Abstract
The dimer initiation site/dimer linkage sequence (DIS/DLS) region of the human immunodeficiency virus type 1 (HIV-1) RNA genome is thought to play important roles at various stages of the virus life cycle. Recently we showed that the DIS/DLS region affects RNA-RNA interaction in intact virus particles, by demonstrating that duplication of the region in viral RNA caused the production of virus particles containing partially monomeric RNAs. We have extended this finding and succeeded for the first time in creating mutant particles which contain only monomeric RNAs without modifying any viral proteins. In terms of RNA encapsidation ability, virion density, and protein processing, the mutant particles were comparable to wild-type particles. The level of production of viral DNA by the mutant virus construct in infected cells was also comparable to that of the constructs that produced exclusively dimeric RNA, indicating that monomeric viral RNA could be the template for strand transfer. These results indicated that the RNA dimerization of HIV-1 could be separated from viral RNA packaging and was not absolutely required for RNA packaging, virion maturation, and reverse transcription.
Retrovirus particles contain single-stranded positive-sense viral RNA as a genome. The genomic RNA always forms dimers in mature virions. These two RNAs are linked noncovalently, since incubation at high temperature (≈70°C) or treatment with denaturing agents such as formamide easily dissociates them (for reviews, see references 8 and 16). Electron microscopy has revealed that the two RNAs are linked symmetrically, and the contact point of the dimer, named the dimer linkage sequence (DLS), is located near the 5′ end of each RNA under partially denaturing conditions. It is likely that the presence of two genomes in one virion is advantageous for survival, providing an extra template than can be used when one RNA molecule is damaged and giving genetic variety to their progeny (9, 21). It has been suggested that the DLS in viral genomic RNA usually overlaps with a packaging signal (E/psi) (16). Therefore, it is difficult to assess whether viral RNA dimerization and packaging are independent events or not.
The partial RNA fragments of the 5′ region of a retrovirus genome transcribed and purified in vitro form dimer molecules upon incubation in buffer without any factors such as proteins or cellular extract (16). Thus, the DLS has been inspected mainly by using in vitro transcription systems. In the case of human immunodeficiency virus type 1 (HIV-1), the 5′ untranslated region just downstream of the splicing signal (ss) was first reported to be a DLS, because in in vitro systems, the mutated RNA fragments lacking this region have an impaired ability to form a dimer (3, 28, 42). Recently, several groups reported another site within the 5′ untranslated region which is important for RNA dimerization in vitro. This site was located upstream of the 5′ss and named the dimer initiation site (DIS) (24, 33, 34, 41). The DIS consists of a stem-loop structure with a conserved palindromic sequence at the top of the loop. Two palindromic sites have been suggested to make contact, forming a “kissing-loop complex,” to initiate dimer formation (24, 33, 34, 41).
On the other hand, several studies on RNA dimerization in vivo have shown different features of the dimer. The viral nucleocapsid protein is suggested to alter the secondary structure of the RNA molecule and stabilize RNA dimers such as molecular chaperons (10–12). We and others have reported that a mutation introduced around the DIS/DLS did not affect dimer stability in vivo (5, 7, 39). We also reported the possibility that some region(s) of the virus genome far from the DIS/DLS destabilizes the dimer (39). Electron microscopic observation revealed that the HIV-1 RNA genome contains a central DLS and additional loop structures within each monomer subunit (18), suggesting that HIV-1 RNA contains more than one contact point. Thus, there are many discrepancies between in vitro and in vivo data on HIV-1 RNA dimerization.
Previously, we generated several mutants (DD mutants) with a duplicated 5′ region (1,000 bases) of viral genome containing E/psi and DIS/DLS (E/DLS) at various ectopic positions of RNA and examined their packaging ability and dimer formation in the particles (38). In the mutant virions, we observed the appearance of monomeric forms of virion RNA, which were not present in the wild-type virions. This suggested that the 5′ region of the genome indeed plays an important role in RNA-RNA interaction during virion formation. Since more than 50% of the packaged RNA still formed dimers in these mutant virions, it was unclear whether the dimerization is dispensable for RNA packaging and particle formation or not. To clarify this point, we constructed an additional mutant, sTNΔPBS. In this mutant, a tandem repeated E/DLS is present in the envelope region of RNA and the authentic E/DLS is eliminated by a mutation. Northern blot analysis showed that the RNA in sTNΔPBS virions appeared to be exclusively monomeric. Protein profiles and the density and the infectivity of the mutant virions were also examined.
MATERIALS AND METHODS
Constructs.
The replication-competent HIV-1 proviral clone pNL4-3 (1) and pMSMBA (30), a derivative of pNL4-3, were used as the progenitors for all mutants described below. The construction of p5′ssEnvEXSV, pMPΔPBS, pDDNΔPBS, and pssNΔPBS has been described previously (38). The mutant pDDNΔPBS contains an ectopic E/DLS in the env region and possesses an intact E/DLS at the original position. The mutant pssNΔPBS has an ectopic E/DLS at the same position as in pDDNΔPBS but has a large deletion at the original E/DLS region to eliminate its function. Fragment ΔP was a 1-kb DNA fragment from the HIV-1 5′ region with a deleted primer binding site (PBS) sequence and was isolated by digesting pMPΔPBS with BamHI. It was blunt ended with T4 DNA polymerase and was ligated into a BsaBI site of pDDNΔPBS as an additional ectopic E/DLS to construct pDTNΔPBS. The StuI-XhoI fragment of p5′ssβglob (31) was replaced with the corresponding fragment of pDTNΔPBS, which contains the duplicated fragment ΔP sequences, to construct psTNΔPBS. The HIV-1 proviral protease mutant pMS172 is a derivative of MSMΔEnv2 (30) that contains a 2-nucleotide substitution at nucleotides 2325 and 2326 to create an amino acid substitution in the active site of the protease. In addition, this substitution creates a novel SnaBI site.
