Abstract
RNA plant viruses use various translational regulatory mechanisms to control their gene expression. Translational enhancement of viral mRNAs that leads to higher levels of protein synthesis from specific genes may be essential for the virus to successfully compete for cellular translational machinery. The control elements have yet to be analyzed for members of the genus Carmovirus, a small group of plant viruses with positive-sense RNA genomes. In this study, we examined the 3′ untranslated region (UTR) of hibiscus chlorotic ringspot virus (HCRSV) genomic RNA (gRNA) and subgenomic RNA (sgRNA) for its role in the translational regulation of viral gene expression. The results showed that the 3′ UTR of HCRSV significantly enhanced the translation of several open reading frames on gRNA and sgRNA and a viral gene in a bicistronic construct with an inserted internal ribosome entry site. Through deletion and mutagenesis studies of both the bicistronic construct and full-length gRNA, we demonstrated that a six-nucleotide sequence, GGGCAG, that is complementary to the 3′ region of the 18S rRNA and a minimal length of 180 nucleotides are required for the enhancement of translation induced by the 3′ UTR.
The 3′ untranslated regions (UTRs) of plant virus RNAs are likely to function in a manner similar to that of the corresponding regions of cellular mRNA in regulating translation and maintaining RNA stability. Due to their great variety in terminal structures, the 3′ UTRs of plus-strand viral RNAs play varied roles in replication and translation (5). For example, the turnip yellow mosaic virus RNA contains a 3′ tRNA mimic believed to regulate minus-strand RNA synthesis (6). For brome mosaic virus, the 3′ UTR harbors a unique promoter element that directs minus-strand RNA synthesis (23). The 3′ UTR of tobacco mosaic virus contains a translation enhancer (8).
Hibiscus chlorotic ringspot virus (HCRSV), a member of the genus Carmovirus, is an isometric monopartite plant virus which measures 28 nm in diameter. The virus is found worldwide where hibiscus is cultivated (16, 41, 42). The symptoms on HCRSV-infected plants range from generalized mottle to chlorotic ringspots and vein-banding patterns, severe stunting, and flower distortion (42, 45). HCRSV possesses a single-stranded positive-sense RNA that is not polyadenylated at the 3′ terminus. The genomic RNA (gRNA) is 3,911 nucleotides (nt) long and has the potential to encode seven open reading frames (ORFs), including two novel ORFs, p23 and p25 (12). Two 3′-coterminated subgenomic RNA (sgRNA) species have been identified.
In this study, we examined the 3′ UTR of HCRSV gRNA and sgRNA for its role in the regulation of translation. Through deletion studies and elimination of putative translation enhancer elements, we were able to identify the prerequisites required for the translational enhancement induced by the 3′ UTR of HCRSV. The results showed that the 3′ UTR of HCRSV differentially enhanced the translation of ORFs on gRNA and sgRNA and an HCRSV gene in a bicistronic construct by 1.5- to 4-fold. Computer predictions and phylogenetic comparisons with other carmoviruses revealed a stable stem-loop structure located at the 3′-terminal region of the HCRSV genome. The influence of this stem-loop on translational enhancement was analyzed by using a series of deletion constructs. In addition, three other putative elements, including a stem-loop structure, a 6-nt element located in the first half of the 3′ UTR, and a CU-rich region located in the second half of the 3′ UTR, were systematically analyzed by deletion and mutagenesis studies. The results revealed that both the stem-loop structure and the CU-rich element were not essential for translational enhancement. Instead, a 6-nt element, GGGCAG, within the first 62 nt of the 3′ UTR was critical. Furthermore, a minimum length of 180 nt was required for the 3′ UTR to function as a translation enhancer.
MATERIALS AND METHODS
PCR and site-directed mutagenesis.
Appropriate primers and template DNA were used in amplification reactions with Pfu DNA polymerase (Strategene) under standard buffer conditions. The PCR conditions were 35 cycles of 94°C for 45 s, 50 to 55°C for 45 s, and 72°C for 1 to 3 min. The annealing temperature and extension time were subjected to adjustments according to the melting temperatures of the primers used and the lengths of the PCR fragments synthesized. Site-directed mutagenesis was carried out with two rounds of PCR and two pairs of primers as previously described (19).
