Abstract
Human telomeric DNA composed of (TTAGGG/CCCTAA)n repeats may form a classical Watson–Crick double helix. Each individual strand is also prone to quadruplex formation: the G-rich strand may adopt a G-quadruplex conformation involving G-quartets whereas the C-rich strand may fold into an i-motif based on intercalated C·C+ base pairs. Using an equimolar mixture of the telomeric oligonucleotides d[AGGG(TTAGGG)3] and d[(CCCTAA)3CCCT], we defined which structures existed and which would be the predominant species under a variety of experimental conditions. Under near-physiological conditions of pH, temperature and salt concentration, telomeric DNA was predominantly in a double-helix form. However, at lower pH values or higher temperatures, the G-quadruplex and/or the i-motif efficiently competed with the duplex. We also present kinetic and thermodynamic data for duplex association and for G-quadruplex/i-motif unfolding.
INTRODUCTION
A eukaryotic chromosome begins and ends with a specific nucleoprotein structure called the telomere. Telomeres are essential for genome integrity and appear to play an important role in cellular aging and cancer (1). They have a unique mode of replication, based on an enzyme called telomerase initially identified in ciliates (2). In contrast to cancer cells, which often express high levels of telomerase (3), this activity is tightly regulated in normal human somatic cells (reviewed in 4). In a large number of organisms, telomeric DNA consists of highly repetitive short sequences. These repeats are characterized by the asymmetry of the guanines and cytosines. Many telomeres are composed of repetitions of two to five adjacent guanines on the same strand with corresponding cytosines on the other strand. These repetitions are generally regular, except in some lower eukaryotes. In humans the telomeric repeat motif is (5′TTAGGG):(5′CCCTAA) (5). Human telomere length, as estimated by the size of the terminal restriction fragments, varies between 7 and 15 kb in somatic cells (6,7). Most of telomeric DNA is double-stranded except for the extreme terminal part where the 3′ region of the G-rich strand is single-stranded (8). For all cells tested, these 3′ overhangs are relatively long (50–210 bases in length) and present on all chromosomal ends (8).
Both G-rich and C-rich telomeric strands may form in vitro unusual DNA structures. The G-rich strand can adopt a four-stranded G-quadruplex structure involving planar G-quartets (Fig. 1A) (9,10), while the C-rich strand can form the so-called i-motif with intercalated C·C+ base pairs (Fig. 1B) (11). Intramolecular G-quadruplex and i-motif structures from human telomeric DNA have been reported recently (12–14). The structures and the stability of the G-quadruplex depend on monocations (15,16). Different G-quadruplex structures exist, depending on the orientation of the strands and the syn/anti conformations of guanines (17,18). The stability of the i-motif depends on pH, and C-rich telomeric repeats may fold into a stable i-motif at slightly acidic pH (19,20). There are different possible i-motif structures with different intercalation and looping topologies (19,21).
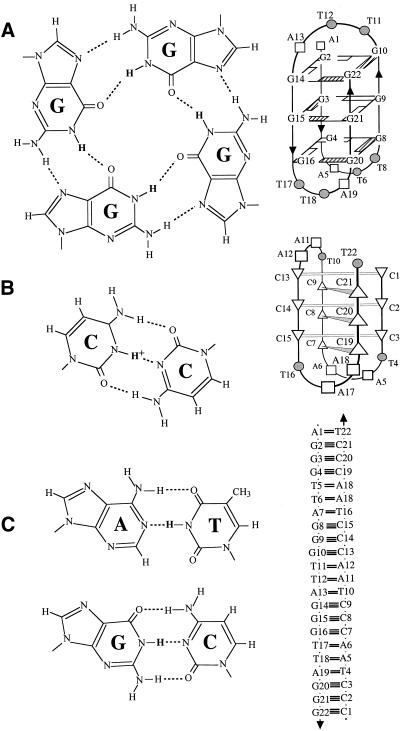
Figure 1.
(A) G-quartet (left) and schematic structure of the intramolecular Na+ G-quadruplex from 22AG (right). (B) C·C+ base pair (left) and schematic structure of the intramolecular i-motif from 22CT (right). (C) Watson–Crick A·T and G·C base pairs (left) and schematic structure of the Watson–Crick duplex 22AG·22CT (right).