Transfection.
293T cells (15) (approximately 7 × 106) were seeded on dishes (diameter, 150 mm) on the day before transfection with plasmid DNA (20 μg) using the calcium phosphate precipitation method (2). On the day after transfection, the supernatant was discarded and fresh medium was added.
Isolation of RNA from cytoplasm and virions.
At 48 to 72 h after transfection, the medium and cytoplasmic RNA were collected concurrently as described elsewhere (30). The viral pellet was resuspended in TSE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, and 1 mM EDTA). The physical virus titer was determined using an enzyme-linked immunosorbent assay kit to quantitate CA-p24 (ZeptoMetrix, Inc.). To isolate RNA from particles, virions were disrupted by the addition of sodium dodecyl sulfate (SDS) to 1% and treated with proteinase K (300 μg/ml) at room temperature for 60 min, followed by Tris-EDTA-saturated phenol-chloroform extraction, chloroform extraction, and ethanol precipitation.
Northern blotting analysis.
Pelleted RNA was resuspended in T-buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% SDS, 100 mM NaCl, and 10% formamide), and the thermostability of dimeric viral RNA was determined by incubating RNA aliquots for 10 min at the temperatures indicated in Fig. 2 (39). The proportions of dimer and monomer were measured by electrophoresis at room temperature on a nondenaturing 0.75% native agarose gel in 0.5× Tris-borate-EDTA buffer (25). Field inversion gel electrophoresis was performed for better separation of large RNA molecules. The conditions for field inversion gel electrophoresis were as follows: forward, 5 V/cm, 0.6 s; reverse, 5 V/cm, 0.1 s. The agarose gel was then treated with 10% formaldehyde at 65°C before being washed with H2O three times, and RNA was blotted electrically onto a Hybond N+ nylon membrane (Amersham Pharmacia Biotech UK Ltd.).
FIG. 2.
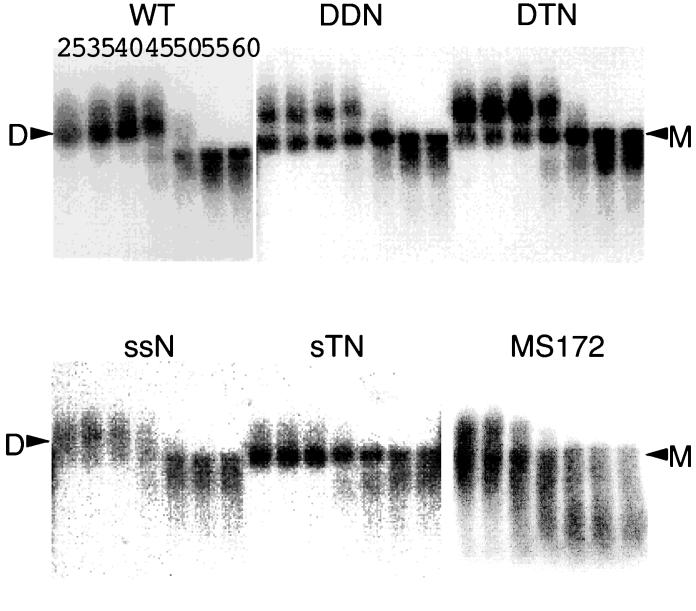
Representative MacBAS image of RNAs detected by Northern blotting. Aliquots of RNA extracted from virions were resuspended in T-buffer, incubated for 10 min in parallel reactions at various temperatures, and then analyzed on a native agarose gel. The membrane was hybridized with an in vitro-synthesized pol region-complementary riboprobe. Positions of dimer (D) and monomer (M) viral RNAs are indicated by arrowheads. The various temperatures (degrees Celsius) at which aliquots were incubated are indicated for the wild-type (WT) lanes. Three or more independent experiments gave similar results.
RNA Northern hybridization analysis was performed as described previously (25). The plasmid T7pol (39) was used to synthesize cRNA probes for Northern hybridization. RNA probes were synthesized using T7 RNA polymerase (New England Biolabs.). Approximately 7 × 106 cpm of riboprobe per blot was used in the hybridization reaction. Hybridization was carried out in the presence of Rapid-Hyb buffer (Amersham Pharmacia). Membranes were washed extensively with 0.1× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate [pH 7.0])-0.1% SDS at 70°C. In experiments designed to assess the conversion of dimers to monomers, the relative amounts of both RNA species were quantitated with a BAS1000 imaging plate system and MacBAS software (Fujifilm Co.).
RNase protection assay.