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) of in vitro-translated products was performed with SDS-15 to 17.5% polyacrylamide gels. The labeled polypeptides were detected by autoradiography of dried gels.
In vitro transcription and translation.
Plasmid DNAs were expressed in wheat germ extracts by using a transcription-coupled translation (TnT) system according to manufacturer’s instructions (Promega). Briefly, 1 μg of uncut plasmid DNA was included in a 50-μl reaction mixture which was incubated at 30°C for 60 min in the presence of 50 μCi of [35S]methionine per ml. Equal amounts of templates were used. Reaction products were separated by SDS-PAGE and detected by autoradiography. For in vitro transcription, equal amounts of linearized plasmids were used as templates.
RNA stability test.
The test constructs were pgM23CITE-Δ3′UTR and pgM23CITE-3′UTR. Equal amounts of templates were linearized with SmaI and used for in vitro transcription in the presence of [α-32P]UTP. Equal amounts of radiolabeled RNA transcripts were then added directly to the in vitro translation reaction mixture. At specific time intervals, a portion was withdrawn from each translation reaction mixture, and the RNA present was extracted by phenol-chloroform extraction and ethanol precipitation. The extracted RNAs were then resolved in a 1% agarose gel containing 0.1% SDS. The gel was dried and exposed to an X-ray film for autoradiography.
Densitometry.
The intensities of the protein bands were measured by using a GS-710 calibrated imaging densitometer (Bio-Rad). The bands were analyzed by using Molecular Analyst computer software (Bio-Rad).
RNA secondary structure prediction.
The computer program MFOLD was used to predict the RNA secondary structure of the 3′ UTR of HCRSV. The program was made available from the Bioinformatics Centre, The National University of Singapore.
Plasmid construction.
The construction of plasmid pHCRSV223, which contains the full-length biologically active cDNA clone of HCRSV, was described previously (12). Plasmid pHCRSV223-Δ3′UTR, which has the 3′ UTR of HCRSV removed, was generated from pHCRSV223 by using primer p5(+) and p38SacII(−) for PCR. The fragment was digested with NcoI and SacII at nt 2148 of the HCRSV sequence and at nt 759 of the pBlueScript vector sequence, respectively. Two fragments were ligated into NcoI/SacII-digested pHCRSV223, giving rise to pHCRSV223-Δ3′UTR.
Plasmid pHCRSV223M23 contains a mutation of the AUG codon for p23 in the full-length HCRSV cDNA (12). An internal ribosome entry site (IRES) from plasmid pCITE (Novagen) was cloned into StuI/NcoI-digested pHCRSV223M23, giving rise to pHCRSV223M23CITE. The extra T7 promoter present was removed when the plasmid was digested with BamHI and ligated into BglII/BamHI-digested PING16 (18), giving rise to pgM23CITE-3′UTR. This plasmid was used to construct all deletion plasmids.
Plasmid pgM23CITE-Δ3′UTR was made by cloning an NcoI/BamHI-digested PCR fragment covering the HCRSV sequence from nt 2148 to 3627 into NcoI/BamHI-digested pgM23CITE-3′UTR. The BamHI restriction site of the PCR fragment was introduced by downstream primer BamHI(−). The upstream primer used was p5(+). Plasmid pgM23CITE-5′half was created by PCR with pHCRSV223 as the template. Primers UTR1(−), which introduced a BamHI site, and p5(+) were used. The NcoI/BamHI-digested PCR fragment was ligated into NcoI/BamHI-digested pgM23CITE-Δ3′UTR. Plasmids pgM23CITE-UTR2(−), pgM23CITE-UTR3(−), and pgM23CITE-UTR4(−) were similarly constructed by using downstream primers UTR2(−), UTR3(−), and UTR4(−), respectively, and upstream primer p5(+). Each downstream primer introduced a BamHI site.