Obviously, the G- and C-rich complementary strands may also form a double helix based on A·T and G·C Watson–Crick base pairs (Fig. 1C). Although the presence of G-quadruplexes in the macronuclei of ciliates was recently demonstrated with a G-quadruplex-specific antibody (22), it is not clear whether G-quadruplex and i-motif are formed and used in higher eukaryotes. Proteins that interact with telomeric G-strand, C-strand and duplexes have been identified and could play a role in the relative stability of these structures (18). Duplex–quadruplex interconversion has been studied for a number of different sequences (16,23–27). However, besides a recent report based on circular dichroism spectroscopy (28), no systematic analysis on the behavior of the human telomeric DNA has been undertaken previously.
The G-quadruplex, i-motif and Watson–Crick double helix have distinct nuclear magnetic resonance (NMR) signatures such as imino proton chemical shift (29). Ultraviolet (UV) absorption at different temperatures provides melting profiles and may be used to monitor the stability of these structures (20,30). Here, using NMR and UV spectroscopy we examine the conversion of a regular double-helix DNA structure (composed of both human telomeric G-rich and C-rich strands) into intramolecular G-quadruplex and i-motif. We have chosen for this investigation human telomeric fragments of 22 nt, d[AGGG(TTAGGG)3] and d[(CCCTAA)3CCCT], whose crystal and solution structures are known (12–14). Duplex–quadruplex equilibrium for an equimolar mixture of these telomeric oligonucleotides was studied under a variety of experimental conditions. Under near-physiological conditions of pH, temperature and salt concentration, telomeric DNA was predominantly in a duplex form. However, at lower pH, the G-quadruplex and the i-motif may compete with the duplex. The same methods also provided information on the kinetics and on the thermodynamics of formation and unfolding of different structures.
MATERIALS AND METHODS
Sample preparation
The oligonucleotides d[AGGG(TTAGGG)3] and d[(CCCT AA)3CCCT] (denoted 22AG and 22CT, respectively) were synthesized and purified as previously described (31) or purchased from Eurogentec (Belgium). The strand concentrations were computed from the absorbance at 260 nm, using a nearest-neighbor model (32). Purity (>90%) was checked by gel electrophoresis.
UV spectroscopy
All experiments were performed, unless otherwise specified, with 1–20 µM oligonucleotide strand concentration in a 10 mM sodium cacodylate (pH 5.5–7.5) or sodium acetate (pH 4–5.5) buffer supplemented with 100 mM NaCl or KCl. Thermal denaturation profiles were obtained with a Kontron Uvikon 940 spectrophotometer as previously described (20,30,33). For each temperature, absorbance was measured at 245, 260, 295 and 405 nm (the control wavelength) using 1 cm pathlength cells. The temperature was measured using a probe immersed in a control cell. Absorbance spectra (220–335 nm) were recorded at high (87°C) or low temperature (4°C) after each experiment.
Nuclear magnetic resonance
The strand concentration of the NMR samples was 0.1 or 0.2 mM. The solutions contained 0.1 mM EDTA, 0.01 mM DSS and 10–100 mM of NaCl (or KCl). The sample pH was measured with a glass microelectrode before and after each NMR experiment and adjusted with HCl and NaOH (or KOH) solutions. NMR experiments were performed on a 500 MHz Varian Unity INOVA spectrometer equipped with a 5 mm Penta probe. For experiments in H2O, the ‘Jump and Return’ sequence (34) was used for solvent signal suppression with the maximum sensitivity set at 13.5 p.p.m. and a repetition time of 1.5 s. For 2D nuclear Overhauser effect spectroscopy (NOESY) and nuclear Overhauser effect spectroscopy in the rotating frame (ROESY) experiments in D2O, the repetition time was 2 s. At high temperatures, the G-quadruplex or i-motif unfolding rate (1 / τ) was derived from the variation versus mixing time (tmix) of the volume of the exchange cross peaks (between folded and melted structures) in NOESY experiments. The exchange cross peaks could be distinguished from the dipolar cross peaks, because they were positive (same sign as diagonal peaks) in ROESY and their intensity in NOESY (and ROESY) increased when the temperature was increased. For tmix << τ (e.g. for tmix < 10% τ), the volumes of the exchange cross peak (Vx) and of the diagonal peak (Vd) are related by:
Vx ≈ (tmix / τ) · Vd
In real-time mixing experiments at low temperatures, the G-strand and C-strand were mixed and rapidly put in the NMR spectrometer. The 1D spectra were then obtained at different periods of time. The formation of the Watson–Crick duplex at time t was followed with the intensity I(t) of imino protons at 12–14 p.p.m. The time constant (τ) was derived from the equation:
I(t) / I0 = 1 – exp(–t / τ)
where I0 is the intensity at equilibrium.
