Abstract
To exert control over RNA folding and catalysis, both molecular engineering strategies and in vitro selection techniques have been applied toward the development of allosteric ribozymes whose activities are regulated by the binding of specific effector molecules or ligands. We now describe the isolation and characterization of a new and considerably versatile RNA element that functions as a communication module to render disparate RNA folding domains interdependent. In contrast to some existing communication modules, the novel 9-nt RNA element is demonstrated to function similarly between a variety of catalysts that include the hepatitis delta virus, hammerhead, X motif and Tetrahymena group I ribozymes, and various ligand-binding domains. The data support a mechanistic model of RNA folding in which the element is comprised of both canonical and non-canonical base pairs and an unpaired nucleotide in the active, effector-bound conformation. Aside from enabling effector-controlled RNA function through rational design, the element can be utilized to identify sites in large RNAs that are susceptible to effector regulation.
INTRODUCTION
The recent development of effector-controlled RNA catalysts through rational design (1–3) and in vitro selection techniques (4–7) has established the possibility of utilizing allosteric ribozymes as molecular diagnostic components for genome (8,9), proteome (10–13) and metabolome analysis (1,2,4–7, 14,15) and drug discovery (12), or as genetic regulatory switches and controllable therapeutic agents (3,16–18) (reviewed in 19–23). Moreover, allosteric ribozymes present an intriguing mechanistic twist to the already daunting kinetic and thermodynamic complexities of RNA folding, the intricacies of which are exemplified by studies of the self-splicing Tetrahymena group I intron (24–28). Such studies support a model of RNA folding in which the native state, as the most thermodynamically favorable conformation, is represented by the lowest position on a multidimensional energy landscape or folding funnel, where RNA folding progresses downhill energetically. Allosteric ribozymes are unique in that they must possess conditional ‘traps’ along the folding pathway represented as effector-dependent crevasses or shelves within the energy landscape, the precise natures of which govern allosteric function. Consideration of these concepts will provide insight to the mechanistic function of certain allosteric ribozymes and will further empower the design of allosteric ribozymes for specified applications.
En route to the goal of generating and characterizing increasingly complex allosteric catalysts, we have discovered a truly modular RNA element that functions as a discrete but conditionally folded unit between various RNA architectures. This so-called communication module (5,7) was isolated by in vitro selection for allosteric hepatitis delta virus (HDV) ribozymes (29,30). However, we demonstrate that the module functions with a variety of structural domains, including those of the Tetrahymena ribozyme, to effect transitions in RNA conformation and to allosterically regulate RNA function. Consequently, the module lends itself to general and facile control of RNA folding and catalysis.
MATERIALS AND METHODS
In vitro selection of allosteric ribozymes
In vitro selection for theophylline-dependent HDV catalysts was performed as previously described (5,7). The initial population was comprised of >1.5 × 106 sequence variants generated by combining three populations that contained 10 or 9 random nucleotide positions, where one position was omitted from either the 5′ or 3′ segment of the random-sequence domain. DNA templates for transcription of the initial population were generated by primer extension of oligonucleotides 5′-TAATACGACTCACTATAGGGAACTGATGGCCGGCATGGTCCCAGCCTCCTCGCTGGCGCCGGCTGGGCAA and 5′-CCTCTAGTGATCTTATTCGC (N4–5)CTGCCAAGGGCCTTTCGGCTGGTAT(N4–5)CCCAGTTAACGCCAGCGAGGAGGCT using Superscript II reverse transcriptase (Invitrogen). 32P-labeled RNA was transcribed using T7 RNA polymerase and purified by denaturing polyacrylamide gel electrophoresis (PAGE). RNA populations (1013 molecules; <1 µM) were preselected against ligand-independent self-cleavage by incubation for 2.5 h at 37°C in reaction buffer [50 mM Tris–HCl (pH 7.5 at 37°C), 250 mM KCl and 5 mM MgCl2]. The uncleaved fraction of each population was purified by denaturing PAGE and subsequently selected for self-cleavage in the presence of 200 µM theophylline in reaction buffer by incubation for 2 min at 37°C. The cleaved fraction of each population was isolated by PAGE, and sequences were reverse transcribed and amplified by PCR using primers A (5′-TAATACGACTCACTATAGGGAACTGATGGTTAACATGATCTTAGCCT) and B (5′-CCTCTAGTGATCTTATTCGC). Individual sequences were isolated from the final population by cloning (TOPO-TA Cloning Kit; Invitrogen) and analyzed by sequencing (ThermalSequenase Kit; Amersham).
Template preparation
Individual 32P-labeled ribozymes were transcribed using T7 RNA polymerase and DNA templates prepared by PCR or primer extension of synthetic oligonucleotides. For theophylline-dependent HDV catalysts, DNA corresponding to the desired RNA sequence was prepared by PCR amplification of 5′-GGTTAACATGATCTTAGCCTCCTCGCTGGCGTTAACTGGG(Xn)ATACCAGCCGAAAGGCCC using primers A and C (5′-CCTCTAGTGATCTTATTCGC(Yn)CTGCCAAGGGCCTTTCGGCTGGTAT). Xn and Yn, respectively, represent nucleotides analogous and complementary to the intended 5′- and 3′-sequence segments that comprise P4′ of the RNA. DNA templates for FMN and ATP-dependent HDV ribozymes were similarly constructed. For hammerhead ribozyme constructs, templates were prepared by primer extension of 5′-TAATACGACTCACTATAGGGCGACCCTGATGAG and 5′-GGGCAACCTACGGCTTTCACCGTTTCGATTG(Xn)CATCCCTCATCAGGGTCGCCC using reverse transcriptase, where Xn represents nucleotides complementary to the desired ligand-binding domain. The theophylline-dependent X motif ribozyme was similarly constructed by primer extension of 5′-TAATACGACTCACTATAGGAGTAGCCAATGAAAATTGAGAGCCTTAAGCTGTA and 5′-GGAGTAGCCAGATTGCTGCCAAGGGCCTTTCGGCTGGTATCATCCCTACAGCTTAAGGCTCTC.
