Abstract
G4 and p13II are accessory proteins encoded by the X region of bovine leukemia virus and human T-cell leukemia virus type 1 (HTLV-1), respectively. Disruption of the G4 and p13II open reading frames interferes with viral spread in animal model systems, indicating that the corresponding proteins play a key role in viral replication. In addition, G4 is oncogenic in primary cell cultures and is absolutely required for efficient onset of leukemogenesis in sheep. To gain insight into the function of these proteins, we utilized the yeast two-hybrid system to identify protein partners of G4. Results revealed that G4 interacts with farnesyl pyrophosphate synthetase (FPPS), a protein involved in the mevalonate/squalene pathway and in synthesis of FPP, a substrate required for prenylation of Ras. The specificity of the interaction was verified by glutathione S-transferase (GST) pull-down assays and by coimmunoprecipitation experiments. Furthermore, confocal microscopy showed that the subcellular localization of G4 was profoundly affected by FPPS. The G4 protein itself was not prenylated, at least in rabbit reticulocyte lysate-based assays. The domain of G4 required for binding to FPPS was restricted to an amphipathic α-helix rich in arginine residues. Subtle mutation of this α-helix abrogated G4 oncogenic potential in vitro, providing a biological relevance for FPPS-G4 complex formation in cells. Finally, HTLV-1 p13II was also found to specifically interact with FPPS (in yeast as well as in GST pull-down assays) and to colocalize with G4 in mitochondria, suggesting a functional analogy between these oncoviral accessory proteins. Identification of FPPS as a molecular partner for p13II and G4 accessory proteins opens new prospects for treatment of retrovirus-induced leukemia.
Bovine leukemia virus (BLV), a naturally occurring B-lymphotropic retrovirus, is a member of the Oncovirinae subfamily and belongs to the Deltaretrovirus genus, which also includes the human T-cell leukemia virus types 1 and 2 (HTLV-1 and -2) and the simian T-cell leukemia viruses. All these viruses share a similar genomic organization and induce related pathologies in their respective host species (reviewed in references 44 and 54). In addition to the gag, pol, and env genes, deltaretroviruses contain an X region, located between the env sequences and the 3′ long terminal repeat. At least four proteins are encoded by this genomic region; perhaps the best-characterized ones are the Tax transactivator and the Rex posttranscriptional regulator. These two proteins are expressed from a single double-spliced RNA and are considered to be essential since their mutation abrogates viral infectivity or pathogenicity (39, 40, 52). Two additional open reading frames are transcribed from the X region, and the corresponding mRNAs potentially encode accessory proteins: R3 and G4 for BLV, p12I/p13II/p30II for HTLV-1, and p10I/p28II/p11V for HTLV-2 (2, 5, 6, 8–10, 27, 28). Among these, HTLV-1 p12I is a highly hydrophobic, very unstable, and poorly immunogenic protein which is localized in cellular endomembranes (28, 49). It has distant homology to the bovine papillomavirus E5 oncoprotein and harbors weak oncogenic potential. p12I binds to the 16-kDa subunit of a vacuolar ATPase, to the β and γ chains of the interleukin 2 receptor, and to the major histocompatibility complex class I heavy chain (18, 23, 29, 34). p12I forms dimers through two putative leucine zipper domains and contains four SH3-binding motifs (PXXP) (1, 49). In cell culture, p12I is required for the infection of primary quiescent lymphocytes, suggesting a role in activation of host cells during early stages of infection (1). Another HTLV-1 accessory protein, p30II (also called Tof), contains a bipartite arginine-rich domain involved in nuclear targeting (14). p30II shares homology with proteins of the POU family and harbors transcriptional function (56). Finally, p13II, which is produced from a distinct singly spliced mRNA, is mainly localized in mitochondria and perturbs their morphology (11). Nuclear localization of p13II has also been reported in a subpopulation of cells (11, 14, 28). p13II interacts in vitro with proteins harboring similarities to members of the nucleoside monophosphate kinase superfamily and actin-binding protein 280 (22). Although expression of p12I, p13II, and p30II proteins has not been directly detected in vivo, evidence for their chronic production was obtained from cellular (cytotoxic T lymphocytes) or humoral immune responses (7, 15, 36).
BLV potentially encodes two accessory proteins called R3 and G4 (2). The R3 amino terminus shares the common nuclear localization signal with Rex and could therefore inhibit its function, although this has not been demonstrated. Sequence analysis of G4 identifies an arginine-rich region in the middle of the protein (amino acids [aa] 58 to 70) and, in the NH2-terminal part, a stretch of hydrophobic amino acids (aa 1 to 24) followed by two potential proteolytic cleavage sites. The two predicted forms of G4 will be referred to here as G4 (the full-length protein) and ΔG4 (the protein after cleavage of the hydrophobic leader). In primary rat embryo fibroblasts (REF cells), G4 exhibits oncogenic potential (26). Indeed, cotransfection of G4 and Ha-ras expression vectors into REF cells generates fully transformed cells able to induce tumors in nude mice.
Initial reports indicated that HTLV and BLV accessory proteins were dispensable for viral expression, replication, and immortalization in cell culture (13, 16, 20, 38, 52). In vivo, however, BLV recombinant proviruses deleted in the R3 and G4 open reading frames are impaired for viral propagation (53, 55). Similarly, selective ablation of the p12I or p13II/p30II protein reduces HTLV-1 (clone ACH) infectivity in rabbits (3, 12). In addition, BLV R3 and G4 are required for the induction of lymphosarcoma or leukemia in sheep, indicating a role for these genes during BLV-associated pathogenesis (26). These observations are presently the best evidence for a biological relevance of the BLV and HTLV accessory proteins. The goal of this study was to further characterize the mechanisms by which these proteins regulate viral spread and pathogenesis.
MATERIALS AND METHODS
Plasmids.
Control yeast vectors purchased from Stratagene were p53 (which expresses the DNA-binding domain of Gal4 fused to aa 72 to 390 of murine p53) and pSV40 (encoding aa 84 to 708 of the SV40 large T-antigen linked to the activation domain of Gal4). pBDG4 was constructed by PCR amplification of the gene g4 cloned into plasmid pRSG4 (2), using primers BDG4RI (5′-TTTGAATTCCTGGCTTGCACCCGCGTTTGT-3′) and 7325SalI (5′-TTTGTCGACCATCGATGGTGACATCATTGG-3′) (at positions 430 and 7325, respectively, according to the numbering of Sagata et al. [43]). The PCR-amplified fragment was digested by EcoRI/SalI and inserted into the corresponding sites of pBD-GAL4 (Stratagene). The same fragment was also introduced into vector pGBT9 (Clontech) digested by EcoRI/SalI, generating pGBTG4. Plasmids pGBTG4S and pGBTG4B were derived from pGBTG4 by 3′-end deletion of the sequences downstream of the SacI or BamHI restriction sites at positions 7196 and 7202, respectively. These two constructs were obtained by PCR using oligonucleotides BDG4RI and 5′-AAAGTCGACTCACAAGGGGTCTCGAAGGGCTCGGCGGGG-3′ (for pGBTG4S) or 5′-AAAGTCGACTCATAATGGATCCCGAAGAGCTCG-3′ (for pGBTG4B). To construct pGBTΔG4S, a fragment corresponding to the second exon of G4 mRNA (positions 7066 to 7310) was PCR amplified using primers 5′-TTTGAATTCCCACATCCAGCAGCATTTGG-3′ and 7325SalI and then inserted into the EcoRI-SalI sites of pGBT9. pGBTΔG4, which harbors DNA sequences derived from BLV strain 344 (53), encodes a G4 variant with four codon substitutions (F35S, P44L, L52H, R67Q) compared to the sequence reported by Sagata et al. (43). Plasmid pGexΔG4 contains the G4 sequences (between positions 7073 and 7931) cloned into the pGEX-2T prokaryotic expression vector. pGBTΔG4 was constructed by cloning the SmaI insert of pGexΔG4 into pGBT9. pGBTΔG4ΔQALR is identical to pGBTΔG4 except for a deletion of four codons (Q67A68L69R70) and was created by PCR using two partially overlapping primers: ΔALR (5′-CCCCGCCGAGCCCTTGATCCATTACCTGATAACGAC-3′) and ΔALRC (5′-ATCAGGTAATGGATCAAGGGCTCGGCGGGGGAG-3′). To generate pHisG4, the EcoRI/SalI insert of pBDG4 was transferred to pcDNA3.1/HisC (Invitrogen). The resultant plasmid was cleaved by KpnI/XbaI, and the G4 fragment was cloned into the pFlag-CMV-2 vector (Sigma), yielding pFlagG4. pHisΔG4 was constructed by cloning the EcoRI/SmaI insert of pGexΔG4 into pcDNA3.1/HisA (Invitrogen). The green fluorescent protein (GFP)-based plasmids pEGFPG4, pEGFPΔG4, and pEGFPΔG4ΔQALR were all derived from pEGFP-C1 (Clontech) and contain the HindIII/EcoRI G4 fragment of pSGG4 (26), the SmaI/SmaI ΔG4 insert of pGexΔG4, or the EcoRI/SalI sequences of pGBTΔG4ΔQALR, respectively. G4 was fused to the NH2-terminal part of GFP after PCR amplification of the g4 gene from pRSG4 using two primers, 5′-TTTAAGCTTCCCGGGAGTATGGCTTGCA-3′ and 5′-TTTAGATCTGATTGGACAAAACCAGGGCCG-3′, and the resulting amplicon was inserted into the HindIII/BglII sites of pEGFP-N1 (Clontech), generating pG4GFP. To construct pSGG4ΔQALR, the g4 fragment from pSGG4 was mutated by a two-step PCR procedure using four primers, ΔALR, ΔALRC, T7 (5′-TAATACGACTCACTATAGGG-3′), and 7325 SalI, digested by HindIII/SalI, and introduced into the HindIII and XhoI sites of pG4MII (50). The bovine fpps gene from pADFPPS, isolated from the two-hybrid screen, was subcloned into mammalian vector pcDNA3.1/HisC (Invitrogen) and pGEX-2T (Amersham-Pharmacia), generating pHisFPPS and pGexFPPS, respectively. The yeast expression vector pGBTP13 contains the p13II open reading frame cloned into plasmid pGBT9 (V. Ciminale, unpublished results). Insertion of the EcoRI insert of pGBTP13 into pcDNA3.1/HisC generated pHisP13. Finally, plasmid pWMras encoding murine M-ras was provided by J. C. Renauld (Ludwig Institute, UCL, Brussels, Belgium), and vector p13II-GFP has been described elsewhere (11). The integrity of all the constructs was verified by restriction analysis and by nucleotide sequencing.
Yeast two-hybrid system.