The antisense probe (≈108 cpm/mg) was synthesized by transcription of pGEM (600–1000) (31) with T7 RNA polymerase or by transcription of pT7HIV-1 410–910 (39) with SP6 RNA polymerase (Promega) following linearization with NotI or SalI, respectively, by a previously described protocol (27). These riboprobes were designed to discriminate between unspliced and spliced HIV-1 RNAs. To serve as size markers for denaturing polyacrylamide gels, HpaII-digested fragments of pGEM3Zf (+) were 32P end labeled (27). One-fifth of the virion-associated RNA or 10 μg of cytoplasmic RNA preparation was mixed with 8 × 104 Cerenkov counts of 32P-labeled antisense RNA and precipitated with ethanol. RNase protection assays were performed by using an RPA III RNase protection assay kit (Ambion, Inc.). After electrophoresis in 5% polyacrylamide-8 M urea gels, quantitation of various protected RNA species was achieved with the BAS1000 imaging plate system and MacBAS software.
Sucrose gradient analysis of virions.
Virions pelleted through a 20% sucrose cushion were resuspended in 200 μl of TSE buffer and layered on 20 to 60% sucrose gradients prepared in the same buffer. The gradients were centrifuged in an SW50.1 rotor at 37,000 rpm for 20 h at 4°C, and 250-μl fractions were collected from the bottom of the tube. The fractions were analyzed for endogenous and exogenous reverse transcription activity and CA-p24.
Western blotting analysis.
Lysates of transfected 293T cells and pelleted virus were prepared as described previously (43), and proteins were resolved on SDS-12% polyacrylamide gels and then electrophoretically transferred to polyvinylidene difluoride membranes. ECL Western blotting detection reagents (Amersham Pharmacia) were used to detect viral proteins on the membrane. Briefly, the membranes were incubated for 1 h at room temperature with serum from an individual infected with HIV-1 and then for another 1 h with horseradish peroxidase-labeled protein A, washed, and visualized by exposure to X-ray film.
Infection.
293T cells (approximately 2 × 106) were transfected with 3 μg each of p5′ssEnvEXSV and MSMBA or the other mutants. At 48 to 72 h posttransfection, the medium was centrifuged and the supernatant was used for infection. Infection was accomplished by incubating cells for 18 to 24 h (M8166 cells) or 72 h (MAGI cells) with the equivalent CA-p24 units of virus. MAGI cell assays were performed as described previously (23).
PCR analysis of viral DNA.
For PCR analysis, extrachromosomal DNA in infected cells was extracted at 16 h after infection by the method described by Hirt (17). DNA samples were pretreated with DpnI restriction endonuclease (New England Biolabs, Inc.) for 24 h to remove contaminating plasmid DNA. PCR amplification was performed with three diagnostic primer pairs. The first pair (P1) was designed to detect nearly completed products of the reverse transcription (432F, 5′-GCAGCTGCTTTTTGCCTGTAC-3′; 783R, 5′-CCTTCTAGCCTCCGCTAGTC-3′). The second pair (P2) was designed to detect the viral strong-stop DNA (462F, 5′-CTGGTTAGACCAGATCT-3′; 633R, 5′-GCTAGAGATTTTCCACAC-3′), and the third pair (P3) was designed to detect aberrant DNA products that arose from homologous recombination (PBS-F15, 5′-GGCGCCCGAACAGGG-3′; 7350R, 5′-GTAGAAAAATTCCCCTCCAC-3′). Samples (0.5 μg of DNA) were subjected to 25 cycles of PCR in a 50-μl reaction mixture. Each cycle consisted of 30 s of denaturation (94°C), 30 s of annealing (60°C), and 30 s of extension (72°C). A series of known amounts of plasmid DNA were amplified along with the Hirt DNA to serve as a standard in each experiment. Amplified products were run through a 1.5% agarose gel and analyzed by Southern blotting hybridization. An antisense probe (≈108 cpm/mg) synthesized by transcription of pT7HIV-1 410–910 (39) was generated using T7 RNA polymerase. Quantitation of amplified DNA fragments was achieved with the BAS1000 imaging plate system and MacBAS software.
RESULTS
Generation of virions that contain exclusively monomeric RNA.
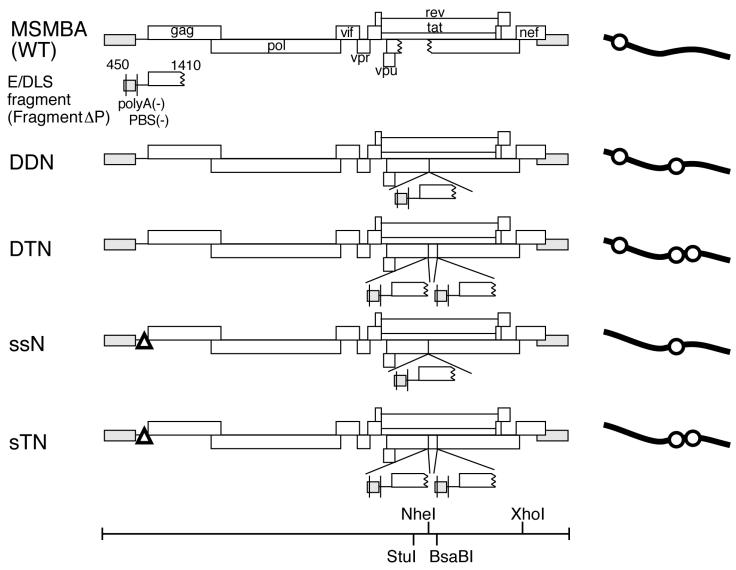