Templates for Tetrahymena group I ribozymes were constructed by enzymatic ligation of four synthetic oligonucleotides: 5′-TCCCGCAATTTGACGGTCTTGCCTTTTAAACCGATGCAATCTATTGGTTTAAAGACTAGCTACCAGGTGCATGCCTGATAACTTTTCCCTCC, 5′-AGGAC CATGTCCGTCAGCTTATTACCATACCCTTTGCAAGGCCATCTCAAAGTTTCCCCTGAGACTTGGTACTGAACGGCTGTTGACCCCTT, 5′-GTGAACTGCATCCATA(Xn) CTTAGGACTTGGCTGCGTGGTT and 5′-AGGTCCGACTATATCTTATG(Yn)CCGACCGACATTTAGTCT that were phosphorylated using T4 polynucleotide kinase (Invitrogen) and ligated using T4 DNA ligase (Invitrogen) and 5′-ACCGTCAAATTGCGGGAAAGGGGTCAACAGCCGT, 5′-CTGACGGACATGGT CCTAACCACGCAGCCAAGTC and 5′-TATGGATGCAGTTCACAGACTAAATGTCGGTCG as oligonucleotide splints. Xn and Yn, respectively, represent sequences complementary to the portions of P6 or P8 that were replaced by cm+theo6 and the theophylline-binding domain. Following ligation and purification by denaturing PAGE, oligonucleotides were amplified by PCR using 5′-TAATACGACTCACTATAGGAGGGAAAAGTTATCAGGCAT and 5′-CGAGTACTCCAACTCCCTTTCGGGAGAGGTCCGACTATATCTTAT as primers.
Characterization of allosteric ribozymes
Rate constants for each ribozyme were determined as described in Figure 1C and represent the average of those derived from two or three replicate assays wherein values differed by <2-fold. Where reported, the error is the standard deviation. For HDV and hammerhead ribozymes, reactions contained <1 µM RNA, 50 mM Tris–HCl (pH 7.5 at 23 or at 37°C), 250 mM KCl and 5 mM MgCl2 with or without 200 µM theophylline, 200 µM FMN or 1 mM ATP and were incubated at the specified temperature in the appropriately buffered solution. For the X motif ribozyme, reactions contained <1 µM RNA, 50 mM Tris–HCl (pH 7.5 at 37°C), 250 mM KCl and 50 mM MgCl2 with or without 200 µM theophylline and were incubated at 37°C. For Tetrahymena group I ribozymes, reactions contained 500 nM ribozyme, trace (<1 pmol) 5′-32P-labeled substrate, 25 mM Tris–HCl (pH 7.5 at 23°C), 150 mM KCl and 20 MgCl2 with or without 500 µM theophylline and were incubated at 23°C. The substrate is 5′-r(CUCUCUAGAAGA)d(GCGTACCGTAGCA)B (Dharmacon Research), where ‘r’ and ‘d ’, respectively, denote ribo- and deoxyribonucleotides, and ‘B’ indicates a 3′-biotin moiety.
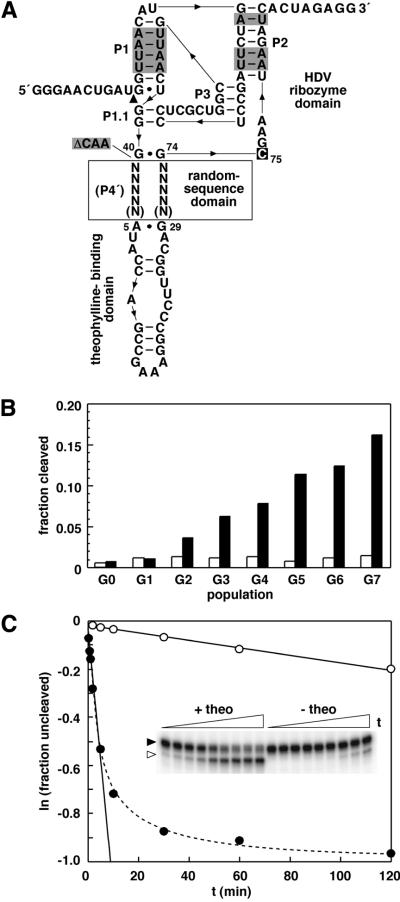
Figure 1.