RESULTS
G-quadruplex, i-motif and Watson–Crick double helix: NMR spectra
Quadruplex and duplex structures are formed based on different base associations, respectively G·G·G·G (G-quartet), C·C+, A·T and G·C base pairs (Fig. 1). Imino protons in these base alignments are involved in different types of hydrogen bonds, and their chemical shifts are characteristic for each structure: 10–12 p.p.m. for G-quartets, 15–16 p.p.m. for C·C+ base pairs, and 12–14 p.p.m. for Watson–Crick A·T and G·C base pairs (29,35). The exchange properties also differ, with exchange times ranging from milliseconds in Watson–Crick base pairs to minutes/hours in C·C+ base pairs and hours/days in G-quartets (29).
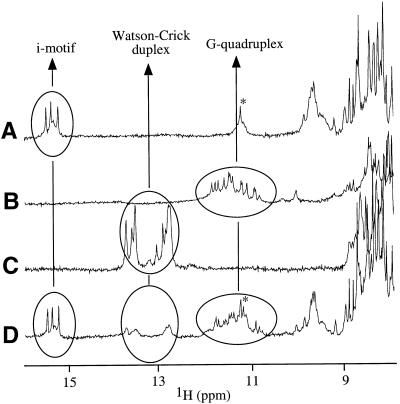
The structures of the separate 22CT (13) and 22AG (12) sequences have been studied by NMR. The 1D spectra obtained here (Fig. 2) are similar to those published previously. The spectrum of 22CT at pH 4.5 is shown in Figure 2A. The chemical shift (15–16 p.p.m.) and exchange properties characterize an i-motif structure. The structure persists up to neutral pH (13). Similarly, the imino protons of 22AG (10–12 p.p.m.) characterize a G-quadruplex (Fig. 2B). The imino proton spectrum is unchanged between pH 4 and 7 (data not shown), except for small shifts in the case of the imino protons assigned to external G-quartets (12). At pH 7, the imino proton spectrum of the equimolar mixture (12–14 p.p.m.) is characteristic of a duplex with Watson–Crick A·T and G·C base pairs, and no other species are present (Fig. 2C). At pH 4.5, all three structures have been observed (Fig. 2D). The structures have been confirmed using 2D NOESY experiments (data not shown), which detected characteristic NOE (short distances) patterns for each structure (29). We conclude that at low pH, the duplex dissociates to form i-motif and G-quadruplex structures. The proportion of each form depends on experimental conditions such as DNA concentration, pH, temperature and counter-ions (see below). Such G-quadruplex and i-motif structures may be stabilized by their interaction via the complementary loops (19). At pH 4.5, we did not observe imino proton peaks that would correspond to Watson–Crick A·T base pairs between the loops of the i-motif and the G-quadruplex structures. However, these loop bindings can exist temporarily in an intermediate or fast (faster than milliseconds) NMR time scale that does not enable the detection of such imino protons. We did not observe changes in chemical shifts of imino protons from the C-strand. However, some slight shifts of imino protons from the G-strand were observed which might suggest such an interaction.
Figure 2.
One-dimensional 500 MHz 1H NMR spectra of 22CT, 22AG and their 1:1 mixture in different conditions. (A) 22CT at pH 4.5; (B) 22AG at pH 4.5; (C) 22CT + 22AG at pH 7; (D) 22CT + 22AG at pH 4.5. Imino protons of thymines in the i-motif structure are labeled with a star. Experimental conditions: strand concentration, 0.1 mM; temperature, 10°C; 100 mM NaCl.
Kinetics of G-quadruplex/i-motif unfolding and of double-helix formation
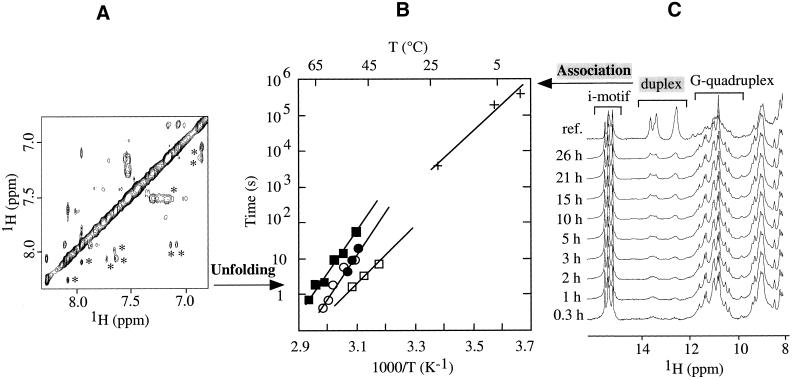
Since each form can be monitored separately by NMR, the kinetics of folding/unfolding and of association processes can also be studied. The lifetimes of the G-quadruplex and of the i-motif were derived from the exchange cross peaks between these structures and the melted high temperature form of each strand. The exchange cross peaks are observed in NOESY-type spectra (13,35) at temperatures close to the melting point where the structured and melted forms exist in comparable concentrations. A NOESY spectrum of the C-strand at pH 5, 50°C is shown in Figure 3A. Some resolved exchange cross peaks (see Materials and Methods) are labeled with stars. Lifetime values of the structured form, as derived from the exchange times of different protons, are plotted as a function of temperature in Figure 3B. The lifetimes of the i-motif do not depend on pH or salt conditions, while those of the G-quadruplex are approximately 10 times longer in 100 mM KCl than in 10 mM NaCl.