Previously, we reported a series of HIV-1 mutant constructs containing an additional E/DLS region at the ectopic position (38). These mutant RNA molecules were packaged into virions as efficiently as the wild-type RNA. Nearly 40% of the mutant RNA molecules in virions, however, appeared to be monomeric, whereas the wild-type RNA molecules in virions were exclusively dimeric. We speculated that an additional E/DLS region at the ectopic position binds to the authentic E/DLS on the same RNA molecules, competitively interfering with intermolecular dimer formation. Since more than 50% of the mutant RNA molecules were still in a dimeric form, it was not clear whether the dimerization of genomic RNA is essential for RNA packaging and maturation of viral particles or not. Presumably, the original E/DLS site on the mutant molecules worked more efficiently than the ectopic ones to form authentic intermolecular dimers. We therefore knocked out the original E/DLS site and placed two ectopic E/DLS sites in the viral genome to determine whether virions completely lacking dimeric RNA can be generated. Four mutant constructs, DDNΔPBS, DTNΔPBS, ssNΔPBS, and sTNΔPBS, were tested for their ability to produce virions containing intermolecular dimeric RNA. There is one ectopic site in DDNΔPBS and ssNΔPBS and two sites in DTNΔPBS and sTNΔPBS (Fig. 1). Fragment ΔP, which was inserted in these mutants, was approximately 1,000 bases long, spanning the TAR, R/U5, U5/L, SL1, SL2, SL3, and SL4 stem-loops and the 5′ half of the gag gene. These stem-loops are located in the 5′ region of the HIV-1 genome and are reported to play important roles in viral genome packaging and dimerization. Polyadenylation signals and PBSs in the ectopic fragment were deleted to abolish undesired polyadenylation and ectopic initiation of reverse transcription. The original E/DLS site in ssNΔPBS and sTNΔPBS was largely deleted and exchanged with a human β-globin splicing donor to maintain the splicing function. As shown in Fig. 1, there is one E/DLS site in the wild type (MSMBA) and ssNΔPBS, two sites in DDNΔPBS and sTNΔPBS, and three sites in DTNΔPBS. All of the constructs carried a deleterious or inactivating mutation at the env gene to prevent multiple rounds of replication.
FIG. 1.
Diagram of the wild type (WT) and mutants containing copies of viral E/DLS sequences. DDN, DTN, ssN, and sTN are DDNΔPBS, DTNΔPBS, ssNΔPBS, and sTNΔPBS, respectively. A polyadenylation signal (polyA) and a PBS on fragment ΔP were mutated as described in Materials and Methods. Open triangles on ssNΔPBS and sTNΔPBS represent mutations introduced in the 5′ leader region. Schematics of the transcripts are shown at the right; the lines and circles represent viral RNA and the E/DLS region, respectively. The positions of the restriction endonuclease recognition sites in the constructs are indicated at the bottom.
We first analyzed the effects of additional E/DLS sites on the efficiency of viral RNA and protein expression and packaging of RNA into virions (Table 1). The amount of viral genome RNA in cytoplasm and purified virions was quantitated by an RNase protection assay using a riboprobe which can detect both the wild-type and mutant unspliced viral genome RNAs. Expression of viral protein in cytoplasm and purified virions was determined using a CA-p24 detection kit. The viral protein production by the mutants lacking authentic E/psi (5′ssβglob, ssNΔPBS, and sTNΔPBS) was severely impaired, whereas viral RNA expression from all of the mutants was reduced only moderately. The encapsidation efficiency was determined by calculating the ratio of virion-associated RNA to virion CA-p24 released from the 293T cells transfected with these constructs. Some variations in packaging efficiency were observed among constructs. The DTNΔPBS mutant construct showed increased packaging efficiency along with multiplication of the E/DLS regions (Table 1), indicating that the presence of additional E/DLS regions in the HIV-1 genome had a positive effect on RNA packaging. DDNΔPBS-producing particles also tended to have increased packaging efficiency. On the other hand, the packaging efficiencies of two other mutant constructs, ssNΔPBS and sTNΔPBS, were approximately half of that of the wild type. Those mutant constructs lacked authentic E/DLS sites. Therefore, the results supported our previous assumption that the authentic E/DLS site is dominant, probably because the ectopic sites could not completely substitute for the packaging ability of the authentic E/DLS.
TABLE 1.
Relative encapsidation and expression efficiency of the mutants from transfected cellsa
| Virus | Packaging efficienyb | Virion productionc | Viral RNA in cytoplasmd | Viral CA-p24 in cytoplasmc |
|---|---|---|---|---|
| Wild type | 1.00 | 1.00 | 1.00 | 1.00 |
| DDNΔPBS | 1.05 ± 0.23 | 0.55 ± 0.13 | 0.62 ± 0.11 | 0.46 ± 0.11 |
| DTNΔPBS | 1.61 ± 0.24 | 0.38 ± 0.14 | 0.50 ± 0.10 | 0.35 ± 0.12 |
| ssNΔPBS | 0.66 ± 0.29 | 0.08 ± 0.05 | 0.27 ± 0.11 | 0.07 ± 0.03 |
| sTNΔPBS | 0.63 ± 0.11 | 0.10 ± 0.05 | 0.36 ± 0.12 | 0.13 ± 0.04 |
| 5′ ssβglob | 0.19 ± 0.05 | 0.19 ± 0.08 | 0.32 ± 0.13 | 0.21 ± 0.08 |
The value for the wild-type control was set as 1.00 in all experiments. All results represent means ± standard errors from at least three independent experiments.
The relative packaging efficiency of each mutant was calculated by dividing the amount of viral RNA in a purified virion by the amount of CA-p24 in the virion.