Isolation and characterization of theophylline-dependent HDV ribozymes. (A) Design of initial populations for in vitro selection. The genomic HDV ribozyme was modified to include random-sequence domains of either 9 or 10 total nucleotides (N) and a theophylline-binding domain in place of P4 (P4′). Other modifications to the ribozyme include the replacement of G-C base pairs with A-U base pairs (shaded) and the inclusion of both 5′- and 3′-terminal extensions to enable selection and facilitate manipulation by RT–PCR. Additionally, 3 nt at positions 41–43 of the genomic HDV ribozyme were deleted (ΔCAA; shaded) as their absence or presence has little effect on ribozyme activity. C75 (boxed) is the nucleotide that functions at the active site of the catalyst as a general acid/base. (B) Progress of selection for theophylline-dependent HDV catalysts. The self-cleavage activity of the initial population (G0) and each subsequent population derived by in vitro selection for theophylline-dependent function is shown for reactions in the absence (open bars) or presence (filled bars) of theophylline under selection conditions incubated for 2 min. (C) Self-cleavage activity of an allosteric HDV ribozyme identified from the final population (G7). The inset depicts the precursor (filled arrowhead) and 3′-cleavage product (open arrowhead) of the theophylline-dependent HDV catalyst containing cm+theo6 as separated by denaturing PAGE following reaction under selection conditions in the absence or presence of theophylline for various lengths of time. The observed rate constants in the absence (open circles) or presence (filled circles) of theophylline were derived by plotting the natural logarithm of the fraction of uncleaved RNA versus time, and establishing the negative slope of the resulting lines (solid lines). The biphasic kinetic profile (dashed line) exhibited by the catalyst in the presence of theophylline indicates that ∼50% of the molecules might be irreversibly misfolded or only slowly convert to the active conformation.
RESULTS
Selection for allosteric HDV ribozymes
To investigate whether a structurally complex and highly stable catalyst might be regulated through a mechanism of conformational transition in response to effector binding, an in vitro selection experiment was performed to isolate allosteric catalysts derived from the genomic HDV ribozyme. The ribozyme features an intricate and exceptionally stable structure (31) that is resistant to either chemical (32) or thermal (33) denaturation, where P4 forms a foundation upon which the core of the catalyst rests. To generate a population for in vitro selection, P4 of the genomic HDV ribozyme was largely replaced by a theophylline-binding domain (34,35) adjoined through random-sequence segments (Fig. 1A). Furthermore, we attempted to constrict the catalytic core by deleting nucleotides 41–43, an alteration that has little effect on the rate constant of the ribozyme (36) and renders the catalytic core more homologous to that of the anti-genomic HDV ribozyme. Other mutations to the genomic HDV ribozyme include A-U base pairs in place of G-C base pairs in P1 and P2 (Fig. 1A). These mutations were not included to reduce the stability of the catalyst per se, but to render the sequence more amenable to reverse transcription and PCR amplification.
In vitro selection for theophylline-dependent self-cleavage was performed by depleting populations of ligand-independent ribozymes, and isolating the fraction of theophylline-dependent catalysts as previously described for related in vitro selection experiments (5,7). Through seven rounds of selection for effector activation (Fig. 1B), several populations of catalysts that exhibit theophylline-dependence were isolated. Members of the final population (G7) were cloned and sequenced for further analyses.
Characteristics of theophylline-dependent HDV ribozymes
The sequence elements isolated by in vitro selection unite effector-binding and catalytic domains of allosteric ribozymes and are termed communication modules (5,7). Interestingly, the communication modules isolated in this study, designated cm+theo6 through cm+theo12 (Table 1), are functionally similar but structurally distinct in that they exhibit each of the three possible configurations for symmetric and asymmetric segment lengths present in the initial population. By far, the most frequently represented asymmetric module in the final population was cm+theo12, while the only symmetric module isolated was cm+theo11. Modules cm+theo6 through cm+theo10 represent a small but unique class of homologous asymmetric modules. Moreover, these modules generally provide for relatively greater effector-mediated ribozyme activation as is evident from the observed rate constants in the absence (kobs–) or presence (kobs+) of theophylline (Table 1 and Fig. 1C).
Table 1. Sequences, kinetic parameters and other attributes of theophylline-dependent HDV ribozymes isolated by in vitro selection.
| Modulea | 5′ Sequence | 3′ Sequence | Frequencyb | kobs+ (min–1)c | kobs– (min–1)d | Fold activatione | Rapid switchingf |
|---|---|---|---|---|---|---|---|
| cm+theo6 | GGAUG | CAAU | 1 | 1.1 × 10–1 | 1.7 × 10–3 | 65 | + |
| cm+theo7 | GGAGG | CCUU | 1 | 4.6 × 10–2 | 1.4 × 10–3 | 33 | + |
| cm+theo8 | AUACG | CGGU | 1 | 1.4 × 10–1 | 3.5 × 10–3 | 40 | + |
| cm+theo9 | AUUUG | CUGU | 1 | 4.3 × 10–2 | 1.3 × 10–3 | 33 | + |
| cm+theo10 | AAAUG | CUGC | 1 | 1.2 × 10–1 | 3.2 × 10–3 | 38 | + |
| cm+theo11 | UCGAG | CUCUA | 1 | 5.7 × 10–2 | 3.0 × 10–3 | 19 | + |
| cm+theo12 | AGGG | CUCUA | 14 | 6.6 × 10–2 | 2.9 × 10–3 | 23 | + |
aThe designation for each communication module (cm) conveys that allosteric activation (+) was originally achieved with an effector specificity for theophylline (theo), and provides an arabic numeral for the sequence class of module in the series (6). These modules are listed 6–12 as a continuation of those derived from a previous study using the hammerhead ribozyme as a catalyst (7).
bNumber of total isolates sequenced.
cInitial observed rate constant for self-cleavage in the presence of ligand.
dObserved rate constant for self-cleavage in the absence of ligand.
ekobs+ / kobs–.