Figure 3.
(A) NOESY spectrum of 22CT at 50°C (mixing time, 50 ms) showing exchange cross peaks between folded and unstructured conformations. Exchange cross peaks are labeled with stars. Experimental conditions: strand concentration, 0.2 mM; pH 5; 10 mM NaCl. (B) Lifetime of the folded 22AG and 22CT, and association time of the duplex (22AG + 22CT). Strand concentration, 0.2 mM. Squares represent the lifetime of folded 22AG at pH 7 in 100 mM KCl (filled) and in 10 mM NaCl (open). Circles represent the lifetime of folded 22CT in 100 mM KCl at pH 6 (filled) and in 10 mM NaCl at pH 5 (open). Crosses represent the association time of the duplex measured in a real-time experiment at pH 5, 100 mM NaCl. The lines are interpolations. In the case of 22AG, multiple structures were present in 100 mM KCl (the sample was first prepared with sodium counterions, then KCl was added) and only the lifetime of the major form was measured. (C) Real-time mixing experiments at 0°C: 22AG and 22CT were mixed and followed with 1D 1H NMR spectra, obtained at different times as indicated. The reference spectrum, labeled ‘ref.’, was obtained at equilibrium (after 1 month). G-quadruplex, i-motif and Watson–Crick duplex structures were followed by imino protons at 10–12, 15–16 and 12–14 p.p.m., respectively. Experimental conditions: strand concentration, 0.2 mM; pH 5; 100 mM NaCl.
The formation (association) of the duplex with Watson–Crick base pairs was followed by NMR at low temperatures (Fig. 3C). The G-strand and C-strand, which form at pH 5 a G-quadruplex and an i-motif, respectively, were mixed and put in the NMR spectrometer. The formation of the Watson–Crick duplex was monitored by the growth of the imino proton peaks at 12–14 p.p.m. The derived time constants at different temperatures are plotted in Figure 3B. Extrapolation to high temperatures of the formation time of the Watson–Crick duplex gives values which are hardly longer than the melting times of the i-motif and of the G-quadruplex. This finding suggests that melting of the structures of the individual strands may be the limiting kinetic step for duplex formation.
Thermodynamics: UV absorption studies
Behavior of the individual strands. The melting profiles of each of the telomeric strands were recorded under various experimental conditions (pH 4.5–7.2, 100 mM NaCl or KCl). The thermal denaturation of the i-motif and of the G-quadruplex gives rise to hyperchromism at 245 nm (Fig. 4A) and to hypochromism at 295 nm (Fig. 4B). The analysis of the 245 and 295 nm profiles led to identical melting temperatures (Tm). All melting profiles were concentration-independent (strand concentration range 1–15 µM), corresponding to monomeric intramolecularly folded species (data not shown).
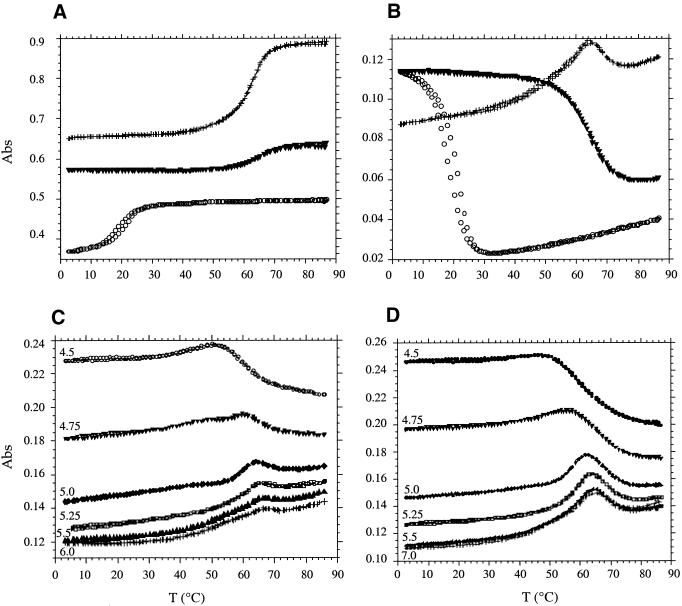
Figure 4.