Plasmid pgM23CITE-3′half and pgM23CITE-UTR2(+) were created by PCR with pgM23CITE-3′UTR as the template. Upstream primers UTR1(+) and UTR2(+), which introduced a BamHI site, respectively, and a downstream primer, sLKP-3 (5′-CACAGGAAACAGCTATGACCATG-3′), 29 to 51 nt downstream of the BamHI site in pING16 (18), were used. The BamHI-digested PCR fragments were ligated into BamHI-digested pgM23CITE-Δ3′UTR, respectively.
Site-directed mutagenesis with two rounds of PCR and two pairs of primers was used to generate plasmids pgM23CITE-M1, pgM23CITE-M2, pgM23CITE-stem, pHCRSV223-M1, and pHCRSV223-M2. For plasmid pgM23CITE-M1, the first round of PCR included templates pHCRSV223 and pgM23CITE-Δ3′UTR and primer sets p7A(+)-mut3660-1(−) and mut3660-1(+)-sLKP-3, respectively. This step was followed by a second round of PCR with primers p7A(+) and sLKP-3. The PCR fragment was digested with EspI and BamHI and cloned into EspI/BamHI-digested pgM23CITE-Δ3′UTR. Plamids pgM23CITE-M2 and pgM23CITE-stem were similarly generated by using primer set p7A(+)-mut3660-2(+) or stem(+) and primer set sLKP-3-mut3660-2(−) or stem(−), respectively. pHCRSV223 was used as a template to generate plasmids pHCRSV223-M1 and pHCRSV223-M2. For pHCRSV223-M1, primer sets p7A(+)-mut3660-1(−) and mut3660-1(+)-nt3911-SacII(−) were used, followed by a second round of PCR with primers p7A(+) and nt3911-SacII(−). The PCR fragment was digested with EspI and SacII and cloned into EspI/SacII-digested pHCRSV223-Δ3′UTR. pHCRSV223-M2 was similarly constructed. The CU-rich region, from nt 3765 to 3867, was deleted in construct pgM23CITE-ΔCU by using two rounds of PCR. The primers used included p7A(+), ΔCU(+), ΔCU(−), and sLKP-3. The sequences of all the primers used are listed in Table 1. All constructs were confirmed by automated sequencing.
TABLE 1.
Primers used for the construction of plasmids
| Name | Nucleotide sequence (5′-3′)a | Position (nt)b |
|---|---|---|
| UTR1(+) | CAATCAAGGATCCGTACACTATTTTGG | 3627–3765 |
| UTR1(−) | CCCAAAGGATTCTACATTATGTTG | 3751–3911 |
| UTR2(+) | CCCCTGGATCCTGTGCTCCGTCTAAG | 3690–3715 |
| UTR2(−) | ATGGCGGATCCGAGGGTATTC | 3900–3874 |
| UTR3(−) | CAAACAAGGATCCGAAATCCAAGCC | 3873–3850 |
| UTR4(−) | GCGTAGAGGATCCGGGAGAGGAG | 3823–3801 |
| p5(+) | GATCAGCACGTTTCTGTGGA | 1372–1391 |
| p7A(+) | GACGTGGACTTGAGAACCCTCCGG | 2416–2436 |
| mut3660-1(+) | CGTGCTGTTAGGCCAGCTGGAAACGTCGG | 3648–3672 |
| mut3660-1(−) | CCGACGTTTCCAGCTGGCCTAACAGCACG | 3672–3648 |
| mut3660-2(+) | CGTGCTGTTAGGGCGAGTGGAAACGTCGG | 3648–3676 |
| mut3660-2(−) | CCGACGTTTCCACTCGCCCTAACAGCACG | 3676–3648 |
| stem(+) | CCTGGGTAGTCAGGTCCGTCTAAGTAC | 3648–3676 |
| stem(−) | GTACTTAGACGGACCTGACTACCCAGG | 3676–3648 |
| CU(+) | CAAAATAGTGTACGAAATACCCTCACAAC | 3753–3889 |
| CU(−) | GTGAGGGTATTTCGTACACTATTTTGGGAG | 3886–3749 |
| 3911SacII(−) | GCTGGAGCTCCACCGCGGGCTGCCTCAC | 3911–3900 |
Underlining indicates an introduced restriction enzyme site. Bold facing indicates a mutated nucleotide.