A notable characteristic of the allosteric HDV ribozymes is that effector binding affords a more modest rate enhancement relative to other allosteric catalysts similarly derived from the hammerhead ribozyme (5,7). In terms of allosteric activation, the thermodynamic stability of the effector-bound, active conformation is expected to exceed that of the inactive conformation or any of the innumerable unfolded states. Furthermore, we propose that the greater the energetic difference between folded states, or the energy barrier separating them, the greater will be the magnitude of allosteric activation. However, if the energy barrier between folded states is so great that it precludes effector-mediated conformational transition, there will be a requirement for ligand binding during folding to avoid entrapment in the inactive state. Consequently, it is interesting to note that each modestly activated theophylline-dependent HDV ribozyme demonstrates rapid conformational switching (5) (Table 1), a qualitative measure of facile ligand-mediated transition from the inactive state(s) to the active state consistent with a relatively low energy barrier. Therefore, we hypothesize that the likely mechanism of allosteric function is one in which catalysts transition from locally unfolded conformations (shelves in the energy landscape along the folding funnel) to the native state rather than between distinct and stable folded states (crevasses in the energy landscape between which the energy barrier is high).
Mutational analysis of communication module function
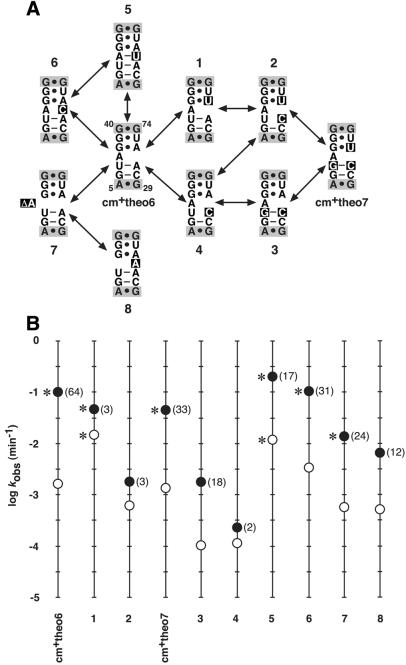
To assess the active state of an allosteric HDV ribozyme with regard to this proposed mechanism, the effects of single-nucleotide mutations on allosteric function of modules cm+theo6 and cm+theo7 were examined (Fig. 2). Based on sequence similarity, cm+theo6 and cm+theo7 are each predicted to form a relatively weak secondary structure that consists of two canonical base pairs, an unpaired nucleotide and two non-canonical base pairs in the active state. In this manner, effector-dependent folding of the theophylline- binding domain (35) is likely to sequentially promote folding of the communication module, followed by folding and activity of the catalyst. It is interesting to note that every mutant construct maintains some degree of allosteric function, albeit meager for certain cases (e.g. constructs 1, 2 and 4). Moreover, the effect of the mutation on the relative values of kobs– and kobs+ between related constructs reveals the relationship between secondary structure and allosteric function. For example, construct 1 (Fig. 2A) contains a mutation that alters the identity of a G·A base pair in cm+theo6 to a G·U base pair like that in cm+theo7. Although the mutation does not obviate base pairing, it has a distinct effect in that it reduces the magnitude of allosteric activation chiefly by increasing the kobs– (Fig. 2B). The effect might be attributed in part to the fact that a G·U base pair is more thermodynamically favorable (37) and could provide a measure of stability to communication module structure in the absence of ligand. A second mutation that eliminates the U-A base pair with a U-C mismatch (construct 2) largely disrupts allosteric function, while a third mutation that leads to cm+theo7 and repairs the mismatch with a G-C base pair restores function (Fig. 2). A correlative effect is observed for the series of mutations that lead from cm+theo7 to cm+theo6 through constructs 3 and 4 (Fig. 2). Obviously, there exists a more subtle aspect to communication module structure and allosteric function than simple base pairing, as is evident from the activities of constructs 1 and 3 (Fig. 2). Namely, there appears to be some requirement for certain base-pair identities flanking the unpaired nucleotide. Consequently, base-stacking interactions might play a crucial role in communication module structure and function.
Figure 2.
Mutational analysis of theophylline-dependent HDV ribozymes. (A) Theophylline-dependent HDV ribozymes and mutant constructs. Shown is the communication module sequence and flanking G40·G74 and A5·G29 base pairs (shaded) of the catalyst and theophylline-binding domain, respectively, for each ribozyme containing cm+theo6 and cm+theo7. The secondary structures depicted are those proposed for the active, ligand-bound conformations of the catalysts. Arrows denote the relationship of each construct to other constructs (arbitrarily numbered 1–8) containing single nucleotide mutations (boxed relative to cm+theo6 sequence). (B) Theophylline-dependent activities of allosteric HDV ribozymes and mutant constructs. The activity of each construct is represented as the logarithm of the initial kobs determined by reaction in the absence (open circle) or presence (filled circle) of theophylline under selection conditions. Numbers in parentheses indicate the fold-activation elicited by effector. Asterisks denote those catalysts and conditions under which the catalysts exhibit biphasic kinetic profiles similar to that shown in Figure 1C.
