulted in a drastic reduction of binding, and binding was inhibited by anti-E2 antibodies directed against conformation-dependent epitopes. Inhibition of HCV-LP binding by defined anti-E2 antibodies is also consistent with the hypothesis that HCV envelope glycoprotein E2 plays an important role in mediating viral attachment and entry (21). Studies are under way to map the envelope epitopes required for HCV-LP-host interactions and to define the role of E1 in HCV-LP binding.
The next step after particle binding is uptake or entry into cells. Since virus binding represents only a first step for the initiation of viral infection, it is of great interest to study whether HCV-LPs can enter the cell after they have bound to the cell surface. Preliminary studies with HCV E2-specific immunofluorescence and confocal laser scanning microscopy have provided evidence for temperature-dependent entry of HCV-LPs into MOLT-4 cells (H. Barth, B. Klumpp, S. Wellnitz, D. Steinmann, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert, presentation P-149, 8th International Meeting on Hepatitis C and Related Viruses, Paris, France, 2 to 5 September 2001). Further studies are under way to explore this important question in detail.
Studies from several laboratories have shown that the binding of C-terminally truncated recombinant glycoprotein E2715 to human target cells can be used as a surrogate model to study virus-host interactions (20, 26, 41, 44). This assay has been used to analyze antibody-mediated neutralization of binding and to isolate CD81 as a E2 binding protein (20, 26, 41, 44). In contrast to individually expressed E2 used in these systems, the envelope glycoproteins of HCV-LPs are expressed as E1-E2 heterodimers (8) and are presumably presented in a native conformation as part of a structure with morphological, biophysical, and antigenic properties similar to those of the virion (5, 8). Indeed, E2715 and HCV-LPs were characterized by different binding profiles. As shown in Fig. 3, E2715 protein did not inhibit the binding of HCV-LPs in direct competition experiments. Since the binding of HCV-LP could be inhibited by anti-E2 antibodies, these observations strongly suggest that the different conformations of HCV-LP envelope proteins and C-terminally truncated E2715 result in binding to different cellular molecules. We therefore propose that HCV-LPs represent an ideal tool to study virus-host interactions, allowing the isolation of HCV receptor candidates and the assessment of antibody-mediated neutralization of binding. This concept is further supported by the successful application of virus-like particles of other viruses for the characterization of virus-host interactions. Virus-like particle-based binding and entry systems have been developed for papillomavirus (17, 24, 28, 37, 42, 53), rotavirus (22, 23), and Norwalk virus (45, 50).
Other models for the study of HCV-host interactions include the binding of unpurified or sucrose gradient-purified virions to human cell lines (1, 51, 54). These systems use virions but require the use of either ultrasensitive detection methods, such as reverse transcription-PCR and in situ hybridization, or flow cytometry-based systems with less standardized reagents, such as polyclonal anti-HCV antibodies derived from human serum, for the detection of viral attachment. In comparison, our flow cytometry-based system is highly standardized by the use of well-defined ligands and MAbs for the detection of binding. Therefore, our system provides a fast, simple, and quantifiable high-throughput assay for the study of virus-host interactions.
The initial step in a virus cycle is the attachment of the virion to the host cell membrane. Very little is known about the nature of the HCV receptor and mechanisms involved in virus entry into host cells. The abolition of binding by pretreatment of MOLT-4 cells with the protease trypsin suggests that HCV-LP binding is mediated by a cellular protein (Fig. 5). The expression of this protein appeared to be dependent on nondefined factors present in the serum used for the cultivation of cells (Fig. 5). As a first step to using our model and gaining novel insights into the biology of HCV-host cell interactions, we studied the functional role of the recently discovered HCV-E2 binding molecule CD81 for the requirement of HCV-LP binding to target cells. CD81 is a member of the tetraspanin family and has been shown to play an important role in signal transduction and adhesion in the immune system (32). The LEL of CD81 has been shown to interact specifically with mammalian and insect cell-derived HCV E2 proteins (12, 20, 41), suggesting a potential role of CD81 as an HCV receptor candidate (21). The E2-CD81 interaction appears to be independent of E2 glycosylation (12). The binding of recombinant E2 to target cells appears to be at least partially dependent on the E2-CD81 interaction, as demonstrated by a lack of E2 binding to cells expressing no CD81 (HepG2) (20) or nonhuman CD81 (COS-7 cells) (20). Furthermore, E2 binding to target cells can be strongly inhibited by soluble CD81 (Fig. 7) and blocked by defined anti-CD81 antibodies (20, 40, 41).