The amount of CA-p24 in purified virions and cytoplasm was determined using a CA-p24 enzyme-linked immunosorbent assay kit.
The amount of viral unspliced genome RNA was determined by RNase protection assay.
We then analyzed the virion conformation of the RNA by native agarose gel electrophoresis followed by Northern blot hybridization. We observed various patterns of viral RNA produced by mutant constructs (Fig. 2). As reported in our previous paper (38), DDNΔPBS produced both monomeric and dimeric virion RNAs, while ssNΔPBS produced exclusively dimeric RNA like the wild-type construct. DTNΔPBS, which carried three E/DLS sites, also produced both monomeric and dimeric virion RNAs, although the amount of monomeric RNA was reduced compared with that of DDNΔPBS-derived virions. This result suggested that the third E/DLS site in DTNΔPBS could work intermolecularly to form dimeric RNA even after the first site bound to the second site on the same RNA molecule. In contrast, there was no apparent dimeric RNA in sTNΔPBS-derived virions. The monomeric RNA signal in sTNΔPBS-derived virions was even clearer than that of the protease mutant MS172 (Fig. 2), which was previously shown to produce virions lacking stable dimeric RNA. Therefore, this result indicated that the duplication of the ectopic E/DLS site without a functional authentic E/DLS site made the RNA dimerization in virions undetectable.
Protein profiles and density of virions containing the monomeric genome.
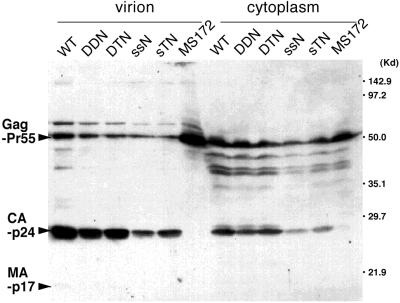
As described above, the HIV-1 protease mutant MS172 produced virions lacking stable dimeric RNA (Fig. 2). To exclude the possibility that the lack of dimeric RNA in sTNΔPBS-derived virions was due to altered viral protein maturation or virion formation like in a protease mutant, we analyzed protein profiles of the virions by Western blotting. In contrast to MS172, which produced virions totally lacking mature Gag CA-p24 or MA-p17, the precursor Gag Pr55 molecules were cleaved into mature proteins in all of the mutant virions as efficiently as in wild-type virions (Fig. 3). Furthermore, all of the mutant and wild-type virions peaked at a density of ca. 1.16g/ml in sucrose gradient centrifugation (data not shown). Therefore, we concluded that the maturation of viral protein and formation of virions was not affected by the mutations introduced in sTNΔPBS.
FIG. 3.
Detection of HIV-1 protein produced in transfected 293T cells and pelleted virions by Western blotting with serum from an HIV-1-infected patient. Positions of Gag precursor Pr55 and Gag products CA-p24 and MA-p17 are indicated.
Genome dimerization is not essential for reverse transcription.
We then compared the infectivities of those mutants with that of the wild type, ΔPBS mutants, and 5′ssβglob. Since all of these constructs carried a deleterious or inactivating mutation in the env gene, HIV-1 Env proteins were supplied by cotransfection of the Env expression vector p5′ssEnvEXSV to produce infectious virions by complementation. At 72 h after transfection, culture supernatants were assayed for the levels of viral CA-p24 protein, and equivalent amounts of virus in CA-p24 were used to infect MAGI cells. At 48 h after infection, cells were fixed and stained, and the cells that were successfully infected as evidenced by bacterial β-galactosidase expression were enumerated. DDNΔPBS produced virions with 100-fold-reduced infectivity compared with the wild type (in one representative experiment, 1.4 versus 187.5 β-galactosidase-inducing units per ng of CA-p24). All of the other mutant constructs, including sTNΔPBS, failed to produce virions with detectable infectivity in this assay. As the MAGI cell assay reflects the magnitude of Tat expression in the sample, this result suggested that the mutations introduced in these constructs affect mainly the step(s) between virus penetration and early gene expression. Since ssNΔPBS carried only one intact E/DLS region and formed dimeric RNA as efficiently as wild-type virus, it seemed unlikely that the loss of infectivity of DDNΔPBS and ssNΔPBS was simply due to the lack of dimeric RNA in virions. Instead, it was more likely that the presence of an ectopic E/DLS site affected one or more steps between virus penetration and gene expression.