A cDNA library was constructed from the BL3 BLV-negative cell line using a cDNA synthesis kit and vectors from Stratagene. Briefly, 5 μg of mRNA, corresponding approximately to 107 BL3 cells, was isolated by using the QuickPrep mRNA purification kit (Amersham-Pharmacia) and was converted to cDNA by MLV-RT and poly(dT) linker-primers. Following RNase H digestion, double-stranded cDNA was synthesized using 100 U of DNA polymerase I, blunt ended, ligated to adaptors, and inserted into HybriZap lambda phages. After packaging using Gigapack Gold extract (Stratagene) and amplification (up to 6 × 109 PFU/ml), the phage library (108 PFU) was converted to plasmids by incubation for 3 h at 37°C in the presence of XL1-Blue MRF′ and 1010 ExAssist helper phages (Stratagene). After culture, the bacteria were lysed at 70°C for 20 min in order to release the phagemids. To amplify the mass-excised library, nonsuppressing Escherichia coli XLOLR cells (2 × 1010), which contain an amber mutation that prevents helper phage replication, were combined with 2 × 109 phagemids (at a 10:1 ratio). After cultivation for 3 h in Luria-Bertani broth supplemented with 50 μg of ampicillin/ml, the phagemids were purified by centrifugation on a cesium chloride gradient and transformed using the LiAc-polyethylene glycol protocol into Saccharomyces cerevisiae strain PJ696 Mata. The genotype of this strain is trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ.
A total of 107 transformants were plated onto 245-cm2 plates containing SD selective medium (0.67% Bacto-yeast nitrogen base, 2% glucose, 1.5% agar supplied with dropout nutrient missing one or several amino acids) without leucine. On the other hand, the PJ696 Matα yeast strain (corresponding to PJ696 Mata transfected with the URA3-containing YEp50 vector) was transformed with pBDG4 and plated onto SD medium lacking both tryptophan and uracil. A total of 8 x 108 Mata yeast containing the cDNA library was thawed in YPAD medium (1% Bacto-yeast extract, 2% Bacto-peptone, 2% glucose, with 0.6% adenine) and mixed with approximately the same amount of grown PJ696 Matα yeast transformed with bait pBDG4. After concentration of the cells on 0.22-μm-pore-size filters by using a Swinnex filter support (Millipore), mating was performed during 4.5 h at 30°C. Yeast cells were plated onto on SD medium lacking uracil, leucine, tryptophan, and histidine and supplemented with 0.25 mM 3-AT (Sigma) and incubated for 5 days. His+ colonies were then screened for β-galactosidase in a qualitative colony-lift filter assay using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate. To this end, colonies were replicated on Whatman no. 1 filters, grown overnight on YPAD plates, permeabilized by immersion in liquid nitrogen, and thawed at room temperature. The replicas were placed onto another filter presoaked in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM Mg2SO4) containing X-Gal (0.33 mg/ml). Positive colonies were isolated and cultivated on SD minimum medium lacking uracil, leucine, tryptophan, and histidine in the presence of 0.25 mM 3-AT and retested for β-galactosidase activity on the basis of three different clones. Plasmid DNA was isolated from positive colonies and transformed by electroporation into E. coli HB101. Bacteria were then plated onto M9 minimal medium agar plates containing ampicillin and supplemented with all amino acids but lacking leucine, in order to select for plasmids bearing the LEU2 marker. After transformation into yeast, the different clones were tested for β-galactosidase activity and sequenced.
Quantitative β-galactosidase assay.
Cultures were grown overnight in appropriate SD selective medium, diluted fivefold in YPAD rich medium, and cultivated until the optical density at 600 nm (OD600) reached 0.5 to 0.8. After a single wash in Z-buffer, yeast cells were disrupted by two freeze-thaw cycles in liquid nitrogen. Lysates were resuspended in 0.7 ml of Z-buffer containing 0.27% β-mercaptoethanol and supplemented with o-nitrophenyl-β-d-galactopyranoside (160 μl of a 4-mg/ml solution in Z-buffer). After incubation at 30°C, the reactions were stopped by addition of 400 μl of 1 M Na2CO3, and the OD420 was determined and compared to a standard curve. For the experiments based on chlorophenol red β-D-galactopyranoside (CPRG), the reactions were performed in a buffer (0.7 ml) containing 0.1 M HEPES (pH 7.3), 0.9% NaCl, 0.065% l-aspartate, 1% bovine serum albumin, 0.05% Tween 20, and 1.355 mg of CPRG/ml, incubated at room temperature, and arrested with 0.5 ml of 3 mM ZnCl2 after different intervals of time. Relative CPRG units (U) were calculated with the formula U = OD578 × 1,000)/(OD600 × Fc × t) (where OD578 is the optical density of the reaction after addition of ZnCl2, OD600 is the optical density of the culture, Fc is the dilution factor, and t is the duration of the assay).
GST pull-down assay.
The prokaryotic expression plasmids pGexFPPS and pGex2T were propagated in the HB101 strain of E. coli. Overnight cultures were diluted 10 times in fresh 2YT-G medium (1.6% tryptone, 1% Bacto-yeast extract, 0.5% NaCl, and 2% glucose) containing 100 μg of ampicillin/ml and incubated until the OD600 reached 0.7. After induction of expression with 1 mM isopropyl-β-d-thiogalactopyranoside, bacteria were allowed to grow for an additional 2.5 h. After being harvested and washed with phosphate-buffered saline (PBS), bacteria were lysed by sonication and incubated at 4°C in the presence of 1% Triton X-100. After centrifugation at 10,000 × g for 15 min at 4°C, the supernatant was mixed with glutathione-Sepharose beads (Pharmacia) for 30 min at room temperature. Finally, the beads were washed three times with PBS and resuspended as a 15% suspension in the presence of a mixture of protease inhibitors (Complete; Roche). The GST-FPPS or GST polypeptides complexed with the beads were quantified using a Bio-Rad assay. Equal amounts of fusion proteins were added to 5 μl of rabbit reticulocyte lysates (TNT in vitro transcription-translation; Promega) programmed with pHisΔG4, pHisG4, or pHisP13 plasmid DNA in the presence of a mixture of [35S]methionine and [35S]cysteine (Promix; Amersham-Pharmacia). After gentle shaking for 2.5 h at 4°C in NETN binding buffer (200 mM NaCl, 20 mM Tris-HCl [pH 8], 1 mM EDTA, and 0.5% NP-40), the beads were washed four times in binding buffer. Bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), soaked in amplifying solution (Enlightening; NEN), and visualized by autoradiography. Rainbow 14C-methylated molecular weight markers were purchased from Amersham-Pharmacia.
Coimmunoprecipitation and confocal microscopy.
HelaTat cells (47) were cultivated at 37°C in a 5% CO2--air humidified atmosphere in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 0.25 μg of amphotericin B/ml, 100 μg of streptomycin/ml, and 1 mM sodium pyruvate. One day before transfection, the cells were divided and seeded at a density of 3.5 × 105 to 5 × 105 cells per 10 to 30 cm2. The cells were next transfected with 2 to 3 μg of plasmid DNA using Lipofectamine reagent (GibcoBRL) as described by the manufacturer.
Thirty hours after transfection, the cells were washed in PBS buffer, scraped, and lysed in NET buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 5 mM EDTA, 0.5% NP-40, and 0.25% sodium deoxycholate) containing proteolytic inhibitors (Complete; Roche). Then, the lysates were immunoprecipitated with anti-GFP antibody (Molecular Probes) coupled with protein A-Sepharose beads (Amersham-Pharmacia). The immunoprecipitates were washed three times in NET buffer and once in TNE (10 mM Tris-HCl [pH 8.3], 150 mM NaCl, and 1 mM EDTA) and migrated on a denaturing polyacrylamide gel (SDS-PAGE). After transfer of the proteins onto polyvinylidene difluoride membranes, the filters were saturated in 1% blocking reagent (Roche), incubated overnight with anti-His antibody (Sigma), washed in TBST (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20), and revealed by chemiluminescence using horseradish peroxidase-conjugated anti-mouse immunoglobulin antibody (essentially as described in the BM Chemiluminescence Western Blotting kit; Roche).
For confocal microscopy examinations, HelaTat cells were seeded onto coverslips and transfected as described above. Cells were fixed in 3.7% formaldehyde, permeabilized with 0.1% Nonidet P-40, incubated with anti-His or anti-Flag antibodies (Sigma), and revealed with fluorescein isothiocyanate (FITC)- or Texas Red-coupled anti-mouse immunoglobulin conjugates. Coverslips with labeled cells were mounted by using the Prolong Antifade kit (Molecular Probes) and examined under a confocal microscope (TCS; Leica).
In vitro prenylation assays.
Rabbit reticulocyte lysates (TNT in vitro transcription-translation; Promega) were programmed with 1 μg of plasmid DNAs in the presence of either [35S]methionine-[35S]cysteine (Promix; Amersham-Pharmacia) or R-[5-3H]mevalonic acid (NEN). After incubation at 30°C for 90 min, the G4 andΔG4 and the M-ras proteins, respectively, were immunoprecipitated with anti-His (Sigma) or with Jal4 (a kind gift from J. C. Renauld, UCL, Brussels, Belgium) antibodies and Staphylococcus aureus protein A. After three washes in NET buffer and one in TNE, the immunoprecipitates were migrated onto an SDS-PAGE gel, fixed, soaked in amplifying solution (Enlightening; NEN), and autoradiographed for 10 to 30 days.
Transformation of primary REF cells.
Primary REF cells were prepared from F344 rats at 14 days of pregnancy (IFFA CREDO) and cultivated for 2 days in modified Eagle’s medium supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). A total of 2 μg of plasmids was transfected into 2 × 106 REF cells by using Effectene reagent (Qiagen) as described by the manufacturer. Forty-eight hours posttransfection, cells were collected, washed with PBS, and either cultured in the presence of 400 μg of G418 per ml to score for foci formation or injected subcutaneously into thymusless nude mice (nu/nu NMRI background). A total of six mice in three independent experiments were injected for each plasmid combination. The tumor volume was calculated by the ellipsoid formula 4/3 π a b2 where a and b are, respectively, the length and width of the tumor.
RESULTS
Identification of FPPS as a ligand for G4.