UV melting profiles, (A) at 245 nm (–0.25 absorbance offset for the 22CT + 22AG mixture) and (B) at 295 nm (–0.02 offset for the mixture). Open circles, 22CT; filled triangles, 22AG; crosses, 22CT + 22AG. Strand concentration, 3 µM; 10 mM sodium cacodylate; 100 mM KCl; pH 6. (C and D) The effect of pH on the 295 nm absorbance of the mixture. Strand concentration, 3 µM; buffer composition: 10 mM sodium cacodylate or acetate; 100 mM NaCl (C) or KCl (D).
The melting of the C-rich strand was strongly pH-dependent, with an optimal pH 4.5 (Tm = 60°C at pH 4.5). Nevertheless, i-motif formation was still clear at pH 6 (Fig. 4A and B, open circles; Tm ∼20°C). The slight hysteresis between the heating and cooling profiles of 22CT at pH 6 and above is the result of slow kinetics of folding and unfolding. The stability of the i-motif was the same in Na+ and K+: the melting temperatures were identical in 100 mM NaCl and 100 mM KCl. The melting temperature of the G-quadruplex was pH-independent in the pH 4.5–7.5 range. The melting profile of 22AG was reversible at all pH values (Fig. 4A and B, triangles). The stability of the G-quadruplex was slightly counterion-dependent, with a Tm of 63°C in 100 mM KCl (Fig. 4A and B, triangles) and 58°C in 100 mM NaCl (data not shown).
Mixing the C- and G-rich telomeric strands. Mixing the two complementary 22CT and 22AG strands could lead to the formation of a 22-bp-long duplex (Fig. 1C), provided that individual strands do not fold into i-motif and G-quadruplex. To test this hypothesis, the two strands were mixed and UV absorbance was recorded as a function of temperature. A typical melting profile is shown in Figure 4A and B (crosses). At 245 nm (Fig. 4A) or 260 nm (data not shown) a clear transition with a large hyperchromism is obtained (Tm = 61°C at pH 6). Raising the strand concentrations led to an increase in the Tm value, as expected for the melting of an intermolecular duplex. The melting temperature was marginally pH-dependent in the pH range 5.25–7.5: 58°C at pH 5.25, 61°C at pH 7.5 (data not shown). In any case, this Tm value is slightly lower than that of the G-quadruplex (63°C under identical conditions).
The 295 nm profile of the mixture (Fig. 4B) is more complex than that of 245 nm (Fig. 4A): the absorbance increases upon cooling from 70 to 64°C and slightly decreases upon cooling below 64°C. To interpret these data, it should be remembered that G-quadruplex formation leads to an increase, whereas duplex formation leads to a slight decrease (or no variation) in absorbance at 295 nm. Therefore, when the mixture is slowly cooled from a high temperature, the increase in absorbance at 295 nm, obtained between 70 and 64°C, corresponds to a partial conversion of the G-rich unfolded strand into a G-quadruplex structure (compare Fig. 4B with A). However, upon cooling below 64°C, absorbance at 295 nm decreases, resulting from the unfolding of the quadruplex and conversion into the duplex. Very similar profiles were obtained in the pH range 5.25–7.0 (Fig. 4C and D). On the other hand, the profiles at pH 4.5 and 4.75 were different: absorbance at 295 nm continues to increase upon lowering the temperature below 64°C, in agreement with the formation of a G-quadruplex below 60°C. Differential absorbance spectra (subtraction of the 4°C absorbance curve from the 87°C absorbance curve) confirmed that duplex formation was complete in the pH range 5.25–7.0 (data not shown).
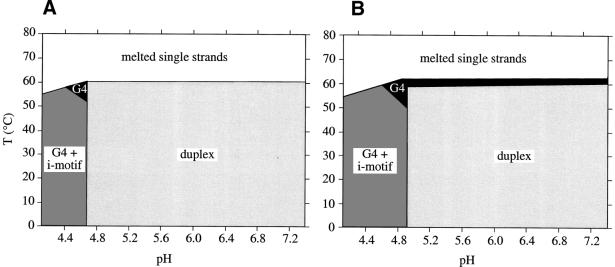
These absorbance curves indicate that a Watson–Crick 22-bp duplex predominates at low or medium temperatures (0–55°C) for pH >5. At pH 4.75 or lower, instead of the duplex, the quadruplex structures predominate. These observations are summarized in Figure 5, where phase diagrams in NaCl and KCl are presented. As shown in Figure 5, the shift from duplex to quadruplex occurs at slightly different values of pH in NaCl and KCl. The slightly higher stability of the G-quadruplex in potassium explains why this transition occurs at pH 4.9 instead of pH 4.7 (in sodium). This small difference in stability values has another consequence for pH >5: in sodium (Fig. 5A) the majority duplex is directly converted to unstructured single strands upon heating the sample, whereas in potassium (Fig. 5B) a quadruplex intermediate is formed.