To investigate the importance of the unpaired adenosine in cm+theo6 structure and function, single nucleotide mutations were made in order to base pair, mismatch, delete or relocate the adenosine nucleotide. Predictably, pairing the adenosine produces a more highly active and less theophylline-responsive catalyst (construct 5) relative to that containing cm+theo6, while mismatching the adenosine produces an approximately equivalent species (construct 6; Fig. 2). These data suggest that the function of the unpaired nucleotide in the communication module is to destabilize the element in the absence of effector and to maximize the effector-mediated rate enhancement. However, deleting the unpaired adenosine (construct 7) or relocating it to the opposite segment of the communication module (construct 8) reduces the rate constants, indicating that the unpaired nucleotide also provides requisite and site-specific flexibility (Fig. 2). Taken together, the mutational analyses of cm+theo6 and cm+theo7 are consistent with the proposed mechanism of allosteric activation and secondary structure model for the active conformation of the effector-bound ribozymes.
Versatile control of RNA folding and catalysis
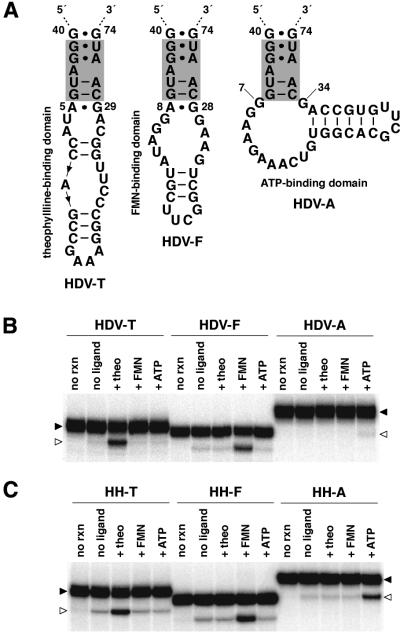
Given that cm+theo6 appears to exist as a discrete folded unit within the effector-bound allosteric ribozyme and likely transitions between unfolded and folded states in response to ligand-mediated structural stabilization, the communication module was predicted to possess a general aptitude for rendering the folding of disparate RNA domains interdependent. As previously demonstrated using certain other communication modules, effector specificity of an allosteric ribozyme can be altered simply by exchanging the ligand-binding domain (5,7). Consistent with those studies, cm+theo6 demonstrates an ability to confer effector-dependent function upon the HDV ribozyme when the theophylline-binding domain is replaced with a domain that binds FMN (38,39) or ATP (40,41), albeit considerably less effectively in the latter case (Fig. 3A and B).
Figure 3.
Versatile control of HDV and hammerhead ribozymes. (A) Allosteric HDV ribozymes. Catalysts are comprised of the modified genomic HDV ribozyme, cm+theo6 (shaded), and theophylline-, FMN- or ATP-binding domains (HDV-T, HDV-F and HDV-A, respectively). Depicted is P4′ for each construct. (B) Effector specificities and activities of allosteric HDV ribozymes. Shown is the uncleaved ribozyme (filled arrowheads) and 3′-cleavage product (open arrowheads) separated by denaturing PAGE following reaction in the absence (no ligand) or presence of theophylline (+theo), FMN (+FMN) or ATP (+ATP) at 23°C for 45 min. Unreacted ribozyme is indicated (no rxn). (C) Effector specificities and activities of allosteric hammerhead ribozymes. The hammerhead ribozyme was similarly modified by replacing all but the G10.1·C11.1 base pair of stem II with cm+theo6 and theophylline-, FMN- or ATP-binding domains (HH-T, HH-F and HH-A, respectively). Each allosteric hammerhead ribozyme was analyzed identically as described for allosteric HDV ribozymes with the exception that open arrowheads denote the 5′-cleavage product.
More striking is the fact that cm+theo6 functions similarly to confer theophylline-, FMN- or ATP-dependent function upon the hammerhead ribozyme (42,43) (Fig. 3C). The function of the FMN- and ATP-dependent hammerhead ribozymes exemplifies the inclusive but interdependent structural organization of the communication module, as the module is effective in contexts wholly distinct from that of the theophylline-dependent HDV ribozyme. This level of versatility was not observed for previous communication modules developed within the context of theophylline-dependent hammerhead ribozymes (7), where cm+theo3 and cm+theo5 failed to confer theophylline-dependent activity to the HDV ribozyme when similarly appended (data not shown). Interestingly, the ability of cm+theo6 to mediate ATP-dependence with the hammerhead ribozyme demonstrates that the relative inactivity of the equivalent HDV ribozyme construct might result from an incompatibility between the aptamer and ribozyme domains rather than the aptamer and communication module (Fig. 3B and C). Additionally, analysis of rate constants determined at 23 and 37°C for the theophylline-dependent HDV and hammerhead ribozymes demonstrates that temperature has little effect on allosteric function, as kobs values remain relatively unchanged (Table 2). The result might reflect counteracting effects of temperature on the chemical rate of the reaction and the stability of the active conformation.