In contrast to the CD81-dependent binding of C-terminally truncated E2, HCV-LPs displayed a different binding profile, as follows. (i) HCV-LPs showed strong binding to HepG2 cells, lacking significant CD81 expression on the cell surface (Table 2). (ii) HCV-LPs also bound to COS-7 cells (Table 2), expressing a CD81 variant with a poor affinity for E2 (27). (iii) HCV-LP binding to HuH-7 cells was not inhibited by soluble CD81-LEL, and that to MOLT-4 cells was only partially inhibited (<50%) by soluble CD81-LEL. (iv) HCV-LP binding to hepatoma cells could not be inhibited by an anti-CD81 antibody shown to block the E2-CD81 interaction (19, 20, 27).
Since HCV-LP E2 bound soluble CD81-LEL with a much lower affinity than recombinant truncated E2 in an ELISA (data not shown), we cannot exclude the possibility that the failure of soluble CD81-LEL to block HCV-LP binding was due to conformational differences between soluble CD81-LEL and CD81. However, all experimental approaches exploring the direct interaction of HCV-LPs with cell surface CD81 clearly indicate the lack of a role of cellular CD81 in mediating HCV-LP binding. These results have two implications, as follows. (i) The binding of HCV-LPs and C-terminally truncated E2 protein appears to be mediated by different target molecules. (ii) Virus attachment appears to require additional surface proteins besides CD81. The latter conclusion is corroborated by recent observations providing indirect evidence that CD81 is not the major HCV receptor: (i) the molecule is expressed ubiquitously on the surface of various cell types not limited to HCV-susceptible cells (32), (ii) the binding of sucrose gradient-purified virions from infected individuals to lymphoma cells and foreskin fibroblasts does not require an HCV-CD81 interaction (51), (iii) the binding of E2 to CD81 in various species does not predict susceptibility to HCV infection (1a, 38), and (iv) the overexpression of human CD81 has not been shown to confer infectivity in transgenic mice (F. Masciopinto, G. Freer, V. Burgio, S. Levy, M. Houghton, L. Galli-Stampino, M. Bendelli, S. Abrignani, and Y. Uematsu, 8th International Meeting on Hepatitis C and Related Viruses, Paris, France, 2 to 5 September 2001).
The human LDLr has been proposed as a cell surface molecule mediating attachment and endocytosis of HCV complexed with plasma very-low-density lipoprotein (VLDL) or LDL (1). In contrast, HCV purified from plasma fractions without VLDL or LDL did not exhibit measurable LDLr-mediated binding or endocytosis in the described experimental systems (1, 51). Whether this proposed route of entry can result in a productive infection in vivo remains to be determined (1). Since HCV complexed with VLDL or LDL also interacted with LDLr-deficient cells (1) and LDL only partially inhibited the binding of HCV-LDL complexes to target cells (51), it is likely that HCV binds cells via additional surface proteins. Our own observations obtained with HCV-LPs as a ligand for cellular binding appear to support this conclusion. HCV-LP binding did not correlate with LDLr expression (Fig. 6A and B) and could not be inhibited by anti-LDLr antibody C7. In contrast to HCV complexed or associated with plasma VLDL or LDL, HCV-LPs are derived from a serum-free insect cell culture system. Therefore, our HCV-LP-based system may allow study of the direct interaction of the viral envelope with the cell membrane and may be used to assess cell surface proteins mediating this interaction.
In summary, we have shown that insect cell-derived HCV-LPs represent a novel model for the study of virus-host interactions, allowing the functional assessment of HCV receptor candidates. This system may allow definition of host and virus determinants required for virus-cell interactions and may be used to characterize the functional properties of anti-HCV antibodies and their ability to neutralize HCV-LP binding to target cells.
Acknowledgments
We thank T. J. Liang (Liver Diseases Section, NIDDK, NIH, Bethesda, Md.), A. H. Patel (MRC Virology Unit, Glasgow, United Kingdom), J. A. McKeating (Rockefeller University, New York, N.Y.), and C. Grüllich (Department of Medicine I, University of Freiburg, Freiburg, Germany) for helpful discussions and P. Liljestrom (Karolinska Institut, Stockholm, Sweden) for critical reading of the manuscript. We thank R. Purcell and J. Bukh for the gift of plasmid pCVH77C; S. Levy for the gift of soluble CD81-LEL and anti-CD81 antibodies; M. Houghton for the gift of recombinant E2 and anti-E2 antibody 3E5; and H. B. Greenberg, M. Da Silva Cardoso, J. Y. N. Lau, and M. Houghton for the gift of anti-HCV antibodies. G. Wolff-Vorbeck (University of Freiburg, Freiburg, Germany) is gratefully acknowledged for advice and support in the FACS analysis. We thank M. Nauck and K. Winkler (Division of Clinical Chemistry, Department of Medicine, University of Freiburg, Freiburg, Germany) for providing lipoprotein-deficient serum and determining FCS LDL concentrations. P. Schürmann is acknowledged for excellent technical assistance.