Figure 5.
Phase diagrams for the 1:1 mixture of 22AG and 22CT in NaCl (A) and in KCl (B). Strand concentration, 3 µM; buffer composition: 10 mM sodium cacodylate or acetate; 100 mM NaCl (A) or KCl (B). The diagram is divided into several areas, which correspond to melted strands (white), duplex (light gray) and quadruplex species (dark gray and black). The dark gray region corresponds to a domain where both quadruplexes [G-quadruplex (G4) and i-motif] coexist, whereas the black region corresponds to a domain where a folded G-quadruplex (G4) coexists with an unfolded C-rich strand. Over 40 independent melting curves were recorded to draw these diagrams. Areas represent only predominant structures and several structures coexist near the boundaries. (For example, at pH 5, 37°C, in KCl the double helix is predominant, but there are also a minority of G-quadruplex and i-motif.) The exact position of some boundaries may also depend on oligonucleotide concentration. At millimolar concentration, the duplex region should be extended towards lower pH and higher temperature.
DISCUSSION
Induction of transition to quadruplexes
At 3 µM strand concentration, the duplex, the Na+ G-quadruplex and the K+ G-quadruplex melt in the same temperature range (61, 58 and 63°C, respectively). This finding suggested that G-quadruplexes could compete with duplex formation, and this assumption was verified at acidic pH, using NMR and UV-absorbance melting experiments. At neutral pH, despite a relatively small difference in ΔG°, the duplex is the predominant species in the 0–55°C temperature range for strand concentrations ranging from 1 to 200 µM. Shifting the equilibrium from a predominant duplex to predominant quadruplexes (G and/or C) may be achieved at neutral pH by G-quadruplex ligands such as a Bisacridine dye (36) for the human telomeric repeat or PIPER for the c-myc promoter and Oxytricha telomere repeat (26).
In accordance with our results, high temperature and high concentration of monovalent ions such as K+ have been reported as favorable conditions for quadruplex transition in G-rich sequences (16,23,25). Monovalent ions have been found to induce a transition from Watson–Crick duplex (and hairpin) to quadruplex (16,23,24). The duplex–quadruplex equilibrium has been also studied for hybrid duplexes, RNA·DNA and peptide nucleic acid (PNA)·DNA (25,27). High temperatures induced a transition of human telomeric 11-bp-long DNA·RNA hybrid duplex to G-quadruplex (25). Inversely, the duplex–quadruplex equilibrium in the case of PNA was shifted in favor of a 7-bp-long PNA·DNA hybrid duplex (27). Here we show that, for a 22-bp-long telomeric duplex, monocations per se are insufficient to promote quadruplex formation: low pH is another factor that may induce such a transition by the contribution of the complementary C-rich strand, which forms an i-motif structure.
Possible structures in vivo
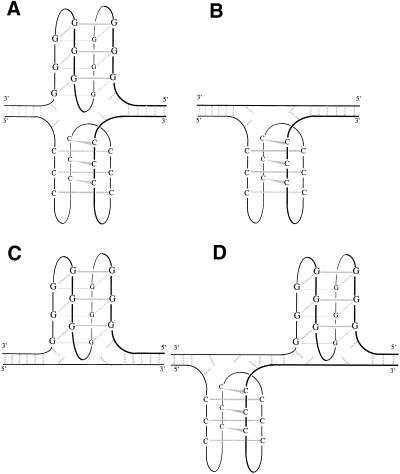
Our results suggest that the Watson–Crick double helix is the predominant form under physiological conditions and that the G-quadruplex and i-motif may be formed under different conditions (high temperatures or low pH). In vivo these structures can occur anywhere along telomeric DNA, permanently or temporarily, either naturally or in a pharmacological context. Stability at neutral or alkaline pH could be enhanced by inter-molecular interactions (proteins–DNA, RNA–DNA, DNA–DNA or drugs–DNA), or by superhelical stress on duplex DNA. The formation of intramolecular G-quadruplex and i-motif can occur separately or together, at the same position or at different locations on a duplex as shown in Figure 6. The existence of some of these structures with Watson–Crick double helices adjacent to i-motif and/or G-quadruplex has been shown (35,37). Intramolecular folding of longer C- and G-rich strands can occur with different i-motif and G-quadruplex topologies (14,21).