As mentioned in the introduction, the G4 protein harbors oncogenic potential in primary cell cultures, and its ablation from the provirus drastically restricts viral spread. G4 is also essential for efficient induction of leukemia by BLV in sheep. To gain insight into the function of this key accessory protein, we adopted a strategy based on the identification of cellular factors interacting with G4. With the aim of identifying the components of the protein complex occurring in infected cells and to understand G4 modes of action, we used a yeast two-hybrid system. To this end, the G4 open reading frame was inserted into the pBDGAL4 vector to generate plasmid pBDG4 expressing a fusion protein between G4 and the DNA-binding domain of the Gal4 transactivator. To screen for G4 partners, a cDNA library was synthesized from a bovine B-lymphocytic cell line (BL3) and inserted into HybriZap lambda phages (Stratagene). Recombinant phagemids, derived from this library by in vivo mass excision, contained a collection of B-cell-specific cDNAs cloned into the pADGal4 vector and expressed target proteins fused to the Gal4 activation domain. The vector (pBDG4) and the target (pADGal4) recombinant phagemids were introduced via a mating procedure into yeast strain PJ696 and plated onto SD selection medium (lacking leucine, tryptophan, and histidine) supplemented with 0.25 mM 3-AT to suppress leaky expression of the HIS3 reporter gene.
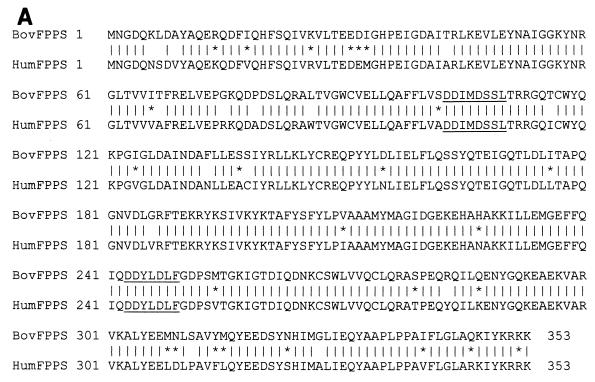
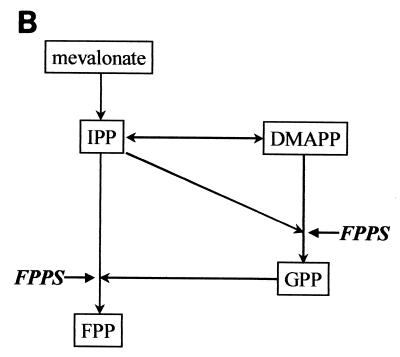
The different transformants were next tested for their ability to specifically activate a β-galactosidase reporter construct that is induced only in the presence of both interacting partners. After completion of these control experiments, we isolated one cDNA encoding a 353-aa polypeptide corresponding to the bovine FPPS protein (Fig. 1A). Bovine FPPS is highly homologous to its counterparts in other species, exhibiting, for example, 88% identity and 95% similarity with human FPPS. Sequence analysis revealed that bovine FPPS belongs to the large family of polyprenyltransferases characterized by two aspartate-rich conserved motifs that are essential for catalytic activity (21, 24, 30, 32, 35, 46, 48). FPPS catalyzes the sequential 1′-4 condensation of IPP (isopentenyl diphosphate) with its isomeric form, dimethylallyl pyrophosphate (DMAPP), to produce a 10-carbon compound, geranylpyrophosphate (GPP). FPPS further adds one molecule of IPP to GPP in order to synthesize 15-carbon FPP (Fig. 1B). FPP is required for the first committed steps in the biosynthesis of cholesterol, farnesylated or geranylated proteins, ubiquinones, dolichols, and heme a. FPPS is, thus, an enzyme that harbors a very specific catalytic activity, generating a major substrate (FPP) involved in a multitude of essential biological pathways.
FIG. 1.
(A) Amino acid sequence of the bovine FPPS and comparison with its human counterpart. The human FPP sequence is derived from GenBank accession number P14324. Vertical bars denote identical residues between both proteins; asterisks show conservatively substituted amino acids; catalytic sites essential for activity are underlined. (B) FPPS catalyzes the sequential 1′-4 condensation of two molecules of IPP (5-carbon) with both DMAPP and the resultant compound, 10-carbon GPP, to produce 15-carbon FPP.
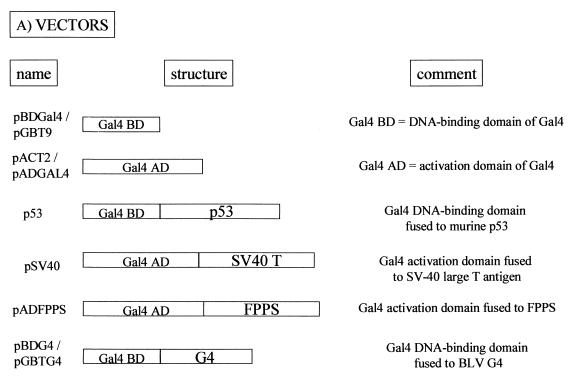
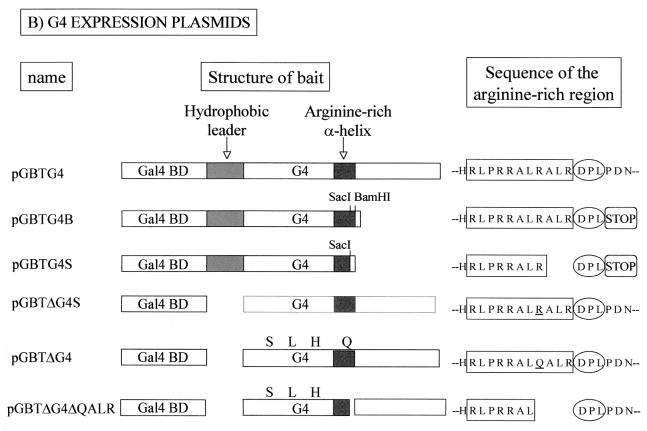
G4 interacts with FPPS via an arginine-rich α-helix.
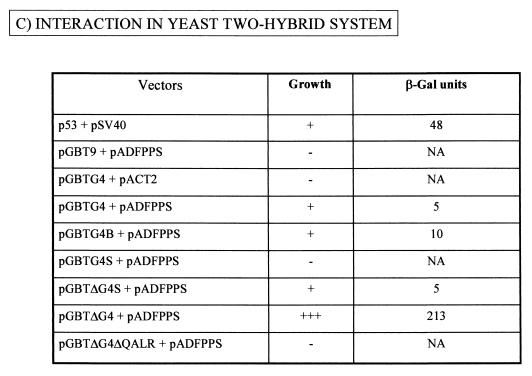
To further characterize the G4-FPPS complex in yeast, we next aimed to identify the residues of G4 required for binding. To this end, we used a series of vectors schematically represented and described in Fig. 2A. For ease of subcloning, the G4 gene was first transferred from pBDG4 (Stratagene) to a very similar vector pGBT9 (Clontech), generating pGBTG4. A series of G4 mutants were derived from this plasmid (Fig. 2B) and cotransformed into yeast strain PJ696 with pADFPPS, which was isolated from the two-hybrid screen. The interactions between FPPS and the G4 mutants were evaluated in terms of cell viability and β-galactosidase enzymatic activity under Leu-, Trp-, His-deficient conditions with 0.25 mM 3-AT selection (Fig. 2C). As expected, yeast transformed with p53 and pSV40 were able to form colonies and expressed high levels of β-galactosidase (48 U). In contrast, yeast containing pGBT9 plus pADFPPS or pGBTG4 plus pACT2 failed to grow, demonstrating that the expression of a single hybrid in the absence of its binding partner was insufficient to support growth. Background enzymatic activity was measured in yeast transformed with pGBTG4 alone and grown on Trp-deficient medium (about 2.5 U [data not shown]). Although this last control was not performed under strictly the same culture conditions (Trp-deficient instead of Leu-, Trp-, and His-deficient conditions for selection), the results indicate that G4, when bound to DNA, can generate weak basal transcriptional activation in this system. Cotransformation of yeast with pGBTG4 and pADFPPS permitted colony outgrowth and yielded a low, but specific and reproducible, β-galactosidase activity (5 U). A twofold increase (10 U) was obtained when the carboxy-terminal end of G4 was deleted (plasmid pGBTG4B). Further deletion of only three residues (ALR) into the central arginine-rich α-helix of G4 abrogated cell growth (pGBTG4S).
FIG. 2.
G4 interacts with FPPS via an arginine-rich α-helix. (A) Plasmids pBDGal4/pGBT9 and pACT2/pADGAL4 used in the two-hybrid system contain, respectively, the Gal4 DNA-binding domain (Gal4 BD) or activation domain (Gal4 AD). As positive controls for interaction, two vectors, p53 (harboring Gal4 BD fused to murine p53) and pSV40 (containing Gal4 AD cloned upstream of SV-40 large T antigen), were cotransfected. Plasmid pADFPPS is a pADGAL4-derivative construct isolated from a two-hybrid screen in which the fpps gene was inserted downstream of the Gal4 activation domain. In the two-hybrid screen, the G4 bait was expressed as a fusion with the Gal4 DNA-binding domain via two closely related vectors, pBDG4 and pGBTG4. (B) Schematic representation of pGBT9-derived constructs, in which the g4 gene (pGBTG4) or its derivatives or variants (pGBTG4B, pGBTG4S, pGBTΔG4S, pGBTΔG4, and pGBTΔG4ΔQALR) were cloned. pGBTG4B and pGBTG4S are 3′-terminal deletants and contain G4 sequences upstream of the BamHI or the SacI restriction sites. Plasmid pGBTΔG4S lacks the hydrophobic leader sequence translated from the first exon of the G4 mRNA and is thought to correspond to the mature form of the protein (ΔG4), generated after proteolytic cleavage. pGBTΔG4, which contains a naturally occurring variant of G4 (from strain 344), is identical to pGBTΔG4S except for four mutations (F35S, P44L, L52H, R67Q). Deletion of four residues (QALR) within the arginine-rich α-helix of ΔG4 yields pGBTΔG4ΔQALR. (C) To identify the G4 domain required for FPPS binding, PJ696 yeast cells were cotransformed with pADFPPS and G4 expression plasmids (pGBTG4, pGBTG4B, pGBTG4S, pGBTΔG4S, pGBTΔG4, and pGBTΔG4ΔQALR). As negative controls for specificity, yeast cells were transformed with pGBT9 and pADFPPS or with pGBTG4 and pACT2 constructs, whereas a positive reaction was obtained by using p53 and pSV40. Four days posttransfection, functional interactions were first evaluated by cell colony formation in selective medium without tryptophan, leucine, or histidine and in the presence of 0.25 mM 3-AT. −, dead; +, viable; +++, already grown at 48 h. Strength of the interaction was next assessed by titration of β-galactosidase activity (results are the average of two representative experiments using six independent transformants). NA, not applicable because of lack of growth.