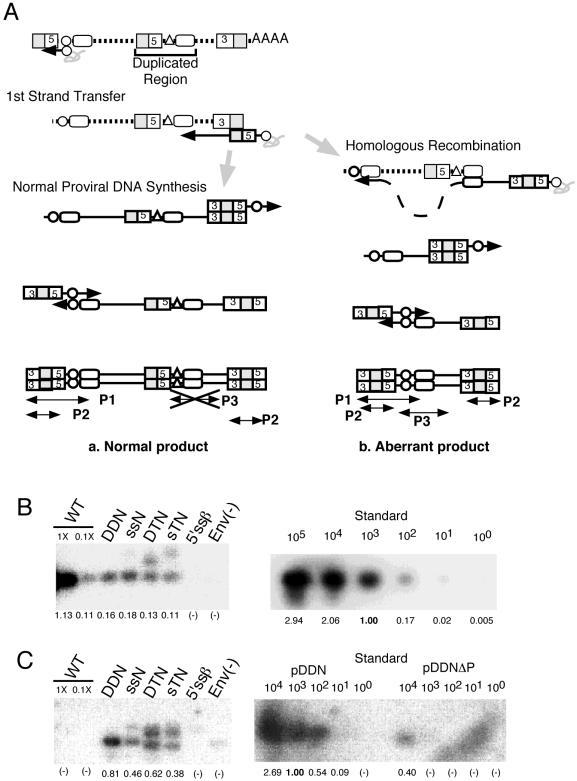
To identify the step affected by the presence of an ectopic E/DLS site, we measured the amount of extrachromosomal viral DNA in infected cells by semiquantitative PCR (Fig. 4). Primer pair P1 would specifically amplify a 351-bp region spanning long terminal repeat U3, R, U5, and a part of the gag gene. In HIV-1-infected cells, this region is reverse transcribed only after negative- and positive-strand transfer occurs. Therefore, this 351-bp fragment represents an HIV-1 cDNA molecule at or near the final step of viral DNA synthesis. On the other hand, the primer pair P2 could detect all of the nascent products of reverse transcription, including minus-strand strong-stop DNA. As shown in Fig. 4B, the amount of P1 products from all of the mutant-infected cells was approximately 1/10 of that from the wild-type-infected cells. Similarly, the amount of P2 products from all of the mutant-infected cells was also approximately 1/10 of that from the wild-type-infected cells (data not shown). These results suggested that the initiation of reverse transcription of the mutants was reduced to 1/10 in mutant virus-infected cells. Since the MAGI cell assay showed that the infectivity of mutant viruses with a duplicated E/DLS site was severely reduced, reduction of provirus synthesis by these mutants could not fully account for the loss of infectivity of the mutants. Therefore, we hypothesized that the duplication of E/DLS results in homologous recombination of viral nucleic acid. To search for and quantitate the amount of such aberrant DNA products, we designed PCR primers that would specifically amplify DNA products derived from homologous recombination (Fig. 4A, P3). This analysis indicated that aberrant proviral DNA products were indeed produced in cells infected with mutant virus (Fig. 4C). The amount of aberrant product was of a level similar to that detected by the P1 primer pair. Since the aberrant product which could be detected by primer pair P3 should be detected by P1 (Fig. 4A), this result indicates that a large portion of the viral DNA in cells infected with these mutant viruses was defective and suggests that the impairment of infectivity of these mutant viruses was in fact caused by homologous recombination.
FIG. 4.
Semiquantitation of viral DNA. (A) Schematic models of the PCR assay. (a) As normal-reverse transcribed products retain the ΔPBS mutation in the duplicated region, the PCR primers designed to anneal to an intact PBS region failed to bind these DNAs. Therefore, the normal reverse-transcribed product was PCR amplified by the P1 or P2 primer pair but not by the P3 primer pair. (b) The duplicated region of the mutants may cause homologous recombination during reverse transcription. Such an error would result in production of aberrant reverse-transcribed products. In the aberrant products, the ΔPBS mutations in the duplicated regions of the mutants are restored, and the P3 primer pair can access the target correctly. Primer pairs P1 and P2 can also amplify this product. Dotted and solid lines represent HIV-1 RNA and DNA, respectively. Boxes numbered 3 and 5 indicate U3 and U5, respectively. Shaded and open boxes indicate R and the duplicated gag region, respectively. Triangles and circles indicate deleted and intact PBSs, respectively. Solid arrows indicate directions of DNA synthesis. Small arrowheads and thin lines indicate PCR primers and their products. (B) Detection of viral DNAs in a subset of mutant viruses. The primer pair P1 was used to amplify a fragment of 351 bp. WT, wild type. (C) Detection of aberrant products by PCR using the primer pair P3. On each blot, wild-type samples (1- and 0.1-fold) were placed as controls. For the HIV-1 DNA positive control, 1 to 100,000 or 10,000 copies of plasmid DNA (indicated above the blots) were amplified in parallel. The plasmid pDDNΔPBS is a negative control. The value of the amplified signal of positive control plasmids (103 molecules) was set as 1 on each blot. The relative value for each sample is indicated below the blots.
DISCUSSION
We successfully constructed a mutant, sTNΔPBS, which produces virions containing only monomeric viral RNA. The mutant sTNΔPBS contained two ectopic E/DLS regions and had its original E/DLS region deleted. The virions produced by this mutant possessed intact viral proteins, were of normal density, and underwent maturation similar to that of wild-type virions (Fig. 3 and data not shown). As far as we know, this is the first report to show the possibility of complete separation of encapsidation from physical dimerization of retroviral RNA. The RNA from DTNΔPBS, which has three E/DLS sites, formed dimers at a reduced level compared to that from DDNΔPBS, which has two E/DLS sites. This result suggested that the additional ectopic site was used mainly for intramolecular interaction and increased the chance of an intermolecular dimer forming at the original site in the case of DTNΔPBS RNA. In addition, the encapsidation ability of ssN/sTNΔPBS RNA lacking the original E/DLS region was partially reduced compared to that of other mutants (Table 1). Taken together, the results indicate that the original E/DLS site was superior to the ectopic ones in mediating encapsidation and dimerization of viral RNA.