Figure 6.
Some possible models of duplex, i-motif and G-quadruplex association for the human telomere repeats. (A) i-motif and G-quadruplex at the same location on duplex. (B) i-motif and duplex. (C) G-quadruplex and duplex. (D) i-motif and G-quadruplex at different places of duplex.
Recently, electron microscopy succeeded in detecting the so-called t-loop structure, formed by strand invasion of the G-rich single strand into the preceding telomeric double-stranded part (38). This invasion would generate a D-loop, which would consist of three strands: two G-strands and one C-strand. More recently, it has been proposed that some terminal duplex portion may also invade the preceding telomeric double-stranded part (39). Therefore, a part of the D-loop would consist of four strands: two G-strands and two C-strands. A number of different nucleic acid structures may be proposed for this three- and/or four-stranded D-loop. Together with double and triple helices, G-quadruplex and i-motif could participate in this structure. For example, in the three-stranded part there could be: (i) a duplex and a G-quadruplex; (ii) an i-motif and a G-quadruplex with two strands; or (iii) an i-motif and two G-quadruplexes.
CONCLUSIONS
The present study revealed that the duplex–quadruplex conversion of human telomeric DNA is under the control of a number of factors, especially pH. Using two complementary techniques (NMR and UV spectroscopy), we could analyze the predominant species under a variety of conditions at millimolar or micromolar strand concentration. An alkali cation switch (Na+ or K+) modulates the position of the transitions. However, even though the Na+ and K+ structures may be significantly different, the phase diagrams obtained in 100 mM KCl or 100 mM NaCl are qualitatively similar, and the phase transition boundaries are marginally affected by the nature of the monocation. In contrast, pH plays a major structural role: low pH stabilizes the folded i-motif structure of the C-rich strand, thus providing a competitor for duplex formation, that acts in concert with G-quadruplex formation on the complementary strand.
Nevertheless, the human telomere repeats are less prone to quadruplex conversion than the c-myc promoter or the Oxytricha telomere motifs (23,26,40): the human telomere motif contains only three consecutive guanines, thus leading to slightly less stable quadruplexes. However, although quadruplexes cannot efficiently compete with duplex formation at pH >5, they may delay the association of two strands, even at millimolar concentration. The relatively small difference in stability between the G-quadruplex and the duplex could explain how quadruplex-specific ligands might prevent duplex formation (36) and/or inhibit telomerase (41).
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Guéron for encouragement during this work and suggestions during the preparation of the manuscript, J. L. Leroy, C. Hélène, T. Garestier, L. Lacroix and P. Alberti for helpful discussions. A.T.P. was supported by the French Ministère des Affaires Etrangères and by the ‘Bourse Varian-Ecole Polytechnique’. This work was partially supported by the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM) and the Association pour la Recherche sur le Cancer (ARC) (grants number 9272 to M. Guéron and 4321 to J.-L.M.).
REFERENCES
- 1.Greider C.W. (1998) Telomerase activity, cell proliferation and cancer. Proc. Natl Acad. Sci. USA, 95, 90–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- 3.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L.C., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.L., Riou,J.F., Mailliet,P., Teulade-Fichou,M.P. and Gilson,E. (2002) Natural and pharmacological regulation of telomerase. Nucleic Acids Res., 30, 839–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyzis R.K., Buckingham,J.M., Cram,L.S., Dani,M., Deaven,L.L., Jones,M.D., Meyne,J., Ratliff,R.L. and Wu,J.R. (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA, 85, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 7.Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 9.Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its applications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 10.Sundquist W.I. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 11.Gehring K., Leroy,J.L. and Guéron,M. (1993) A tetrameric structure with protonated cytosine.cytosine base pairs. Nature, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y. and Patel,D.J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- 13.Phan A.T., Guéron,M. and Leroy,J.L. (2000) The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol., 299, 123–144. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson G.N., Lee,M.P.H. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 15.Hardin C.C., Henderson,E., Watson,T. and Prosser,J.K. (1991) Monovalent cation induced structural transitions in telomeric DNAs: G-DNA folding intermediates. Biochemistry, 30, 4460–4472. [DOI] [PubMed] [Google Scholar]