At its amino terminus, the G4 protein contains a hydrophobic sequence translated from the first exon and separated from the rest of the polypeptide by theoretical proteolytic cleavage sites (2). To analyze the role of this putative leader, we designed a construct called pGBTΔG4S in which the sequences derived only from the second exon of the G4 mRNA were inserted downstream of the Gal4 DNA-binding domain. The levels of β-galactosidase obtained with this vector were similar to those induced by the parental pGBTG4 (5 U), indicating that the leader peptide is not required for binding to FPPS. It thus appears that neither the amino- nor the carboxy-terminal ends are required for G4-FPPS interaction but that the arginine-rich α-helix plays a key role in complex formation. Finally, although the cell growth data were quite convincing, we were concerned about the low levels of β-galactosidase activities obtained with the G4 variant originally isolated by Alexandersen et al. (2). Therefore, we analyzed another variant of G4 (plasmid pGBTΔG4) that was derived from an infectious and pathogenic provirus, BLV strain 344 (52). Both G4 sequences differ by only four amino acids, one of them (R/Q) being located in the arginine-rich α-helix. As expected, cotransformation of yeast with pGBTΔG4 and pADFPPS allowed colony formation and subsequent transcriptional activity, but the extent of the reaction was surprisingly high. Indeed, colonies were fully developed at 2 days instead of 4, and induction of β-galactosidase activity was very high (213 U), exceeding the values obtained with the positive control (p53 plus pSV40) (Fig. 2C). In addition, these yeast cells were viable even under very stringent selection conditions (2 mM 3-AT) (data not shown). As the best-defined structure within G4 is its central arginine-rich motif, we decided to introduce a discrete deletion of only four residues within that region (pGBTΔG4ΔQALR) and, indeed, the G4-FPPS interaction was lost, underlining the key role of this particular α-helix.
Altogether, these data confirm the specificity of the G4-FPPS interaction in yeast, reveal the importance of strain-specific variations, and define the arginine-rich α-helix as an essential region required for binding.
G4 interacts with FPPS in vitro.
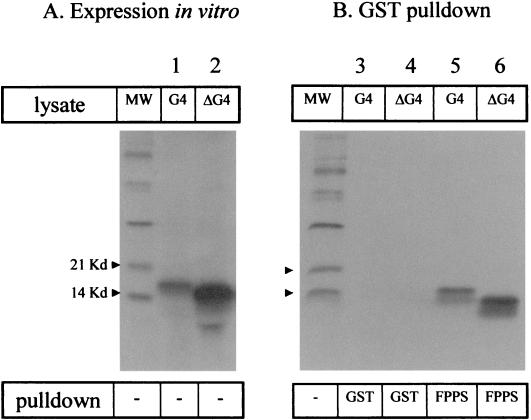
To further verify the specificity of the G4-FPPS binding, we next analyzed the ability of both proteins to physically interact, using GST pull-down experiments. To this end, FPPS was expressed as a fusion protein with GST by means of the pGex-2T prokaryotic expression vector. On the other hand, G4 and its processed form, ΔG4, were expressed via transcription-translation in rabbit reticulocyte lysates using plasmids pHisG4 and pHisΔG4, respectively. The G4 and ΔG4 proteins were synthesized in the presence of 35S-labeled methionine and cysteine and resolved by gel electrophoresis (Fig. 3A, lanes 1 and 2). With the polypeptides being correctly expressed in vitro, the lysates were mixed with purified GST-FPPS fusion protein bound to glutathione-Sepharose beads. As a control for specificity, samples were incubated in parallel with the same amount of GST protein. After extensive washes in high-stringency buffer containing 200 mM NaCl, the proteins were electrophoresed and revealed by autoradiography (Fig. 3B). Under these conditions, both G4 and ΔG4 proteins were pulled down by GST-FPPS (lanes 5 and 6) but not by GST alone (lanes 3 and 4). We conclude that recombinant purified FPPS protein specifically interacts in vitro with both isoforms of G4 under high stringency conditions.
FIG. 3.
In vitro binding of G4 to FPPS in GST pull-down assays. (A) G4 and ΔG4 proteins were synthesized in vitro by using rabbit reticulocyte lysate in the presence of 35S-labeled methionine and cysteine. Aliquots of the lysates were migrated onto an SDS-20% PAGE gel, soaked in amplifying solution, dried, and exposed to X-ray film. MW, molecular weight markers. (B) Five microliters of G4 or ΔG4 protein lysates was mixed with purified GST-FPPS fusion protein bound to glutathione-Sepharose beads or, as a control, with the same amount of GST. After extensive washes, the bound proteins were electrophoresed and revealed by autoradiography. A representative experiment out of three performed is shown.
FPPS redistributes G4 within the cytoplasm.
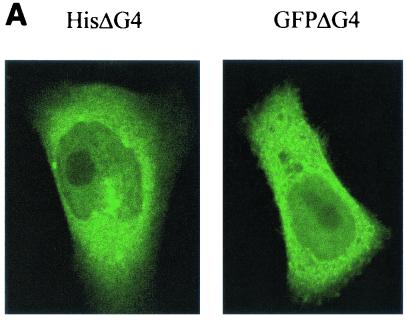
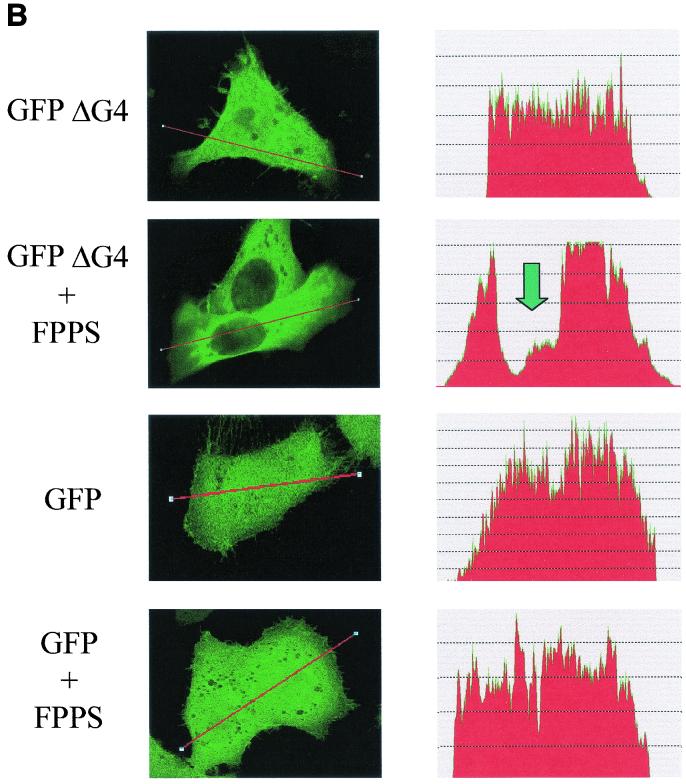
The physical binding between G4 and FPPS, as revealed by GST pull-down experiments, does not imply that both proteins interact in living cells. To test this possibility, the cleaved ΔG4 isoform was expressed as a fusion polypeptide together with Aequorea victoria GFP (pEGFPΔG4). As a preliminary control, the subcellular localization of the ΔG4-GFP hybrid was compared with that of the histidine-tagged ΔG4 protein. Therefore, HelaTat cells were transfected with either pEGFPΔG4 or pHisΔG4, a vector that also allows the addition of an amino-terminal polyhistidine tag and, thereby, examination by fluorescence microscopy. Subsequent labeling of the pHisΔG4-transfected cells with an antipolyhistidine antibody revealed the same pattern of protein distribution as for pEGFPΔG4-HelaTat (Fig. 4A). Quantification of the mean fluorescence indicated that ΔG4 was equally distributed in the different cell compartments (Fig. 4B, GFPΔG4). Surprisingly, coexpression with FPPS induced a drastic reduction of ΔG4 in the nucleus that was associated with a disappearance of the highly fluorescent bodies in the cytoplasm (Fig. 4B, GFPΔG4+FPPS). In contrast, GFP alone is not susceptible to this type of protein redistribution (Fig. 4B, compare GFP with GFP+FPPS). Of note, this phenotype was independent of GFP, since relocalization also occurred with histidine- or FLAG-tagged ΔG4 (data not shown). We conclude that coexpression of FPPS redistributes ΔG4 out of the nucleus, providing good evidence for functional interaction within living cells.
FIG. 4.
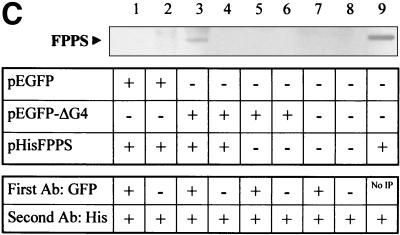
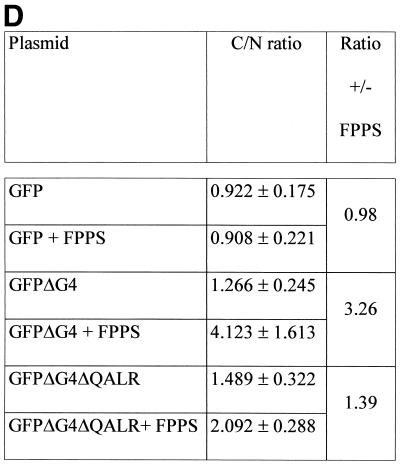
(A) Subcellular localization of G4. HelaTat cells were transfected with either pEGFPΔG4 (GFPΔG4) or pHisΔG4 (HisΔG4) expression vectors. Thirty hours posttransfection, GFPΔG4 cells were fixed and examined under a fluorescence microscope, while cells expressing HisΔG4 were permeabilized, stained with anti-His antibody, and revealed with FITC-coupled anti-mouse immunoglobulin conjugates. (B) FPPS induces redistribution of G4 in the cytoplasm. HelaTat cells were transfected with pEGFPΔG4 (GFPΔG4), pEGFP-C1 (GFP), and pHisFPPS (FPPS) expression vectors in a 1:3 ratio. Thirty hours posttransfection, cells were fixed and examined under a fluorescence microscope (left panels). The intensity of fluorescence along the line was quantified and is represented in a profile plot (right panels). The arrow corresponds to the nucleus compartment. (C) ΔG4 coimmunoprecipitates with FPPS in HelaTat cell lysates. HelaTat cells were transfected with pEGFP-C1, pEGFPΔG4, and pHisFPPS as indicated. The amount of transfected DNA was held constant by adding control vectors (pcDNA3.1/HisC in lanes 5 and 6 and pSG5 in lanes 7, 8, and 9). Thirty hours posttransfection, the cells were recovered and lysed under mild conditions. A first immunoprecipitation was performed using rabbit anti-GFP (+) or with a control antiserum (anti-Rex) (−). After extensive washes, the immunoprecipitates were migrated onto an SDS-PAGE gel, blotted onto a nylon membrane, and revealed with an antihistidine antibody (His). In lane 9, the immunoprecipitation step was omitted (No IP) as a control for FPPS recognition. One representative experiment out of three is presented. Equal amounts of proteins (EGFP, EGFPΔG4, and His-FPPS) were present in the lysates before the coimmunoprecipitation experiment, as demonstrated by Western blotting (data not shown). (D) Subcellular redistribution of G4 induced by FPPS. HelaTat cells were transfected with pEGFP-C1 (GFP), pEGFPΔG4 (GFPΔG4), pEGFPΔG4ΔQALR (GFPΔG4ΔQALR), and pHisFPPS (FPPS) expression vectors. Thirty hours posttransfection, the mean fluorescence intensities were determined from seven representative cells using the Leica quantification program. The C/N ratio is shown. A second ratio (± FPPS) was calculated from values obtained in the presence and in the absence of FPPS.