Nucleotide position in the HCRSV genome.
RESULTS
The 3′ UTR enhances the expression of gRNA and sgRNA of HCRSV.
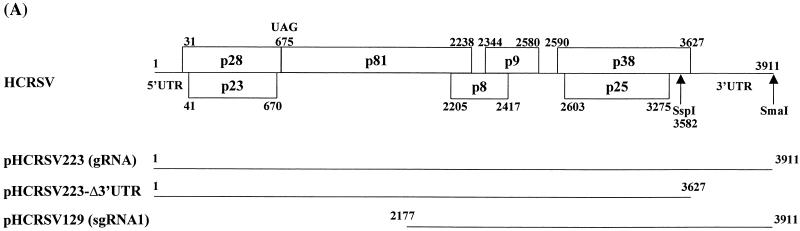
During the course of studying HCRCV gene expression, we found that the 3′ UTR could differentially enhance the translation of various ORFs on gRNA and sgRNA. As shown in Fig. 1B, the expression of pHCRSV223, the full-length cDNA clone with the 3′ UTR, in wheat germ extracts resulted in the detection of four major protein species with apparent molecular masses of 38, 28, 25, and 23 kDa, representing the translation products of ORFs p38, p28, p25, and p23, respectively (Fig. 1A). The expression of pHCRSV223-Δ3′UTR (Fig. 1A), the full-length cDNA of HCRSV without the 3′ UTR, resulted in the detection of the same four protein species with a lower efficiency (Fig. 1B). There was a 1.5- to 2-fold reduction in the expression of p23 and p28, which were located at the 5′ portion of gRNA. Interestingly, the expression of p25 and p38, the two ORFs located at the 3′ portion of gRNA, was reduced by four- to fivefold, indicating that the influence of the 3′ UTR on ORFs located at the 3′ portion of the genome was greater (Fig. 1C). These results confirmed that the 3′ UTR may exert translational enhancement on various ORFs. The mechanism that controls the differential translational enhancement is currently under investigation. Preliminary results showed that the expression of p38 and p25 may be initiated by an alternative mechanism independent of the 5′ end (data not shown).
FIG. 1.
(A) Schematic representation of the cDNA clones of gRNA and sgRNA of HCRSV. Numbers indicate nucleotide positions. (B) Analysis of in vitro translation products of RNAs cotranscribed from nonlinearized templates, pHCRSV223 (3′UTR) and pHCRSV223-Δ3′UTR (Δ3′UTR), encoding the full-length viral sequence in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-15% polyacrylamide gels and visualized by autoradiography. Equal amounts of nonlinearized templates were used. Viral proteins are indicated on the right. (C) Enhancement effects of the 3′ UTR on the translation of ORFs p23, p28, p38, and p25. Percentages are presented as the mean and standard error of the mean (n = 4). The expression of the four proteins from pHCRSV223 (3′UTR) is represented as 100%, and the expression of the same four proteins from pHCRSV223-Δ3′UTR (Δ3′UTR) is shown as a percentage of the expression from pHCRSV223. (D) Analysis of in vitro translation products of RNAs cotranscribed from linearized pHCRSV129/SmaI and pHCRSV129/SspI in a time course experiment (times given above the lanes in minutes) with wheat germ extracts and the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-17.5% polyacrylamide gels and visualized by autoradiography. Molecular markers are indicated on the left, and viral proteins are indicated on the right. Equal amounts of linearized templates were used.