Table 2. Kinetic parameters of various theophylline-dependent ribozymes.
| RNA catalyst | T (°C) | kobs+ (min–1) | kobs– (min–1) | Fold activation |
|---|---|---|---|---|
| HDV-T | 37 | 1.1 × 10–1 ± 9.9 × 10–3 | 1.7 × 10–3 ± 1.4 × 10–4 | 65 ± 0.8 |
| 23 | 6.9 × 10–2 ± 4.2 × 10–3 | 6.0 × 10–4 ± 1.3 × 10–4 | 115 ± 32 | |
| HH-T | 37 | 1.0 × 10–2 ± 1.2 × 10–3 | 9.5 × 10–4 ± 6.4 × 10–5 | 11 ± 0.5 |
| 23 | 2.3 × 10–2 ± 4.2 × 10–3 | 9.8 × 10–4 ± 1.8 × 10–4 | 23 ± 0.1 | |
| P6/P8 | 23 | 3.0 × 10–3 ± 4.4 × 10–4 | 2.9 × 10–3 ± 5.9 × 10–4 | 1 ± 0.1 |
| P6-T | 23 | 2.0 × 10–3 ± 4.6 × 10–4 | 6.9 × 10–4 ± 2.2 × 10–4 | 3 ± 0.5 |
| P8-T | 23 | 1.2 × 10–3 ± 5.8 × 10–5 | 3.3 × 10–4 ± 3.6 × 10–5 | 4 ± 0.5 |
| P6-T/P8-T | 23 | 5.2 × 10–4 ± 4.9 × 10–5 | 2.0 × 10–5 ± 3.2 × 10–6 | 26 ± 2 |
Identification of effector-dependent sites in other RNAs
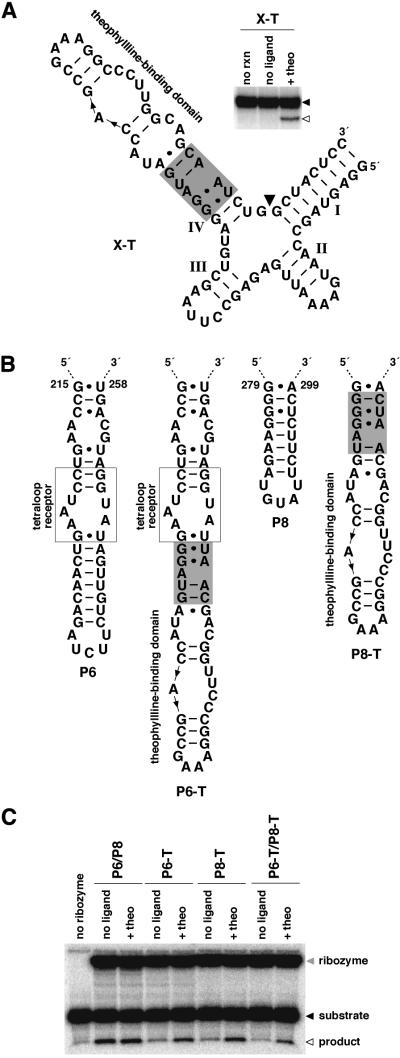
To further examine the ability of cm+theo6 to control RNA folding and catalysis, two additional ribozymes for which no proven mode of modification has previously been demonstrated were altered to contain the communication module and theophylline-binding domain. The X motif ribozyme (44) consists of a four-helix junction for which no tertiary structural model exists. However, judicious modification of stem IV generates a theophylline-dependent variant (Fig. 4A), while various alterations at stem II failed to produce active catalysts (data not shown). More interestingly, the Tetrahymena group I ribozyme (45,46) is a large, multi-domain catalyst that exhibits considerable complexity in the folding pathway required to achieve the active conformation (24–28). Modification of P6 or P8 produced catalysts that exhibit small but significant allosteric affects (Fig. 4B and C; Table 2). However, modification at both P6 and P8 has a synergistic effect on allosteric function and generates an allosteric catalyst comparable with others in this study (Fig. 4B and C; Table 2). Similar modification of P9.0 produced a relatively active catalyst that is unresponsive to effector (data not shown). Consequently, cm+theo6 demonstrates a broad capacity to couple diverse RNA domains to effect control over folding and catalysis, and to identify sites within RNAs where modification might generally render effector-dependent function.
Figure 4.
Identification of effector-dependent sites in X motif and Tetrahymena group I ribozymes. (A) An allosteric X motif ribozyme. An X motif ribozyme was modified by replacing all but two base pairs of stem IV with cm+theo6 and a theophylline-binding domain (X-T). The inset depicts the activity of the allosteric ribozyme. Shown is the uncleaved ribozyme (filled arrowhead) and 5′-cleavage product (open arrowhead) separated by denaturing PAGE following reaction in the absence (no ligand) or presence of theophylline (+theo) at 37°C for 45 min. Unreacted ribozyme is indicated (no rxn). (B) Design of allosteric Tetrahymena group I ribozymes. Group I ribozymes correspond to the L-21 NheI RNA which lacks the P9.1 and P9.2 extensions, and contain CGAAA in place of nucleotides 322–326 in P9. The ribozyme was modified by replacing a portion of P6 or P8 with cm+theo6 and a theophylline-binding domain (P6-T or P8-T, respectively). Alternatively, the ribozyme was modified at both positions (P6-T/P8-T). (C) Activities of unmodified and allosteric Tetrahymena group I ribozymes. Shown is ribozyme (shaded arrowhead), substrate (filled arrowhead) and 5′-cleavage product (open arrowhead) resulting from ribozyme catalysis of the reverse of the second step of splicing (3′-exon ligation). Products were separated by denaturing PAGE following reaction in the absence (no ligand) or presence of theophylline (+theo) at 23°C for 30 min. Unmodified ribozyme is labeled P6/P8. Reaction containing substrate alone is indicated (no ribozyme).