To further characterize this phenotype, we aimed to determine whether both proteins were able to form a complex within mammalian cells. The methodology was based on immunoprecipitation of the complex using antibodies specific for one protein (ΔG4) and detection with an antiserum recognizing its partner (FPPS). HelaTat cells were transfected with the pEGFPΔG4 and pHisFPPS expression vectors and cultivated for 30 h. Then, immunoprecipitation was performed with rabbit anti-GFP (Fig. 4C, + lanes) or with an irrelevant control antiserum (− lanes). After extensive washes, the immunoprecipitates were analyzed by SDS-PAGE, blotted onto a nylon membrane, and revealed with an antihistidine antibody (His). Under these experimental conditions, a 40-kDa protein appeared on the Western blot (Fig. 4C, lane 3), indicating that coimmunoprecipitation of FPPS did occur. As expected, FPPS was not pulled down when the GFP antibody was omitted (lane 4) or when the ΔG4 sequences were absent (empty pEGFP plasmid, lanes 1 and 2). In addition, coimmunoprecipitation was also strictly dependent on transfection of the FPPS vector (pHisFPPS, lanes 5 to 8) and on the use of the antihistidine antibody (data not shown). Since the coimmunoprecipitated 40-kDa band comigrated with FPPS (lane 9), we conclude that ΔG4 indeed drags its partner along during immunoprecipitation. It thus appears that FPPS and ΔG4 are physically associated within the cell lysates, providing further evidence for functional interactions between these two proteins.
To quantify ΔG4 redistribution upon coexpression of FPPS (as illustrated in Fig. 4B), the mean fluorescence expressed in the nuclear and cytoplasmic compartments was measured by confocal microscopy from seven representative double-positive cells (Fig. 4D). The C/N ratio (corresponding to the mean fluorescence in the cytoplasm versus the nucleus) indicated that the ΔG4 protein was indeed concentrated in the cytosol in the presence of FPPS (Fig. 4D, compare GFPΔG4 with GFPΔG4+FPPS). In terms of induction, these values represent a 3.26-fold increase when FPPS is coexpressed (ratio ± FPPS). No relocalization by FPPS occurred in the absence of G4 sequences (Fig. 4D, compare GFP with GFP+FPPS). Interestingly, a mutant of G4 lacking only four residues required for FPPS binding (GFPΔG4ΔQALR) was only weakly, if at all, translocated to the nucleus (C/N ratio, 1.39). The fact that the subcellular localization of ΔG4 can be modulated by FPPS provides very convincing evidence for the biological relevance of the interactions observed in yeast or in vitro.
G4 is not prenylated and does not alter geranylation of M-ras in rabbit reticulocyte lysates.
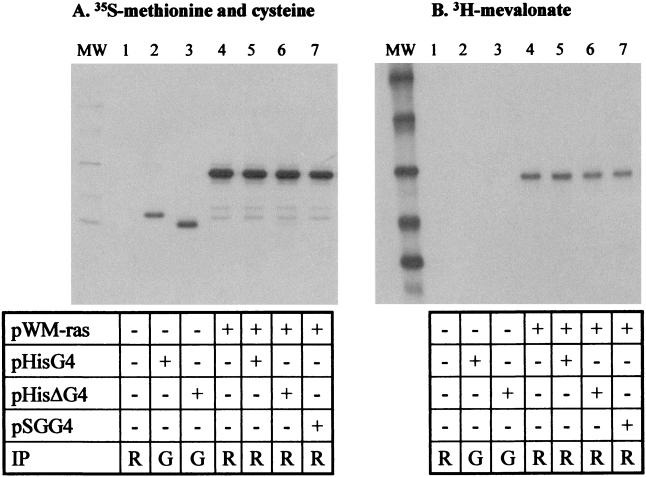
Why does the viral accessory protein G4 target FPPS in cells? To answer this question, we considered two alternate, but not exclusive, possibilities. First, G4 could bind to FPPS because this viral accessory protein is itself modified by farnesylation or geranylation. Alternatively, expression of the viral accessory protein could modify the normal pathways involving prenylation. Both alternatives can in fact be tested in a single in vitro transcription-translation assay based on the incorporation of [3H]mevalonate, the upstream substrate in this cellular pathway (Fig. 1B). Farnesylation occurs on the defined consensus motifs CAAX (where C is a cysteine, A is an aliphatic residue, and X is either a methionine or a serine, whereas geranylation requires a leucine). This type of sequence is present at the carboxy terminus of Ras, a major prenylated protein, but it is absent from G4. Rabbit reticulocyte lysates were programmed with expression vectors coding for G4 (pHisG4 or pHisΔG4) or, as a control protein, M-ras, known to be efficiently prenylated in this system (plasmid pWM-ras). The correct expression of the different proteins was assessed by incorporation of [35S]methionine and [35S]cysteine and immunoprecipitation with their corresponding antisera (Fig. 5A, lanes 2 to 7). As a control for specificity, no product was synthesized in unprogrammed lysates (Fig. 5A, lane 1). In the presence of [3H]mevalonate, only M-ras and not G4 or ΔG4 was metabolically labeled (Fig. 5B, compare lanes 2, 3, and 4). We conclude that, as expected, M-ras is correctly prenylated in vitro, whereas G4 and ΔG4 are not.
FIG. 5.
G4 is not prenylated and does not alter geranylation of M-ras in rabbit reticulocyte lysates. Rabbit reticulocyte lysates (TNT; Promega) were programmed with different plasmids as indicated. In vitro transcription-translation was performed in the presence of [3H]mevalonate to assess prenylation (B), or in the presence of 35S-labeled methionine and cysteine as a control for protein synthesis (A). The G4 and ΔG4 and the M-ras proteins were immunoprecipitated (IP) using anti-six-histidine (G; lanes 2 and 3) or Jal4 (R; lanes 1 and 4 to 7) antibodies. The purified proteins were then migrated onto an SDS-PAGE gel and autoradiographed. MW, molecular weight markers.
We next investigated the possibility that the G4-FPPS interaction might interfere with metabolic pathways leading to prenylation, e.g., by inducing conformational modifications of the enzyme and subsequently modifying the synthesis of FPP or geranyl-GPP (Fig. 1B). A variation in the concentration of the substrate for farnesyl- or geranyl-transferase could then alter prenylation of cellular proteins. To test this possibility, in vitro prenylation of M-ras was performed in the presence of three different types of G4 constructs (Fig. 5, lanes 5, 6, and 7). Neither synthesis of M-ras (panel A) nor its geranylation was affected by the presence of three different G4 expression vectors.
Altogether, these data demonstrate that G4 is not prenylated in vitro and does not alter geranylation of M-ras in rabbit reticulocyte lysates.
A G4 mutant unable to bind FPPS is impaired in its ability to immortalize and transform primary REFs.
G4 harbors oncogenic potential in cell culture due to its ability to transform primary REFs in cooperation with the Ha-ras oncogene (26). Since FPPS provokes a profound alteration of the G4 subcellular localization, it is possible that complex formation between both proteins modifies the outcome of the REF cell transformation assay or, alternatively, that the transforming potential of G4 requires its interaction with this cellular partner. Therefore, we conducted REF cell transformation assays using primary REFs transfected with various combinations of expression vectors for G4 and FPPS. These plasmids were cotransfected with pSV2neoEJ, a construct conferring resistance to neomycin and expressing the Ha-ras oncogene. The goal of this cotransfection with pSV2neoEJ is to fully transform the REF cells, which are then able to induce tumors in nude mice (26). At 48 h posttransfection, cells were harvested by trypsinization and half of them were cultured in the presence of G418 to score drug-resistant colonies as having either normal or transformed morphology. The other collected cells were injected subcutaneously into the flanks of thymusless nude mice. A total of six mice in three independent experiments were injected for each combination of plasmids, and the tumor volume was determined at 1 month postinjection (Table 1). As a positive control, cotransfection of Myc and Ras expression vectors efficiently induced the formation of foci (n = 50) in cell culture and generated large tumors in nude mice (5,000 to 7,000 mm3). The background of the assay was provided by transfection of the empty plasmid pSG5 and pSV2neoEJ (<150 mm3). Coexpression of G4 with Ras induced transformation of REF cells (45 foci and tumors of 500 to 1,100 mm3 in half of the injected mice), confirming our previous results (26). Deletion of the amino-terminal end of G4 (ΔG4) slightly reduced the number of foci (23 instead of 45), but it did not alter tumor formation in nude mice. It thus appears that the putative hydrophobic leader of G4 is dispensable for its oncogenic potential in vitro. In contrast, deletion of only four residues (QALR) located in the arginine-rich α-helix almost completely abrogated transformation of REF cells (1 focus; <150 mm3 tumor volume). Since these residues are also required for FPPS binding, these experiments demonstrate a direct correlation between the oncogenic potential of G4 and its ability to interact with a cellular partner.
TABLE 1.
The G4 α-helix arginine-rich region is required for transformation of primary REFsa
| DNA source | No. of foci (SD) | Tumor vol (mm3) |
|---|---|---|
| Myc | 50 (±7) | 5,000–7,000 |
| pSG5 | 0 (±1) | <150 |
| G4 | 45 (±5) | 500–1,100 (50%)b |
| ΔG4 | 23 (±9) | 500–1,100 (50%)b |
| G4ΔQALR | 1 (±1) | <150 |
Primary REF cells were transfected with plasmids: pSV2NeoEJ (a vector expressing the Ha-ras oncogene), pSVMyc (expressing Myc), the empty pSG5, and G4 constructs (pSGG4, pSGΔG4, and pSGG4ΔQALR). After transfection, cells were cultivated for 2 days, harvested, and either cultured for 1 month under G418 selection to score for foci formation or injected subcutaneously into the flanks of thymusless nude mice (nu/nu NMRI background). A total of six mice in three independent experiments were injected for each plasmid combination. Mean tumor volumes (in cubic millimeters) were calculated at 1 month postinjection.