The effect of the 3′ UTR on the translation of sgRNA 1 was then tested by evaluating the expression of SmaI- and SspI-linearized pHCRSV129 in wheat germ extracts in a time course experiment. Five proteins with molecular masses of approximately 38, 25, 24, 22.5, and 9 kDa were synthesized (Fig. 1D). In the absence of the 3′ UTR, the synthesis of each protein was reduced by at least fourfold (Fig. 1D, lanes 5 to 8). These results, which indicated the enhancement of protein expression by the 3′ UTR, were comparable to those obtained for turnip crinkle virus (TCV) (29) and barley yellow dwarf virus (BYDV) (40). The results prompted a systematic analysis of the role of the 3′ UTR in the enhancement of translation.
The 3′ UTR enhances the expression of HCRSV proteins under the control of an IRES in bicistronic constructs.
For a detailed analysis of the enhancement effect of the 3′ UTR, we designed an in vitro system that could faithfully reflect the translational enhancement of the 3′ UTR of gRNA and sgRNA of HCRSV. We reasoned that if we were to insert an IRES downstream of p28, the expression of p28 would be less influenced, if at all, by the 3′ UTR. Hence, the expression of this ORF could serve as an internal control for the translational enhancement effect of the 3′ UTR and for the stability of RNA templates during translation. The expression of downstream ORF p38, which would be initiated by the IRES and influenced by the 3′ UTR, was normalized to the expression of upstream ORF p28 when quantitative data were presented.
Bicistronic construct pgM23CITE-3′UTR was made by introducing the IRES of encephalomyocarditis virus (EMCV) (15) between nt 551 and 2148 in the region containing part of ORF p28 and ORF p81 in pgHCRSV223M23, which contains the cDNA sequence corresponding to gRNA with a mutation of the ATG codon for ORF p23 (Fig. 2A) (12). pgM23CITE-Δ3′UTR was constructed by removing the 3′ UTR from pgM23CITE-3′UTR.
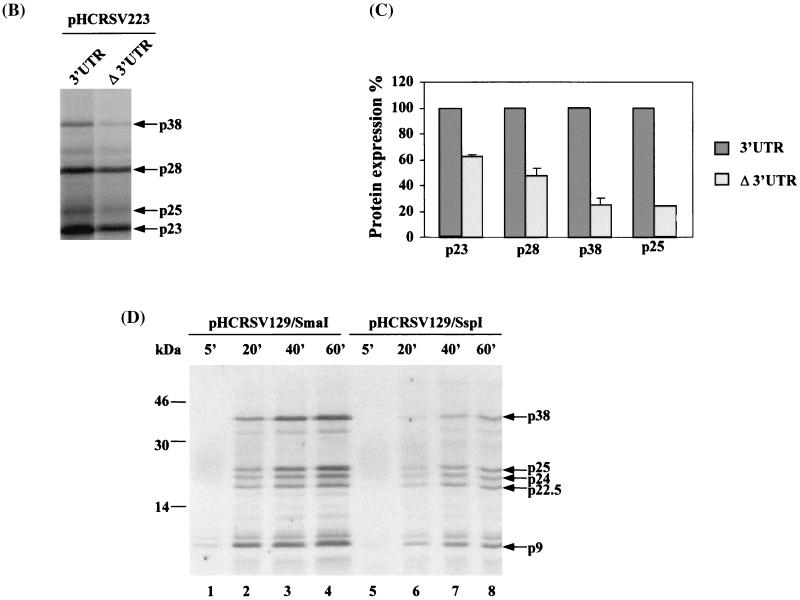
FIG. 2.
(A) Schematic representation of pHCRSV223M23 and the bicistronic constructs pgM23CITE-3′UTR and pgM23CITE-Δ3′UTR. (B) Analysis of in vitro translation products of RNAs cotranscribed from nonlinearized templates, pgM23CITE-3′UTR and pgM23CITE-Δ3′UTR, in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-15% polyacrylamide gels and visualized by autoradiography. Lane M, molecular markers. Proteins are indicated on the right. (C) Enhancement effects of the 3′ UTR on the translation of ORF p38 from pgM23CITE-3′UTR and pgM23CITE-Δ3′UTR. Percentages are presented as the mean and standard error of the mean (n = 4). The expression of p38 from pgM23CITE-3′UTR is represented as 100%, and the expression of p38 from pgM23CITE-Δ3′UTR is shown as a percentage of the expression of p38 from pgM23CITE-3′UTR.