DISCUSSION
Through the isolation and characterization of allosteric HDV ribozymes, we have identified a 9-nt RNA element that functions as a distinct but conditionally folded domain and demonstrates general utility in engineering effector-dependent folding and function of various RNA catalysts. Consideration of recent concepts in the kinetic and thermodynamic complexity of RNA folding provides additional insight into the mechanistic function of these and other allosteric ribozymes. Models of allosteric activation are proposed for which the facility of the allosteric transition and the relative magnitude of the allosteric effect are indicative of mechanism. The models distinguish inactive conformations as distinct folded states or crevasses in an energy landscape from locally unfolded conformations or shelves in an energy landscape. While the versatile communication module identified and characterized in this study produces relatively modest allosteric effects for various catalysts, the simplicity and general applicability of the module enable unprecedented ease in generating new allosteric catalysts with various effector specificities. Moreover, the module lends itself to the assessment of candidate sites in larger RNAs that might be susceptible to effector-mediated regulation, as is demonstrated for the Tetrahymena group I ribozyme. This unique and versatile RNA element is expected to impact the further development of allosteric ribozymes for specified applications.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to express our gratitude to E. DeRose for in vitro selection, C. Weiss for sequencing and R. R. Breaker for valuable guidance. We also thank R. R. Breaker, G. A. M. Emilsson and L. J. Maher III for critical reading of the manuscript. This work was supported by grants to R. R. Breaker from the NIH and to G.A.S. from Nebraska NSF EPSCoR and the Health Future Foundation.
REFERENCES
- 1.Tang J. and Breaker,R.R. (1997) Rational design of allosteric ribozymes. Chem. Biol., 4, 453–459. [DOI] [PubMed] [Google Scholar]
- 2.Tang J. and Breaker,R.R. (1998) Mechanism for allosteric inhibition of an ATP-sensitive ribozyme. Nucleic Acids Res., 26, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuwabara T., Warashina,M., Tanabe,T., Kenzaburo,T., Asano,S. and Taira,K. (1998) A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell, 2, 617–627. [DOI] [PubMed] [Google Scholar]
- 4.Robertson M.P. and Ellington,A.D. (1999) In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol., 17, 62–66. [DOI] [PubMed] [Google Scholar]
- 5.Soukup G.A. and Breaker,R.R. (1999) Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA, 96, 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koizumi M., Soukup,G.A., Kerr,J.N.Q. and Breaker,R.R. (1999) Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol., 6, 1062–1071. [DOI] [PubMed] [Google Scholar]
- 7.Soukup G.A., Emilsson,G.A.M. and Breaker,R.R. (2000) Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol., 298, 623–632. [DOI] [PubMed] [Google Scholar]
- 8.Burke D.H., Ozerova,N.D. and Nilsen-Hamilton,M. (2002) Allosteric hammerhead ribozyme TRAPs. Biochemistry, 41, 6588–6594. [DOI] [PubMed] [Google Scholar]
- 9.Wang D.Y., Lai,B.H., Feldman,A.R. and Sen,D. (2002) A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res., 30, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson M.P. and Ellington,A.D. (2001) In vitro selection of nucleoprotein enzymes. Nat. Biotechnol., 19, 650–655. [DOI] [PubMed] [Google Scholar]
- 11.Wang D.Y. and Sen,D. (2002) Rationally designed allosteric variants of hammerhead ribozymes responsive to the HIV-1 Tat protein. Comb. Chem. High Throughput Screen., 5, 301–312. [DOI] [PubMed] [Google Scholar]
- 12.Hartig J.S., Najifi-Shoushtari,S.H., Grune,I., Yan,A., Ellington,A.D. and Famulok,M. (2002) Protein-dependent ribozymes report molecular interactions in real time. Nat. Biotechnol., 20, 717–722. [DOI] [PubMed] [Google Scholar]
- 13.Vaish N.K., Dong,F., Andrews,L., Schweppe,R.E., Ahn,N.G., Blatt,L. and Seiwert,S.D. (2002) Monitoring post-translational modifications of proteins with allosteric ribozymes. Nat. Biotechnol., 20, 810–815. [DOI] [PubMed] [Google Scholar]
- 14.Seetharaman S., Zivarts,M., Sudarsan,N. and Breaker,R.R. (2001) Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol., 19, 336–341. [DOI] [PubMed] [Google Scholar]
- 15.Wang D.Y., Lai,B.H. and Sen,D. (2002) A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J. Mol. Biol. 318, 33–43. [DOI] [PubMed] [Google Scholar]
- 16.Tanabe T., Kuwabara,T., Warashina,M., Tani,K., Taira,K. and Asano,S. (2000) Oncogene inactivation in a mouse model. Nature, 406, 473–474. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe T., Takata,I., Kuwabara,T., Warashina,M., Kawasaki,H., Tani,K., Ohta,S., Asano,S. and Taira,K. (2000) Maxizymes, novel allosterically controllable ribozymes, can be designed to cleave various substrates. Biomacromolecules, 1, 108–117. [DOI] [PubMed] [Google Scholar]
- 18.Kuwabara T., Tanabe,T., Warashina,M., Xiong,K.X., Tani,K., Taira,K. and Asano,S. (2001) Allosterically controllable maxizyme-mediated suppression of progression of leukemia in mice. Biomacromolecules, 2, 1220–1228. [DOI] [PubMed] [Google Scholar]
- 19.Soukup G.A. and Breaker,R.R. (2000) Allosteric nucleic acid catalysts. Curr. Opin. Struct. Biol., 10, 318–325. [DOI] [PubMed] [Google Scholar]
- 20.Soukup G.A. and Breaker,R.R. (2000) Allosteric ribozymes. In Krupp,G. and Gaur,R.K. (eds), Ribozyme Biochemistry and Biotechnology. Eaton Publishing, Natick, MA, pp. 149–170.