Cotransfection of G4 and ΔG4 with Ha-ras yielded tumors in 50% of the injected mice.
Finally, FPPS itself also displays transformation potential in REF cells (7 foci; tumors of 500 to 2,800 mm3), and coexpression of FPPS with different G4 constructs induces intermediate phenotypes in nude mice (data not shown). Since FPPS and Ras already cooperate during REF cell transformation, we were unable to draw a clear conclusion concerning a possible additional role of G4, and interpretation of these data requires further experimentation.
From these transformation assays, however, we did confirm that G4 harbors oncogenic potential in primary cell cultures and in nude mice. Furthermore, we proved that the processed form ΔG4 is also able to transform REF cells in cooperation with Ras. And most importantly, it appeared that the integrity of the arginine-rich α-helix of G4, shown to be involved in FPPS interaction (Fig. 2B), is essential to confer its oncogenicity.
BLV G4 colocalizes with HTLV-1 p13II in mitochondria, and both proteins interact with FPPS.
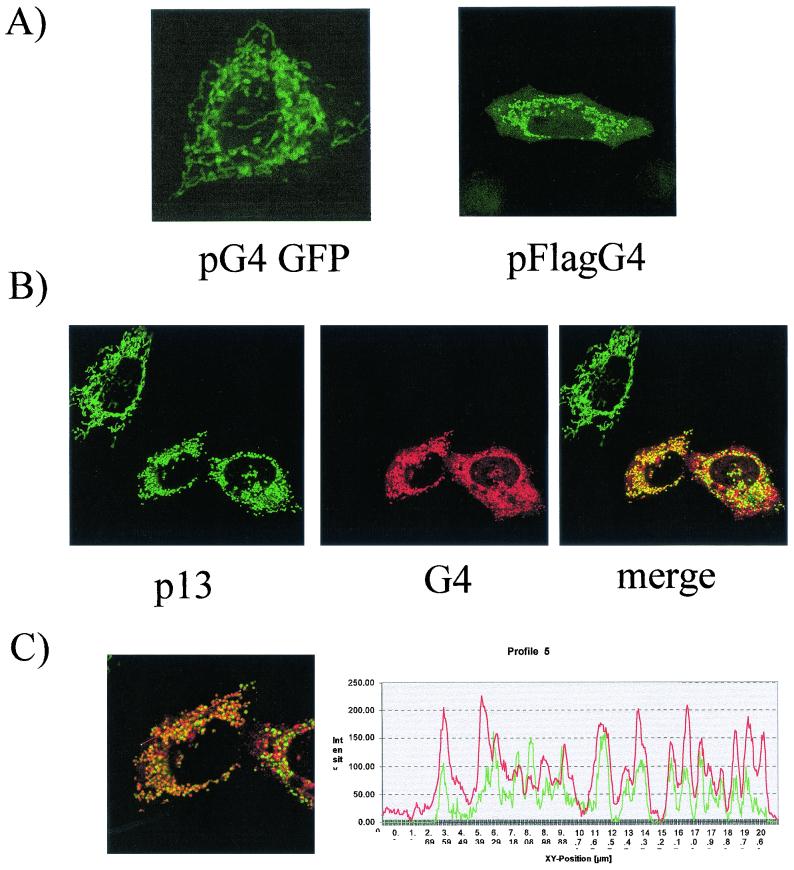
The ΔG4 processed form, which lacks the amino-terminal hydrophobic leader, is localized in the nucleus as well as in the cytoplasm (Fig. 4B). This subcellular localization is consistent with the presence of a theoretical nuclear localization signal (2). Careful sequence analysis also suggests that G4 contains mitochondria-specific motifs within its amino-terminal leader or within the arginine-rich α-helix (L. Lefebvre, unpublished data). To assess the subcellular localization of the uncleaved G4 protein, the complete open reading frame was inserted upstream and in frame with the GFP sequence (vector pG4GFP). The G4 gene was also fused to the Flag epitope, yielding plasmid pFlagG4. Both constructs were transfected into HelaTat cells and examined by confocal microscopy, either directly (for pG4GFP) or after labeling with the anti-Flag antibody (pFlagG4). The subcellular localization of G4 was exclusively cytoplasmic and was characterized by reticulated structures, independently of the type of vector used (Fig. 6A). Since a similar pattern has been described for the p13II protein of HTLV-1 (11), we next aimed to investigate the ability of both accessory factors to colocalize within cells. To this end, HelaTat cells were cotransfected with two plasmids, pFlagG4 and p13II-GFP, and labeled with anti-Flag antibody in association with a secondary Texas red-coupled conjugate. As illustrated in Fig. 6B, the fluorescence patterns of p13II (green) and G4 (red) perfectly coincided in the double-stained cells (yellow) as a result of overlapping fluorochromes (merge). Furthermore, quantification of the induced fluorescence intensities exhibited a nearly perfect parallelism (Fig. 6C). Together, these data demonstrate that p13II and G4 colocalize in the same subcellular compartment. Since HTLV-1 p13II is a mitochondrial protein (11), we conclude that G4 is also targeted to this organelle. Of note, G4 mitochondrial targeting was confirmed by staining with Mitotracker (Molecular Probes) (data not shown). Interestingly, both p13II and G4 accessory factors appear to modify the morphology of the mitochondria, suggesting a functional parallelism between these viral proteins.
FIG. 6.
BLV G4 protein colocalizes with HTLV-1 p13II. (A) HelaTat cells were transfected with vectors pG4GFP or pFlagG4. Thirty hours posttransfection, cells were fixed, permeabilized, and stained with the anti-Flag monoclonal antibody and anti-mouse immunoglobulin-FITC conjugate. (B) Cells were transfected with p13II-GFP and pFlagG4 and labeled as described for panel A except for the secondary antibody, which was Texas red fluorochrome. Cells were analyzed by confocal microscopy with GFP for p13II and Texas red for G4; the third panel shows an overlay with the merged images. (C) Quantification (right panel) of the fluorescence intensities of both fluorochromes was assessed along the line (left panel).
Based on its homologies with G4, we next addressed the question of the ability of p13II to interact with FPPS. To this end, we fused p13II to the DNA-binding domain of the Gal4 transactivator (plasmid pGBTP13). As a control, p13II by itself was unable to induce transactivation of reporter genes when cotransformed with the neutral pACT2 construct (no growth in selective medium) (Table 2). In contrast, yeast cotransformed with plasmids pGBTP13 and pADFPPS could easily be rescued from histidine selection, the strength of the interaction being six times above background levels (18 versus 3 U of CPRG). Compared to G4, these values are in the same order of magnitude (pGBTG4+pADFPPS; 7 U of CPRG). We conclude that, like G4, p13II interacts with FPPS in yeast.
TABLE 2.
FPPS interacts with HTLV-1 p13IIa
| Vectors | Growth | U of CPRG |
|---|---|---|
| pGBTP13 + pADFPPS | + | 18 |
| pGBTP13 + pACT2 | − | 3b |
| pGBTG4 + pADFPPS | + | 7 |
| pGBTG4 + pACT2 | − | 4b |
| pGBTΔG4 + pADFPPS | +++ | 668 |
| p53 + pSV40 | + | 50 |
PJ696 yeast cells were cotransformed with bait vector (pGBTP13, pGBTG4, or pGBTΔG4) and pADFPPS or control pACT2. As a positive control, yeast cells were transformed with p53 and pSV40. To assess growth ability, selection was performed in minimal medium lacking tryptophan, leucine, and histidine (0.25mM 3-AT), and the strength of the interaction (−, +, or +++) was evaluated by titration of CPRG. Mean values of one representative experiment out of three are shown.
Yeast cells were grown in medium deprived only of tryptophan, allowing CPRG assay.
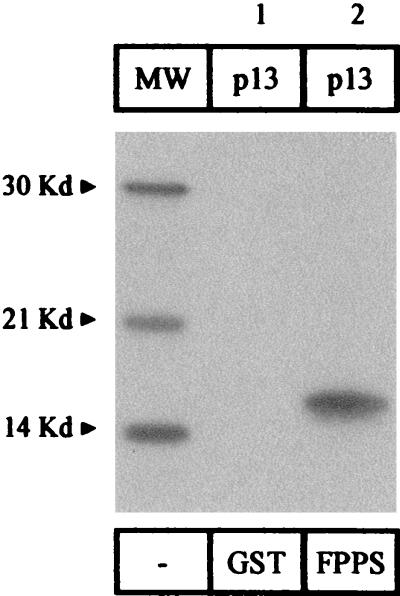
To further verify the specificity of this interaction, we performed GST pull-down experiments using in vitro-translated p13II protein. Under high-stringency conditions, 35S-labeled p13II specifically bound to GST-FPPS fusion protein (Fig. 7, lane 2) but not to GST alone (lane 1). We conclude that recombinant purified FPPS protein specifically interacts in vitro with HTLV-1 p13II. Together, our data demonstrate that, like G4, p13II interacts with FPPS, further supporting the structural and functional homologies between these two oncoviral proteins.
FIG. 7.
HTLV-1 p13II interacts in vitro with FPPS. p13II was synthesized in rabbit reticulocyte lysate in the presence of 35S-labeled methionine and cysteine and mixed with purified GST-FPPS fusion protein bound to glutathione-Sepharose beads or, as a control, with the same amount of GST. After extensive washes, the bound proteins were electrophoresed and revealed by autoradiography. MW, molecular weight markers.
DISCUSSION
Being dispensable for viral infectious potential, BLV g4 might be considered a facultative gene (52). However, deletion of g4 from the provirus profoundly alters its growth properties and restricts pathogenicity in sheep, a host highly susceptible to BLV. In vitro, G4 exhibits oncogenic potential in primary rat cell cultures, a function shared with the transcriptional activator Tax and with the myc oncogene (51). G4 thus appears to play a key role in viral pathogenesis, and the goal of this report is to cast light onto its modes of action. We have shown here that G4 specifically interacts with FPPS, an enzyme involved in the pathway leading to prenylation of a variety of proteins including nuclear lamins, Ras and a multitude of GTP-binding proteins (G proteins), several protein kinases, and phosphatases.