The expression of the two constructs led to the detection of two major protein species with apparent molecular masses of 28 and 38 kDa (Fig. 2B). The 28-kDa protein was the product of upstream ORF p28*, and the 38-kDa protein represented the viral coat protein encoded by ORF p38. It was apparent that the 3′ UTR had no obvious effect on the expression of the 28-kDa protein, as efficient expression of the protein was observed from both constructs. In fact, a higher level of expression of the 28-kDa protein was consistently observed from the translation of pgM23CITE-Δ3′UTR (Fig. 2B). The expression of the 38-kDa protein, however, was under the control of the 3′ UTR; in the absence of the 3′ UTR, much less efficient expression of this protein was observed (Fig. 2B). Quantification and normalization of the two protein bands showed that the expression of the 38-kDa protein from pgM23CITE-Δ3′UTR was 4.2 times less efficient than that from pgM23CITE-3′UTR (Fig. 2C). This expression pattern is consistent with the expression profile expected for ORFs located upstream and downstream of the IRES. These two constructs were then used in subsequent experiments to define the elements within the 3′ UTR required for translational enhancement.
The absence of the 3′ UTR does not significantly affect RNA stability.
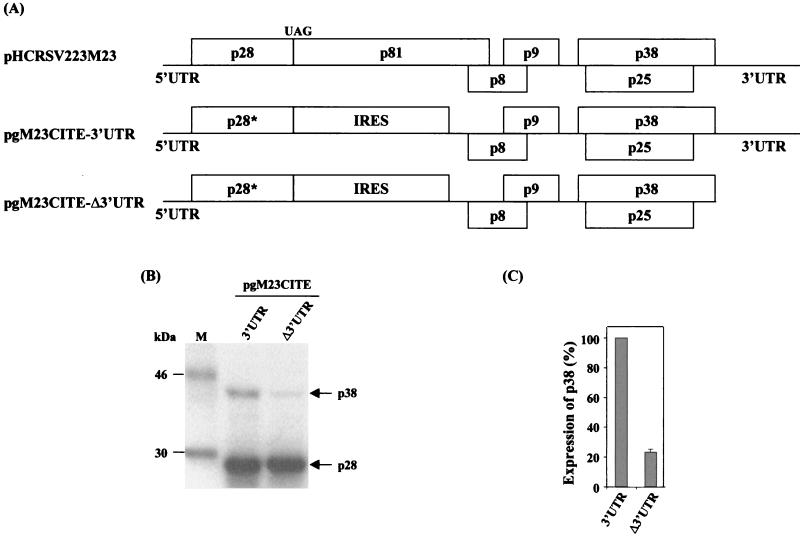
To rule out the possibility that the more efficient expression of the p38 protein from pgM23CITE-3′UTR was due to different stabilities of mRNA during translation, an RNA stability test was carried out. This test showed that the RNA degradation profiles of both transcripts were similar (Fig. 3). The percentage of RNA transcript pgM23CITE-3′UTR decreased from 100 to 26.5% over a period of 60 min, and the percentage of RNA transcript pgM23CITE-Δ3UTR decreased from 100 to 25.8% over the same period. Since very similar degradation rates were observed for both transcripts, it is suggested that the absence of the 3′ UTR did not significantly alter RNA stability in vitro. These results confirmed that the differences in the amounts of p38 protein synthesized were due to the translational enhancement effect exerted by the 3′ UTR.
FIG. 3.
RNA stability tested with bicistronic constructs pgM23CITE-3′UTR and pgM23CITE-Δ 3′UTR. Aliquots of 32P-labeled RNAs were extracted from an in vitro translation reaction mixture at 0, 5, 15, 30, and 60 min, resolved on a 0.1% SDS-1% agarose gel, and visualized by autoradiography. The intensity of each band, as measured by densitometry, is represented as a percentage.