This work was supported by grants from the European Union, Brussels, Belgium (QLRT-PL1999-00356); the Wilhelm Sander-Stiftung, Munich, Germany (99041.1); and the Deutsche Forschungsgemeinschaft, Bonn, Germany (Ba 1417/6-1). S.I. was supported by the Harvard Digestive Disease Center, Boston, Mass.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 98:12766–12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358–367. [DOI] [PubMed] [Google Scholar]
- 2.Alter, H. J., R. H. Purcell, J. W. Shih, J. C. Melpolder, M. Houghton, Q.-L. Choo, and G. Kuo. 1989. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N. Engl. J. Med. 321:1494–1500. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J., D. Kruszon-Moran, O. Nainan, G. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States 1988–1994. N. Engl. J. Med. 341:556–562. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631–1648. [DOI] [PubMed] [Google Scholar]
- 5.Baumert, T. F., S. Ito, D. Wong, and T. J. Liang. 1998. The hepatitis C virus structural proteins assemble into virus-like particles in insect cells. J. Virol. 72:3827–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert, T. F., A. Marrone, J. Vergalla, and T. J. Liang. 1998. Naturally occurring mutations define a novel function of the hepatitis B virus core promotor in core protein expression. J. Virol. 72:6785–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumert, T. F., S. A. Rogers, K. Hasegawa, and T. J. Liang. 1996. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Investig. 98:2268–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397–1407. [DOI] [PubMed] [Google Scholar]
- 9.Baumert, T. F., S. Wellnitz, S. Aono, J. Satoi, D. Herion, J. Tilman Gerlach, G. R. Pape, J. Y. Lau, J. H. Hoofnagle, H. E. Blum, and T. J. Liang. 2000. Antibodies against hepatitis C virus-like particles and viral clearance in acute and chronic hepatitis C. Hepatology 32:610–617. [DOI] [PubMed] [Google Scholar]
- 10.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1975. [DOI] [PubMed] [Google Scholar]
- 11.Celada, F., A. J. Macario, and E. C. De Macario. 1973. Enzyme activation by antibodies: a method to determine the binding constant of the activating antibody towards one determinant of E. coli beta-d-galactosidase. Immunochemistry 10:797–804. [DOI] [PubMed] [Google Scholar]
- 12.Chan-Fook, C., W. R. Jiang, B. E. Clarke, N. Zitzmann, C. Maidens, J. A. McKeating, and I. M. Jones. 2000. Hepatitis C virus glycoprotein E2 binding to CD81: the role of E1E2 cleavage and protein glycosylation in bioactivity. Virology 273:60–66. [DOI] [PubMed] [Google Scholar]
- 13.Choo, Q.-L., A. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362. [DOI] [PubMed] [Google Scholar]
- 14.da Silva Cardoso, M., K. Siemoneit, D. Sturm, C. Krone, D. Moradpour, and B. Kubanek. 1998. Isolation and characterization of human monoclonal antibodies against hepatitis C virus envelope glycoproteins. J. Med. Virol. 55:28–34. [PubMed] [Google Scholar]
- 15.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubuisson, J. 1999. Folding, assembly and subcellular localization of HCV glycoproteins. Curr. Top. Microbiol. Immunol. 242:135–148. [DOI] [PubMed] [Google Scholar]
- 17.Evander, M., I. H. Frazer, E. Payne, Y. M. Qi, K. Hengst, and N. A. McMillan. 1997. Identification of the alpha 6 integrin as a candidate receptor for papillomaviruses. J. Virol. 71:2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farci, P., H. J. Alter, S. Govindarajan, D. C. Wong, R. Engle, R. R. Lesniewski, I. K. Mushahwar, S. M. Desai, R. H. Miller, N. Ogata, and R. H. Purcell. 1992. Lack of protective immunity against reinfection with hepatitis C virus. Science 258:135–140. [DOI] [PubMed] [Google Scholar]
- 19.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flint, M., and J. A. McKeating. 2000. The role of the hepatitis C virus glycoproteins in infection. Rev. Med. Virol. 10:101–117. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, J. M., and H. B. Greenberg. 1998. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72:5323–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, J. M., and H. B. Greenberg. 1997. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J. Virol. 71:4555–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giroglou, T., L. Florin, F. Schafer, R. E. Streeck, and M. Sapp. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75:1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heile, J. M., Y. L. Fong, D. Rosa, K. Berger, G. Saletti, S. Campagnoli, G. Bensi, S. Capo, S. Coates, K. Crawford, C. Dong, M. Wininger, G. Baker, L. Cousens, D. Chien, P. Ng, P. Archangel, G. Grandi, M. Houghton, and S. Abrignani. 2000. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J. Virol. 74:6885–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawana, Y., K. Kawana, H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 2001. Human papillomavirus type 16 minor capsid protein L2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J. Virol. 75:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574. [DOI] [PubMed] [Google Scholar]