To screen for G4 partners, we utilized a cDNA library which was synthesized from a bovine B-lymphocytic cell line (BL3) derived from a cow with sporadic leukemia. The use of these cells thus ensured both species and expression specificities to the FPPS clone. Our initial screening trials under stringent conditions (2 mM 3-AT) did not allow selection of any candidate protein. Therefore, a milder selection procedure (0.25 mM 3-AT) was applied, and it allowed us to specifically isolate FPPS as a binding partner for G4. Interaction of G4 with FPPS does not require the amino-terminal hydrophobic leader, which is absent from the putative mature form of G4 (called ΔG4). In contrast, the central hydrophilic region, which is structurally organized as an α-helix (HRLPRRAL-R/Q-ALR at positions 59 to 70 of the G4 protein), is essential for binding. Of particular interest, we have determined that another variant of ΔG4 containing, among other substitutions, a glutamine at position 67 instead of an arginine, is a far better ligand for FPPS. Basically, this observation enlightens the importance of using natural and pathogenic variants (i.e., strain 344) during in vitro investigations. Although the sequences within FPPS required for binding are unknown, we can hypothesize that the α-helix of G4 interacts via the arginine residues with the two aspartate-rich conserved motifs that are essential for catalytic activity of the enzyme. In fact, among a series of FPPS mutants in which large deletions were introduced, none of them remained competent for G4 binding (our unpublished observations). This is not so surprising, since FPPS is a highly structured enzyme composed of 10 α-helices surrounding a central cavity in which are buried the catalytic sites (48). Experiments based on site-directed mutations of FPPS, as well as competition assays with synthetic peptides corresponding to the binding sites, are currently under investigation. Conversely, the residues within G4 required for FPPS binding were determined by means of a series of limited mutations and have been localized in a region surrounding amino acids Q67A68L69R70. Interestingly, these codons overlap exactly a theoretical mitochondrial cleavage site at positions 68 and 69 (L. Lefèbvre et al., unpublished results). Localization of the G4 interaction site has been further confirmed by means of specific peptides. Our preliminary assays indicate that FPPS activity can be inhibited by up to 30% in the presence of the oligopeptide 58RHRLPRRALQALRDPLPDNDK78 overlapping the G4 binding motif (data not shown).
Convincing evidence for a functional interaction between G4 and FPPS was also obtained in mammalian cells, with the redistribution of the truncated polypeptide within a different cellular compartment from the nucleus into the cytoplasm being one of the key experiments of this report. Of note, this phenotype is slightly dependent on quantities of the different expression vectors (optimal GFP-ΔG4 to FPPS ratio of 1:3) and is cell-type-specific (HelaTat but not OVK, for example). However, we think that FPPS-induced ΔG4 relocalization is specific and relevant, as (i) both proteins partially overlap, as attested by double-staining experiments, and (ii) different expression vectors for FPPS (pSGFPPS, coding for the untagged protein, pFlagFPPS, and pHisFPPS) or ΔG4 (fused to GFP, FLAG, and His) induce the same phenotype (Fig. 4A and data not shown). It thus appears that the processed form of G4 is localized in the cytoplasm as well as in the nucleus. The redistribution phenotype associated with the coexpression of FPPS suggests that ΔG4 could in fact shuttle between different cell compartments.
Subcellular localization of the complete G4 protein (105 aa) significantly differed from the distribution of the processed ΔG4 form. When fused to GFP, G4 indeed perfectly colocalized with a mitochondrial protein, HTLV-1 p13II (Fig. 6). It should be mentioned, however, that when the GFP protein was attached at the amino terminus of G4, the hybrid polypeptide was essentially localized into the nucleus. Differential localization depending on the tagging strategy, either in the nucleus or in the mitochondria, was also reported for HTLV p13II (11, 14, 28). Definitive answers to these discrepancies will require the recognition of the native G4 and p13II proteins in freshly isolated lymphocytes.
Since p13II and G4 do not share primary sequence homologies (below 20% identity), colocalization of these proteins into the same cellular compartment is somehow an unexpected result. The similarities between both accessory proteins are further reinforced by their ability to profoundly alter the morphology of the mitochondria (11; also Fig. 6). Expression of G4 indeed results in specific alterations of mitochondrial shape (Fig. 6A, pFlagG4) and in the disruption of the typical interconnected network, as described for p13II. These phenotypes further suggested functional homologies shared by both accessory proteins and indeed, we have demonstrated here that p13II, like G4, interact with FPPS in vitro.
Concerning the functional consequences of the G4-FPPS interaction, our assay based on the rabbit reticulocyte lysate system demonstrated that G4 is not prenylated. In this system, G4 did not alter geranylgeranylation of the ras proto-oncogene, a major downstream target of FPPS. However, FPPS catalyzes the synthesis of FPP and does not directly anchor this substrate to target proteins. If the levels of precursors are not restricted within rabbit reticulocyte lysates (mevalonate, IPP, or DMAPP, for example), the effect of G4 will not be revealed. To circumvent this type of objection, a more specific in vitro assay based on the synthesis of prenyl diphosphates from the IPP substrate was performed (as described in reference 35). Although this assay confirmed the functionality of our FPPS clone, we did not observe an effect of recombinant G4 on the activity of FPPS (data not shown). However, as stated before, G4 is an unstable protein that is very difficult to express at high levels in bacteria, and it is presently not possible to address its influence on FPPS activity, perhaps because of technical limitations. Besides these practical restrictions, there is probably another straightforward explanation for the failure of G4 to modulate the prenylation metabolic pathway in vitro. It has indeed been reported that the limiting enzyme in isoprenoid biosynthesis is 3-hydroxy-3-methylglutaryl coenzyme A reductase, which catalyzes mevalonate production (33, 37). For example, inhibition of the mevalonate pathway was not associated with the appearance of nonprenylated Ras isoforms in rat AR 4-2J cells, indicating that the levels of isoprenoid precursors were indeed not limiting steps in this system (31). In support of this hypothesis, coexpression of FPPS alone did not increase Ras prenylation in our in vitro reticulocyte-based assay, indicating that indeed this enzyme was not rate limiting (data not shown).
Demonstration of the functional importance of the G4-FPPS interaction was, however, obtained by means of a primary cell immortalization assay. One of the most interesting issues of this work indeed concerns the biological relevance of the G4-FPPS interaction during cellular transformation. We have demonstrated here that a very subtle mutation within the arginine-rich α-helix of G4 abrogates primary cell immortalization (Table 1). It thus appears that G4 oncogenic potential correlates with its ability to interact with FPPS. Since addition of either a farnesyl or geranylgeranyl isoprenoid group to the carboxy terminus of Ras or other G proteins is strictly required for insertion into the plasma membrane and biological function (recently reviewed in references 4, 19, and 45), these metabolic pathways could constitute the molecular bases for the cooperation between G4 and Ras during transformation in primary cell culture. Although these results enlighten a key role of the arginine-rich G4 helix, whether this region is required for pathogenesis remains to be determined. To address this question, a straightforward experiment would be to assess the leukemogenic potential of a recombinant provirus harboring a mutation in the central α-helix of G4. Of note, one of our original proviral constructs, pBLVIG4 (52), contains a stop codon just downstream of the α-helix and is not pathogenic in sheep (26, 53), indicating an essential role for the carboxy terminus of G4 during this process.
The interactions of oncoviral p13II and G4 accessory proteins with FPPS open new prospects for therapeutic treatment. In fact, all three proteins have been localized, at least in part, in the mitochondrial compartment of the cell (references 11 and 42 and this report). Mitochondria play a vital role in the cell, providing most of the energy and participating in Ca2+, redox, and pH homeostases (17). In addition, this organelle also controls the life-or-death decision by releasing cytochrome C into the cytosol, thereby activating caspases. The interplay between p13II or G4 and FPPS could potentially modulate all of these metabolic processes and ultimately lead to cellular transformation. Our results (Table 1) have demonstrated that the G4-FPPS binding site is required to achieve primary cell immortalization in vitro and induction of tumors in nude mice. If indeed a similar situation holds true during the onset of leukemia in vivo, disruption of the interaction between G4 and FPPS could interfere with the process of oncogenesis. As a corollary, if prenylation is required for leukemogenesis, specific inhibition of enzymes participating in this pathway (such as FPPS or farnesyl transferase) (25, 41) could possibly be of therapeutic interest in bovine and, more importantly, human leukemias.
Acknowledgments
L.L. (Télévie Fellow), A.V. (Research Associate), R.K. (Research Director), and L.W. (Research Director) are members of the Fonds national de la Recherche scientifique (FNRS). We thank the Belgian Federation against Cancer, the Fortis Bank Assurance, the FNRS, the Service de Programmation pour la Politique scientifique (SSTC P4/30), and the Action de Recherche Concertée du Ministère de la Communauté Française for financial support.
We are grateful to I. Callebaut (Jussieu, Paris, France), F. Dequiedt (University of California, San Francisco), C. Erneux (IRIBHN, Erasme, Brussels, Belgium), and R. Poirey (DKFZ, Heidelberg, Germany) for helpful discussions. The Jal4 antibody and Mras2 vector were kindly provided by J. Louahed and J. C. Renauld (Ludwig Institute, UCL, Brussels, Belgium). We thank C. Dillen and T. Peremans for excellent technical help.