It is possible that the insertion mutation we generated may affect viral gene expression, since the gag coding region of HIV-1 is reported to form an internal ribosomal entry sequence (6). In fact, all of the mutants showed reduced levels of RNA expression, indicating that the insertion mutation introduced had a negative effect on RNA transcription (Table 1). Deletion in authentic E/psi introduced in the ssNΔPBS, sTNΔPBS, and 5′ssβglob mutants also affected RNA transcription severely. The combination of two kinds of mutations, the deletion of the 5′ E/psi and the insertion of an ectopic E/DLS site(s), might synergistically affect protein translation, since protein expression by the ssNΔPBS and sTNΔPBS mutants was much more severely impaired than RNA expression. On the other hand, reductions in protein expression levels did not greatly differ from those in RNA expression levels in the DDNΔPBS and DTNΔPBS mutants, which contain insertion of ectopic E/DLS but not the deletion in the 5′ E/psi. Mutations introduced did not affect virion budding, since similar levels of protein expression were observed in cytoplasm and in virions. The variation in expression levels of mutants might affect RNA encapsidation to a certain extent. Nevertheless, there should be enough viral RNAs to be encapsidated in cytoplasm, because the reduction in RNA expression was always less than that in protein expression.
The RNA from the protease-defective mutant MS172 also migrated as a monomer on native gels under these experimental conditions (Fig. 2). This result was consistent with previous reports describing genome dimerization of protease-defective retrovirus (13, 14). However, the profiles of the monomers from sTNΔPBS and MS172 were not similar. The sample from MS172 contained molecules of various sizes, seen as a broadly smeared signal throughout the lane. We sometimes observed a mixture of monomers and fragile dimers of RNA from the MS172 sample (data not shown). In addition, the heated MS172 RNA has tended to degrade more than other RNAs (see lanes 60 in Fig. 2). On the other hand, RNA from sTNΔPBS clearly migrated as a monomer, and no smaller molecules were detected. The electrophoretic profile of sTNΔPBS RNA under native conditions is quite similar to that of the wild type, which shows only one peak signal, except in mobility. After denaturation by heating, both the sTNΔPBS and wild-type RNAs were melted and displayed the same smeared signals in the gel. This implies a difference in the process of monomer formation in each case. The RNA from the protease mutant MS172 might form fragile dimers in virions, but they would gradually dissociate and fragment during isolation and electrophoresis. The stability of the RNA itself might not be fully acquired in MS172 virions because the maturation process was lacking. In the case of sTNΔPBS, RNA would undergo maturation and form a stable tertiary structure, which is tolerant of lysing or phenol extraction.
The early-phase infectivity was markedly reduced in sTNΔPBS compared to the wild type (see Results). This, however, was not because of a change in status of its RNA, since the infectivities of other mutants that showed various RNA profiles also displayed the same magnitude of reduction. PCR analysis of viral DNA in infected cells (Fig. 4) indicated that these mutants synthesized reverse-transcribed products. It should be emphasized that all mutants retained the ability to produce late reverse-transcribed products (Fig. 4A), even though a large proportion of them were aberrant. These results suggest that sTNΔPBS virions retain endogenous reverse transcription activity and that monomeric viral RNA could be the template for reverse transcription and strand transfer. One might claim that an undetectable amount of dimeric RNA exists in sTNΔPBS RNA and that such a dimer is a template for reduced reverse transcription. Although we could not completely exclude this possibility, this is highly unlikely since even the RNA from ssNΔPBS, which exclusively forms dimers, could not be a template for efficient reverse transcription. There are several reports describing homologous recombination during PCR (22, 29, 32). In those reports, recombination occurred at less than 6% even after 35 reaction cycles. In our PCR experiment, only 25 cycles were performed, and thus PCR recombination was not expected to be a major problem.
The duplication of the 5′ region of HIV-1 causes a forced strand transfer at the duplicated sequence during reverse transcription (4), and this phenomenon made our attempt to measure virus DNA very complicated. The results of infection experiments would be clearer if we could exclude the aberrant strand transfer by minimizing the duplicated target sequences. We are now actively working to determine more precisely the region responsible for mediating intramolecular interaction of the viral genome. From our present results, TAR is not required but the region from SL1 to SL4 is required for this interaction (unpublished data). If we can determine the region clearly, it may be possible to construct a mutant with minimum repeat sequences and to assay the infectivity of the mutant virus without undesired strand transfer and homologous recombination.
The replication events that the MAGI cell assay reflects include particle binding, penetration, uncoating, reverse transcription, nuclear transport, integration, and gene expression. It is highly unlikely that the mutation affected events other than reverse transcription, as our mutant virion showed only a modified RNA profile and showed normal protein structures. This assumption should be tested with further experiments, however. Our results showed that the main defect of the mutants would be in reverse transcription, not because the RNA does not form dimers but because it has broadly repeated sequences which could be a target for strand transfer or homologous recombination. We even observed abundant production of another aberrant reverse-transcribed product, caused by the strand transfer to the inserted sequence, by these mutants (data not shown).
Basically, the retroviral genome packaged in intact virions always forms a dimer. In previous reports by us and others, even RNAs from DIS/DLS mutants formed dimers in vivo like the wild type only if encapsidated in matured virions (5, 7, 39). We demonstrated here that viral genome RNA could be packaged, encapsidated, matured, and reverse transcribed without detectable physical dimerization in mature virions and thus dissociated packaging from dimerization by genetic engineering. A recent study with a Rous sarcoma virus MA mutant revealed that monomeric RNA was packaged in the mutant virion (36). In addition, studies have found that HIV-1 protease mutants and particles with the gag/gag-pol ratio modified also contained unstable dimeric RNA (13, 40). These reports suggested the dissociation of packaging from the dimerization of viral RNA. In both cases, however, the emergence of monomeric viral RNA was caused by the modification of viral protein. The maturation of virions is essential for the formation of a stable RNA dimer, and the modification of viral protein might affect this maturation directly or indirectly. Thus, their data might not reflect the behavior of viral RNA under native conditions.