- 16.Hardin C.C., Watson,T., Corregan,M. and Bailey,C. (1992) Cation-dependent transition between the quadruplex and Watson–Crick hairpin forms of d(CGCG3GCG). Biochemistry, 31, 833–841. [DOI] [PubMed] [Google Scholar]
- 17.Williamson J.R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 18.Kerwin S.M. (2000) G-quadruplex DNA as a target for drug design. Curr. Pharm. Design, 6, 441–471. [DOI] [PubMed] [Google Scholar]
- 19.Leroy J.L., Guéron,M., Mergny,J.L. and Hélène,C. (1994) Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res., 22, 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergny J.L., Lacroix,L., Han,X., Leroy,J.L. and Hélène,C. (1995) Intramolecular folding of pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc., 117, 8887–8898. [Google Scholar]
- 21.Phan A.T. and Leroy,J.L. (2000) Intramolecular i-motif structures of telomeric DNA. J. Biomol. Struct. Dyn., S2, 245–252. [DOI] [PubMed] [Google Scholar]
- 22.Schaffitzel C., Berger,I., Postberg,J., Hanes,J., Lipps,H.J. and Plückthun,A. (2001) In vitro generated antibodies specific for telomeric guanine quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA, 98, 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura T. and Thomas,G.J. (1994) Structural polymorphism of telomere DNA: interquadruplex and duplex–quadruplex conversions probed by Raman spectroscopy. Biochemistry, 33, 7848–7856. [DOI] [PubMed] [Google Scholar]
- 24.Deng H. and Braunlin,W.H. (1995) Duplex to quadruplex equilibrium of the self-complementary oligonucleotide d(GGGGCCCC). Biopolymers, 35, 677–681. [DOI] [PubMed] [Google Scholar]
- 25.Salazar M., Thompson,B.D., Kerwin,S.M. and Hurley,L.H. (1996) Thermally induced DNA.RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry, 35, 16110–16115. [DOI] [PubMed] [Google Scholar]
- 26.Rangan A., Fedoroff,O.Y. and Hurley,L.H. (2001) Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem., 276, 4640–4646. [DOI] [PubMed] [Google Scholar]
- 27.Datta B. and Armitage,B.A. (2001) Hybridization of PNA to structured DNA targets: quadruplex invasion and the overhang effect. J. Am. Chem. Soc., 123, 9612–9619. [DOI] [PubMed] [Google Scholar]
- 28.Li W., Wu,P., Ohmichi,T. and Sugimoto,N. (2002) Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett., 526, 77–81. [DOI] [PubMed] [Google Scholar]
- 29.Phan A.T., Guéron,M. and Leroy,J.L. (2001) Investigations of unusual DNA motifs. Methods Enzymol., 338, 341–371. [DOI] [PubMed] [Google Scholar]
- 30.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 31.Leroy J.L., Gehring,K., Kettani,A. and Guéron,M. (1993) Acid multimers of oligodeoxycytidine strands: stoichiometry, base pair characterization and proton exchange properties. Biochemistry, 32, 6019–6031. [DOI] [PubMed] [Google Scholar]
- 32.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 33.Mills M., Arimondo,P., Lacroix,L., Garestier,T., Hélène,C., Klump,H.H. and Mergny,J.L. (1999) Energetics of strand displacement reactions in triple helices: a spectroscopic study. J. Mol. Biol., 291, 1035–1054. [DOI] [PubMed] [Google Scholar]
- 34.Plateau P. and Guéron,M. (1982) Exchangeable proton NMR without base-line distortion, using strong pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 35.Phan A.T. (1999) Structures and motions of nucleic acids in solution: telomeres and centromeres; water and ions associated to nucleic acids; NMR methods. PhD Thesis, Ecole Polytechnique, Palaiseau, France.
- 36.Alberti P., Ren,J., Teulade-Fichou,M.P., Guittat,L., Riou,J.F., Chaires,J.B., Hélène,C., Vigneron,J.P., Lehn,J.M. and Mergny,J.L. (2001) Interaction of an acridine dimer with DNA quadruplex structures. J. Biomol. Struct. Dyn., 19, 505–513. [DOI] [PubMed] [Google Scholar]
- 37.Ren J., Qu,X., Trent,J.O. and Chaires,J.B. (2002) Tiny telomere DNA. Nucleic Acids Res., 30, 2307–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 39.Stansel R.M., de Lange,T. and Griffith,J.D. (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J., 20, 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddiqui-Jain A., Grand,C.L., Bearss,D.J. and Hurley,L.H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neidle S. and Parkinson,G. (2002) Telomere maintenance as a target for anticancer drug discovery. Nature Rev. Drug Discov., 1, 383–393. [DOI] [PubMed] [Google Scholar]