REFERENCES
- 1.Albrecht, B., N. D. Collins, M. T. Burniston, J. W. Nisbet, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Human T-lymphotropic virus type 1 open reading frame I p12I is required for efficient viral infectivity in primary lymphocytes. J. Virol. 74:9828–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandersen, S., S. Carpenter, J. Christensen, T. Storgaard, B. Viuff, Y. Wannemuehler, J. Belousov, and J. A. Roth. 1993. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J. Virol. 67:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartoe, J., B. Albrecht, N. Collins, M. Robek, L. Ratner, P. Green, and M. Lairmore. 2000. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 74:1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaupre, D. M., and R. Kurzrock. 1999. Ras and leukemia: from basic mechanisms to gene-directed therapy. J. Clin. Oncol. 17:1071–1079. [DOI] [PubMed] [Google Scholar]
- 5.Berneman, Z. N., R. B. Gartenhaus, M. S. Reitz, Jr., W. A. Blattner, A. Manns, B. Hanchard, O. Ikehara, R. C. Gallo, and M. E. Klotman. 1992. Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc. Natl. Acad. Sci. USA 89:3005–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereseto, A., Z. Berneman, I. Koralnik, J. Vaughn, G. Franchini, and M. E. Klotman. 1997. Differential expression of alternatively spliced pX mRNAs in HTLV-1-infected cell lines. Leukemia 11:866–870. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. M., S. H. Chen, C. Y. Fu, J. Y. Chen, and M. Osame. 1997. Antibody reactivities to tumor-suppressor protein p53 and HTLV-1 Tof, Rex and Tax in HTLV-1-infected people with differing clinical status. Int. J. Cancer 71:196–202. [DOI] [PubMed] [Google Scholar]
- 8.Ciminale, V., G. N. Pavlakis, D. Derse, C. P. Cunningham, and B. K. Felber. 1992. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J. Virol. 66:1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciminale, V., D. M. D’Agostino, L. Zotti, G. Franchini, B. K. Felber, and L. Chieco-Bianchi. 1995. Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphotropic virus type II. Virology 209:445–456. [DOI] [PubMed] [Google Scholar]
- 10.Ciminale, V., D. M. D’Agostino, L. Zotti, and L. Chieco-Bianchi. 1996. Coding potential of the X region of human T-cell leukemia/lymphotropic virus type II. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:S220–S227. [DOI] [PubMed] [Google Scholar]
- 11.Ciminale, V., L. Zotti, D. M. D’Agostino, T. Ferro, L. Casareto, G. Franchini, P. Bernardi, and L. Chieco-Bianchi. 1999. Mitochondrial targeting of the p13II protein coded by the x-II ORF of human T-cell leukemia/lymphotropic virus type I (HTLV-1). Oncogene 18:4505–4514. [DOI] [PubMed] [Google Scholar]
- 12.Collins, N. D., G. C. Newbound, B. Albrecht, J. L. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 91:4701–4707. [PubMed] [Google Scholar]
- 13.Collins, N. D., C. D’Souza, B. Albrecht, M. D. Robek, L. Ratner, W. Ding, P. L. Green, and M. D. Lairmore. 1999. Proliferation response to interleukin-2 and Jak/Stat activation of T cells immortalized by human T-cell lymphotropic virus type 1 is independent of open reading frame I expression. J. Virol. 73:9642–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Agostino, D. M., V. Ciminale, L. Zotti, A. Rosato, and L. Chieco-Bianchi. 1997. The human T-cell lymphotropic virus type 1 Tof protein contains a bipartite nuclear localization signal that is able to functionally replace the amino-terminal domain of Rex. J. Virol. 71:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekaban, G. A., A. A. Peters, J. C. Mulloy, J. M. Johnson, R. Trovato, E. Rivadeneira, and G. Franchini. 2000. The HTLV-1 OrfI protein is recognized by serum antibodies from naturally infected humans and experimentally infected rabbits. Virology 274:86–93. [DOI] [PubMed] [Google Scholar]
- 16.Derse, D., J. Mikovits, and F. Ruscetti. 1997. X-I and X-II open reading frames of HTLV-1 are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237:123–128. [DOI] [PubMed] [Google Scholar]
- 17.Desagher, S., and J. C. Martinou. 2000. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10:369–377. [DOI] [PubMed] [Google Scholar]
- 18.Franchini, G., J. C. Mulloy, I. J. Koralnik, A. Lo Monico, J. J. Sparkowski, T. Andresson, D. J. Goldstein, and R. Schlegel. 1993. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J. Virol. 67:7701–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, H. W., and P. J. Casey. 1999. Enzymology and biology of CaaX protein prenylation. Recent Prog. Horm. Res. 54:315–343. [PubMed] [Google Scholar]
- 20.Green, P. L., T. M. Ross, I. S. Chen, and S. Pettiford. 1995. Human T-cell leukemia virus type II nucleotide sequences between env and the last exon of tax/rex are not required for viral replication or cellular transformation. J. Virol. 69:387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, S. D., R. S. Mehan, T. R. Tansey, H. T. Chen, G. Goping, I. Goldberg, and I. Shechter. 1999. Differential binding of proteins to peroxisomes in rat hepatoma cells: unique association of enzymes involved in isoprenoid metabolism. J. Lipid Res. 40:1572–1584. [PubMed] [Google Scholar]
- 22.Hou, X., S. Foley, M. Cueto, and M. A. Robinson. 2000. The human T-cell leukemia virus type I (HTLV-1) X region encoded protein p13II interacts with cellular proteins. Virology 277:127–135. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. M., J. C. Mulloy, V. Ciminale, J. Fullen, C. Nicot, and G. Franchini. 2000. The MHC class I heavy chain is a common target of the small proteins encoded by the 3′ end of HTLV type 1 and HTLV type 2. AIDS Res. Hum. Retrovir. 16:1777–1781. [DOI] [PubMed] [Google Scholar]
- 24.Joly, A., and P. A. Edwards. 1993. Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J. Biol. Chem. 268:26983–26989. [PubMed] [Google Scholar]
- 25.Keller, R. K., and S. J. Fliesler. 1999. Mechanism of aminobisphosphonate action: characterization of alendronate inhibition of the isoprenoid pathway. Biochem. Biophys. Res. Commun. 266:560–563. [DOI] [PubMed] [Google Scholar]
- 26.Kerkhofs, P., H. Heremans, A. Burny, R. Kettmann, and L. Willems. 1998. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J. Virol. 72:2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koralnik, I. J., A. Gessain, M. E. Klotman, A. Lo Monico, Z. N. Berneman, and G. Franchini. 1992. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I. Proc. Natl. Acad. Sci. USA 89:8813–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koralnik, I. J., J. Fullen, and G. Franchini. 1993. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J. Virol. 67:2360–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koralnik, I. J., J. C. Mulloy, T. Andresson, J. Fullen, and G. Franchini. 1995. Mapping of the intermolecular association of human T cell leukaemia/lymphotropic virus type I p12I and the vacuolar H+-ATPase 16 kDa subunit protein. J. Gen. Virol. 76:1909–1916. [DOI] [PubMed] [Google Scholar]
- 30.Krisans, S. K., J. Ericsson, P. A. Edwards, and G. A. Keller. 1994. Farnesyl-diphosphate synthase is localized in peroxisomes. J. Biol. Chem. 269:14165–14169. [PubMed] [Google Scholar]
- 31.Lambert, M., and N. D. Bui. 1999. Dexamethasone-induced decrease in HMG-CoA reductase and protein-farnesyl transferase activities does not impair ras processing in AR 4-2J cells. Mol. Cell. Biochem. 202:101–108. [DOI] [PubMed] [Google Scholar]
- 32.Marrero, P. F., C. D. Poulter, and P. A. Edwards. 1992. Effects of site-directed mutagenesis of the highly conserved aspartate residues in domain II of farnesyl diphosphate synthase activity. J. Biol. Chem. 267:21873–21878. [PubMed] [Google Scholar]
- 33.Matar, P., V. R. Rosados, E. A. Roggero, and O. G. Scharovsky. 1998. Lovastatin inhibits tumor growth and metastasis development of rat fibrosarcoma. Cancer Biother. Radiopharm. 13:387–393. [DOI] [PubMed] [Google Scholar]
- 34.Mulloy, J. C., R. W. Crownley, J. Fullen, W. J. Leonard, and G. Franchini. 1996. The human T-cell leukemia/lymphotropic virus type 1 p12I proteins bind the interleukin-2 receptor β and γc chains and affects their expression on the cell surface. J. Virol. 70:3599–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohnuma, S.-I., K. Hirooka, C. Ohto, and T. Nishino. 1997. Conversion from archaeal geranylgeranyl diphosphate synthase to farnesyl diphosphate synthase. J. Biol. Chem. 272:5192–5198. [DOI] [PubMed] [Google Scholar]
- 36.Pique, C., A. Ureta-Vidal, A. Gessain, B. Chancerel, O. Gout, R. Tamouza, F. Agis, and M. C. Dokhelar. 2000. Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides. J. Exp. Med. 191:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao, K. N. 1995. The significance of the cholesterol biosynthetic pathway in cell growth and carcinogenesis. Anticancer Res. 15:309–314. [PubMed] [Google Scholar]
- 38.Robek, M. D., F. H. Wong, and L. Ratner. 1998. Human T-cell leukemia virus type 1 pX-I and pX-II open reading frames are dispensable for the immortalization of primary lymphocytes. J. Virol. 72:4458–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robek, M. D., and L. Ratner. 1999. Immortalization of CD4+ and CD8+ T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 73:4856–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross, T. M., S. M. Pettiford, and P. L. Green. 1996. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J. Virol. 70:5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowinsky, E. K., J. J. Windle, and D. D. Von Hoff. 1999. Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. J. Clin. Oncol. 17:3631–3652. [DOI] [PubMed] [Google Scholar]
- 42.Runquist, M., J. Ericsson, A. Thelin, T. Chojnacki, and G. Dallner. 1994. Isoprenoid biosynthesis in rat liver mitochondria. Studies on farnesyl pyrophosphate synthase and trans-prenyltransferase. J. Biol. Chem. 269:5804–5809. [PubMed] [Google Scholar]
- 43.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semmes, O. J., and M. L. Hammarskjold. 1999. Molecular pathogenesis of HTLV-1: a current perspective. ABI Professional Publications, Arlington, Va.
- 45.Seymour, L. 1999. Novel anti-cancer agents in development: exciting prospects and new challenges. Cancer Treat. Rev. 25:301–312. [DOI] [PubMed] [Google Scholar]
- 46.Song, L., and C. D. Poulter. 1994. Yeast farnesyl-diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc. Natl. Acad. Sci. USA 91:3044–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fënyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarshis, L. C., M. Yan, C. D. Poulter, and J. C. Sacchettini. 1994. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-Å resolution. Biochemistry 33:10871–10877. [DOI] [PubMed] [Google Scholar]
- 49.Trovato, R., J. C. Mulloy, J. M. Johnson, S. Takemoto, M. P. de Oliveira, and G. Franchini. 1999. A lysine-to-arginine change found in natural alleles of the human T-cell lymphotropic/leukemia virus type 1 p12I protein greatly influences its stability. J. Virol. 73:6460–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster, N. J., S. Green, J. R. Jin, and P. Chambon. 1988. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54:199–207. [DOI] [PubMed] [Google Scholar]
- 51.Willems, L., H. Heremans, G. Chen, D. Portetelle, A. Billiau, A. Burny, and R. Kettmann. 1990. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 9:1577–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems, L., R. Kettmann, F. Dequiedt, D. Portetelle, V. Vonèche, I. Cornil, P. Kerkhofs, A. Burny, and M. Mammerickx. 1993. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 67:4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willems, L., P. Kerkhofs, F. Dequiedt, D. Portetelle, M. Mammerickx, A. Burny, and R. Kettmann. 1994. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc. Natl. Acad. Sci. USA 91:11532–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willems, L., A. Burny, D. Collete, O. Dangoisse, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, D. Portetelle, J. C. Twizere, and R. Kettmann. 1999. Bovine leukemia virus as a model for human T-cell leukemia virus. Curr. Top. Virol. 1:139–167 [DOI] [PubMed] [Google Scholar]
- 55.Willems, L., A. Burny, D. Collete, O. Dangoisse, F. Dequiedt, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, T. Peremans, J. C. Twizere, and R. Kettmann. 2000. Genetic determinants of bovine leukemia virus pathogenesis. AIDS Res. Hum. Retrovir. 16:1787–1795. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, W., J. W. Nisbet, J. T. Bartoe, W. Ding, and M. D. Lairmore. 2000. Human T-lymphotropic virus type 1 p30II functions as a transcription factor and differentially modulates CREB-responsive promoters. J. Virol. 74:11270–11277. [DOI] [PMC free article] [PubMed] [Google Scholar]