- 21.Kuwabara T., Warashina,M. and Taira,K. (2000) Allosterically controllable ribozymes with biosensor functions. Curr. Opin. Chem. Biol., 4, 669–677. [DOI] [PubMed] [Google Scholar]
- 22.Breaker R.R. (2002) Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol., 13, 31–39. [DOI] [PubMed] [Google Scholar]
- 23.Iyo M., Kawasaki,H. and Taira,K. (2002) Allosterically controllable maxizymes for molecular gene therapy. Curr. Opin. Mol. Ther., 4, 154–165. [PubMed] [Google Scholar]
- 24.Sclavi B., Sullivan,M., Chance,M.R., Brenowitz,M. and Woodson,S.A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science, 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- 25.Treiber D.K., Rook,M.S., Zarrinkar,P.P. and Williamson,J.R. (1998) Kinetic intermediates trapped by native interactions in RNA folding. Science, 279, 1943–1946. [DOI] [PubMed] [Google Scholar]
- 26.Pan J., Thirumalai,D. and Woodson,S.A. (1999) Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics and kinetics. Proc. Natl Acad. Sci. USA, 96, 6149–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang X., Bartley,L.E., Babcock,H.P., Russell,R., Ha,T., Herschlag,D. and Chu,S. (2000) A single-molecule study of RNA catalysis and folding. Science, 288, 2048–2051. [DOI] [PubMed] [Google Scholar]
- 28.Russell R., Zhuang,X., Babcock,H.P., Millett,I.S., Doniach,S., Chu,S. and Herschlag,D. (2002) Exploring the folding landscape of a structured RNA. Proc. Natl Acad. Sci. USA, 99, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H.N., Lin,Y.J., Lin,F.P., Makino,S., Chang,M.F. and Lai,M.M (1989) Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl Acad. Sci. USA, 86, 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P.K.R., Suh,Y.A., Taira,K. and Nishikawa,S. (1993) Point and compensation mutations to evaluate essential stem structures of genomic HDV ribozyme. FASEB J., 7, 124–129. [DOI] [PubMed] [Google Scholar]
- 31.Ferré-D’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 32.Rosenstein S.P. and Been,M.D. (1990) Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry, 29, 8011–8016. [DOI] [PubMed] [Google Scholar]
- 33.Duhamel J., Liu,D.M., Evilia,C., Fleysh,N., Dinter-Gottlieb,G. and Lu,P. (1996) Secondary structure content of the HDV ribozyme in 95% formamide. Nucleic Acids Res., 24, 3911–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenison R.D., Gill,S.C., Pardi,A. and Polisky,B. (1994) High-resolution molecular discrimination by RNA. Science, 263, 1425–1429. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann G.R., Jenison,R.D., Wick,C.L., Simorre,J.P. and Pardi,A. (1997) Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol., 4, 644–649. [DOI] [PubMed] [Google Scholar]
- 36.Wadkins T.S. and Been,M.D. (1997) Core-associated non-duplex sequences distinguishing the genomic and antigenomic self-cleaving RNAs of hepatitis delta virus. Nucleic Acids Res., 25, 4085–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra M.J. and Turner,D.H. (1995) Predicting thermodynamic properties of RNA. Methods Enzymol., 259, 242–261. [DOI] [PubMed] [Google Scholar]
- 38.Burgstaller P. and Famulok,M. (1994) Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew Chem. Int. Ed. Engl., 33, 1084–1087. [Google Scholar]
- 39.Fan P., Suri,A.K., Fiala,R., Live,D. and Patel,D.J. (1996) Molecular recognition in the FMN–RNA aptamer complex. J. Mol. Biol., 258, 480–500. [DOI] [PubMed] [Google Scholar]
- 40.Sassanfar M. and Szostak,J.W. (1993) An RNA motif that binds ATP. Nature, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- 41.Jiang F., Kumar,R.A., Jones,R.A. and Patel,D.J. (1996) Structural basis of RNA folding and recognition in an AMP–RNA aptamer complex. Nature, 382, 183–186. [DOI] [PubMed] [Google Scholar]
- 42.Forster A.C. and Symons,R.H. (1987) Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell, 49, 211–220. [DOI] [PubMed] [Google Scholar]
- 43.Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- 44.Tang J. and Breaker,R.R. (2000) Structural diversity of self-cleaving ribozymes. Proc. Natl Acad. Sci. USA, 97, 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 46.Golden B.L., Gooding,A.R., Podell,E.R. and Cech,T.R. (1998) A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science, 282, 259–264. [DOI] [PubMed] [Google Scholar]