The nature of the dimerization of retroviral RNA is still unclear. Previous studies revealed that retroviruses could utilize both copies of genome RNA in virions for reverse transcription (19, 21, 35, 44), and the presence of two genomes in each virion contributes to genetic variation via recombination and provides an extra template that can be used in instances where one RNA is damaged (20, 26, 37). However, it is not clear whether the formation of dimers is essential for retroviral infection. In this report, we revealed that RNA dimerization might not be essential to the viral life cycle in a single round of replication. Moreover, our results strongly suggest that a monomeric viral RNA can be packaged if it can form the dimeric linkage structure intramolecularly. This finding is completely consistent with the hypothesis that the linkage structures and/or their interaction is required for packaging. Further studies will give us more clues to prove this assumption.
Acknowledgments
We thank Michael Schwartz for providing the plasmid pMS172, Antonito T. Panganiban for helpful discussion and advice, and Sayuri Sakuragi for encouragement.
J.S. was supported by the Organization for Pharmaceutical Safety and Research. This work was supported by grants from the Organization for Pharmaceutical Safety and Research; the Ministry of Education, Culture, Sports, Science and Technology; and the Ministry of Health, Labour, and Welfare, Japan.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini, A., and B. D. Walker. 1990. Techniques in HIV research. Stockton Press, New York, N.Y.
- 3.Awang, G., and D. Sen. 1993. Mode of dimerization of HIV-1 genomic RNA. Biochemistry 32:11453–11457. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B., J. van Wamel, and B. Klaver. 1995. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J. Mol. Biol. 252:59–69. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., and J. L. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. 1984. Genome structure, p.261–368. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumor viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1–26. [DOI] [PubMed] [Google Scholar]
- 10.Darlix, J. L., C. Gabus, M. T. Nugeyre, F. Clavel, and F. Barre-Sinoussi. 1990. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 216:689–699. [DOI] [PubMed] [Google Scholar]
- 11.De Rocquigny, H., C. Gabus, A. Vincent, M.-C. Fournie-Zaluski, B. Roques, and J.-L. Darlix. 1992. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 89:9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 67:5443–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen Virol. 36:59–74. [DOI] [PubMed] [Google Scholar]
- 16.Greatorex, J., and A. Lever. 1998. Retroviral RNA dimer linkage. J. Gen Virol. 79:2877–2882. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369. [DOI] [PubMed] [Google Scholar]
- 18.Höglund, S., Å. Öhagen, J. Goncalves, A. T. Panganiban, and D. Gabuzda. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233:271–279. [DOI] [PubMed] [Google Scholar]
- 19.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227–1233. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. S., R. W. Allan, and H. M. Temin. 1993. Alteration of location of dimer linkage sequence in retroviral RNA: little effect on replication or homologous recombination. J. Virol. 67:3151–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, J. S., R. W. Allan, and H. M. Temin. 1994. One retroviral RNA is sufficient for synthesis of viral DNA. J. Virol. 68:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, H. S., B. W. Popovich, W. R. Shehee, E. G. Shesely, and O. Smithies. 1991. Problems encountered in detecting a targeted gene by the polymerase chain reaction. Gene 103:227–233. [PubMed] [Google Scholar]
- 23.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464–13474. [DOI] [PubMed] [Google Scholar]
- 25.Lear, A. L., M. Haddrick, and S. Heaphy. 1995. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology 212:47–57. [DOI] [PubMed] [Google Scholar]
- 26.Luo, G. X., and J. Taylor. 1990. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 64:4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook (ed.). 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Marquet, R., F. Baudin, C. Gabus, J. L. Darlix, M. Mougel, C. Ehresmann, and B. Ehresmann. 1991. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 19:2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marton, A., L. Delbecchi, and P. Bourgaux. 1991. DNA nicking favors PCR recombination. Nucleic Acids Res. 19:2423–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyerhans, A., J. P. Vartanian, and S. Wain-Hobson. 1990. DNA recombination during PCR. Nucleic Acids Res. 18:1687–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muriaux, D., P. M. Girard, B. Bonnet-Mathoniere, and J. Paoletti. 1995. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 270:8209–8216. [DOI] [PubMed] [Google Scholar]
- 34.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486–27493. [PubMed] [Google Scholar]
- 35.Panganiban, A. T., and D. Fiore. 1988. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science 241:1064–1069. [DOI] [PubMed] [Google Scholar]
- 36.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74:164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112–1118. [DOI] [PubMed] [Google Scholar]
- 38.Sakuragi, J., T. Shioda, and A. T. Panganiban. 2001. Duplication of the primary encapsidation and dimer linkage region of HIV-1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 75:2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakuragi, J. I., and A. T. Panganiban. 1997. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J. Virol. 71:3250–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundquist, W. I., and S. Heaphy. 1993. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic. Proc. Natl. Acad. Sci. USA 90:3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, H., A. E. Jetzt, Y. Ron, B. D. Preston, and J. P. Dougherty. 1998. The nature of human immunodeficiency virus type 1 strand transfers. J. Biol. Chem. 273:28384–28391. [DOI] [PubMed] [Google Scholar]