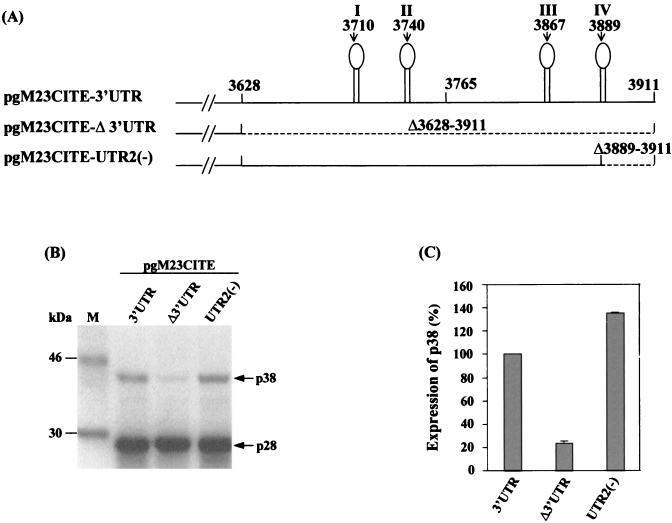
The terminal stem-loop of the 3′ UTR is not essential for translational enhancement.
In order to define the region or potential RNA structures that might be responsible for translational enhancement, a computer analysis of RNA secondary structures was carried out to identify putative elements present within the 3′ UTR. The results showed that the 3′ UTR sequence might form four potential stem-loops (Fig. 4A). The fourth, terminal stem-loop, formed by nt 3876 to 3911, was equivalent to the 3′-terminal stem-loop of TCV, which was identified as the promoter for minus-strand RNA transcription (34). Deletion of nt 3889 to 3911, resulting in the disruption of the stem-loop, was performed to study whether this terminal stem-loop was responsible for the enhancement effect of the 3′ UTR. When the deletion construct pgM23CITE-UTR2(−) was expressed, the level of the p38 protein was comparable to that obtained with pgM23CITE-3′UTR, the construct containing the full-length 3′ UTR (Fig. 4B and C). This result confirmed that this terminal stem-loop was not responsible for the translation enhancement activity of the 3′ UTR. A similar observation was made for TCV; removal of the terminal hairpin structure in its 3′ UTR had a relatively minor effect on translation (29).
FIG. 4.
(A) Schematic representation of pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, and pgM23CITE-UTR2(−). (B) Analysis of the in vitro translation products of RNAs cotranscribed from nonlinearized pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, and pgM23CITE-UTR2(−) in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-15% polyacrylamide gels and visualized by autoradiography. Lane M, molecular markers. Proteins are indicated on the right. (C) Enhancement effects of the 3′ UTR on the translation of ORF p38 from pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, and pgM23CITE-UTR2(−). Percentages are presented as the mean and standard error of the mean (n = 4). The expression of p38 from pgM23CITE-3′UTR is represented as 100%, and the expression of p38 from pgM23CITE-Δ3′UTR and pgM23CITE-UTR2(−) is shown as a percentage of the expression of p38 from pgM23CITE-3′UTR.
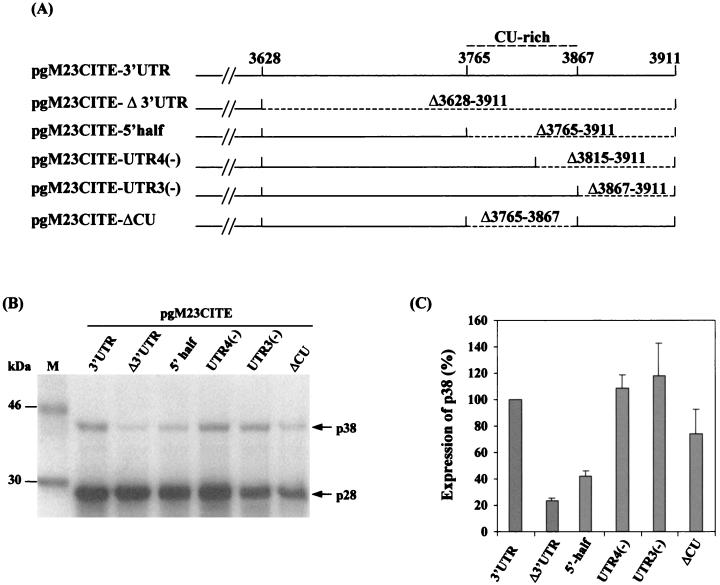
The CU-rich region is not essential for translational enhancement, but a minimum length is required.