- 30.Lechmann, M., J. Satoi, J. Vergalla, K. Murata, K., T. F. Baumert, and T. J. Liang. 2001. Hepatitis C virus-like particles induce virus-specific humoral and cellular immune responses in mice. Hepatology 34:417–423. [DOI] [PubMed] [Google Scholar]
- 31.Lemon, S. M., and D. L. Thomas. 1997. Vaccines to prevent viral hepatitis. N. Engl. J. Med. 336:197–203. [DOI] [PubMed] [Google Scholar]
- 32.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89–109. [DOI] [PubMed] [Google Scholar]
- 33.Liang, T. J. 1998. Combination therapy for hepatitis C virus infection. N. Engl. J. Med. 339:1549–1550. [DOI] [PubMed] [Google Scholar]
- 34.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296–305. [DOI] [PubMed] [Google Scholar]
- 35.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. [DOI] [PubMed] [Google Scholar]
- 36.Major, M. E., and S. M. Feinstone. 1997. The molecular virology of hepatitis C. Hepatology 25:1527–1538. [DOI] [PubMed] [Google Scholar]
- 37.McMillan, N. A., E. Payne, I. H. Frazer, and M. Evander. 1999. Expression of the alpha 6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology 261:271–279. [DOI] [PubMed] [Google Scholar]
- 38.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873–2883. [DOI] [PubMed] [Google Scholar]
- 40.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. [DOI] [PubMed] [Google Scholar]
- 42.Qi, Y. M., S. W. Peng, K. Hengst, M. Evander, D. S. Park, J. Zhou, and I. H. Frazer. 1996. Epithelial cells display separate receptors for papillomavirus VLPs and for soluble L1 capsid protein. Virology 216:35–45. [DOI] [PubMed] [Google Scholar]
- 43.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p.931–959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 44.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. Weiner, J. Y. N. Lau, Q.-L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura, M., K. Natori, M. Kobayashi, T. Miyamura, and N. Takeda. 2000. Interaction of recombinant Norwalk virus particles with the 105-kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J. Virol. 74:11589–11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong, M. J., N. S. El-Farra, A. R. Reikes, and R. L. Co. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463–1466. [DOI] [PubMed] [Google Scholar]
- 47.Vidalin, O., A. Fournillier, N. Renard, M. Chen, E. Depla, D. Boucreux, C. Brinster, T. F. Baumert, I. Nakano, Y. Fukuda, P. Liljestrom, C. Trepo, and Inchauspe. 2000. Use of conventional or replicating nucleic acid-based vaccines and recombinant Semliki forest virus-derived particles for the induction of immune responses against hepatitis C virus core and E2 antigens. Virology 276:259–270. [DOI] [PubMed] [Google Scholar]
- 48.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166–175. [DOI] [PubMed] [Google Scholar]
- 49.Weiner, A. J., H. M. Geysen, C. Christopherson, J. E. Hall, T. J. Mason, G. Saracco, F. Bonino, K. Crawford, C. D. Marion, K. A. Crawford, et al. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA 89:3468–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Winkler, K., M. Nauck, R. Siekmeier, W. März, and H. Wieland. 1995. Determination of triglycerides in lipoproteins separated by agarose gel electrophoresis. J. Lipid Res. 36:1839–1847. [PubMed] [Google Scholar]
- 51.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055–10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeager, M. D., M. Aste-Amezaga, D. R. Brown, M. M. Martin, M. J. Shah, J. C. Cook, N. D. Christensen, C. Ackerson, R. S. Lowe, J. F. Smith, P. Keller, and K. U. Jansen. 2000. Neutralization of human papillomavirus (HPV) pseudovirions: a novel and efficient approach to detect and characterize HPV neutralizing antibodies. Virology 278:570–577. [DOI] [PubMed] [Google Scholar]
- 54.Zibert, A., E. Schreier, and M. Roggendorf. 1995. Antibodies in human sera specific to hypervarable region 1 of hepatitis C virus can block viral attachment. Virology 208:653–661. [DOI] [PubMed] [Google Scholar]