We next made three additional constructs with deletions in the second half of the 3′ UTR to further dissect the enhancement effect of the 3′ UTR. Plasmid pgM23CITE-5′half contains a deletion of nt 3765 to 3911, pgM23CITE-UTR4(−) has a deletion of nt 3815 to 3911, and pgM23CITE-UTR3(−) has a deletion of nt 3867 to 3911 (Fig. 5A). The expression of pgM23CITE-5′half resulted in a twofold reduction in the synthesis of the p38 protein compared with that of the control (Fig. 5B and C). The expression of pgM23CITE-UTR4(−) restored the enhancement effect, as p38 protein expression was comparable to that of the control (Fig. 5B and C). Interestingly, the expression of pgM23CITE-UTR3(−) resulted in a 1.4-fold higher level of p38 protein expression than did that of the control, pgM23CITE-3′UTR (Fig. 5B and C). From these deletion studies, we were able to delineate the region of the 3′ UTR responsible for translational enhancement to its first 188 nt; subsequent data further narrowed this region to 180 nt. This observation was consistent with results obtained for TCV, for which enhancement activity was localized to the first 155 nt of the 3′ UTR (29).
FIG. 5.
(A) Schematic representation of pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-5′half, pgM23CITE-UTR4(−), pgM23CITE-UTR3(−), and pgM23CITE-ΔCU. (B) Analysis of the in vitro translation products of RNAs cotranscribed from nonlinearized pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-5′half, pgM23CITE-UTR4(−), pgM23CITE-UTR3(−), and pgM23CITE-ΔCU in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-5% polyacrylamide gels and visualized by autoradiography. Lane M, molecular markers. Proteins are indicated on the right. (C) Enhancement effects of the 3′ UTR on the translation of ORF p38 from pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-5′half, pgM23CITE-UTR4(−), pgM23CITE-UTR3(−), and pgM23CITE-ΔCU. Percentages are presented as the mean and standard error of the mean (n = 4). The expression of p38 from pgM23CITE-3′UTR is represented as 100%, and the expression of p38 from pgM23CITE-Δ3′UTR, pgM23CITE-5′half, pgM23CITE-UTR4(−), pgM23CITE-UTR3(−), and pgM23CITE-ΔCU is shown as a percentage of the expression of p38 from pgM23CITE-3′UTR.
A CU-rich sequence similar to a pyrimidine-rich domain identified in the 3′ UTR of hepatitis C virus (HCV) was also found in the 3′ UTR of HCRSV, at nt 3765 to 3880. The CU-rich sequence of HCV was implicated in high-affinity binding to a host factor called pyrimidine tract binding (PTB) protein, which enhances translation initiated from the 5′ IRES (13). Construct pgM23CITE-UTR3(−), which contains the entire CU-rich region, enhanced the expression of the p38 protein by 1.5 times that seen with the control, pgM23CITE-3′UTR; however, with construct pgM23CITE-UTR4(−), in which half of the CU-rich region is removed, translational enhancement was comparable to that of the control (Fig. 5B and C). Deletion of the CU-rich region in construct pgM23CITE-ΔCU did not result in a reduction in p38 protein expression, indicating that it had no obvious effect on translational enhancement. Therefore, the CU-rich region is not essential for translational enhancement, but a minimum length of 180 nt is needed.
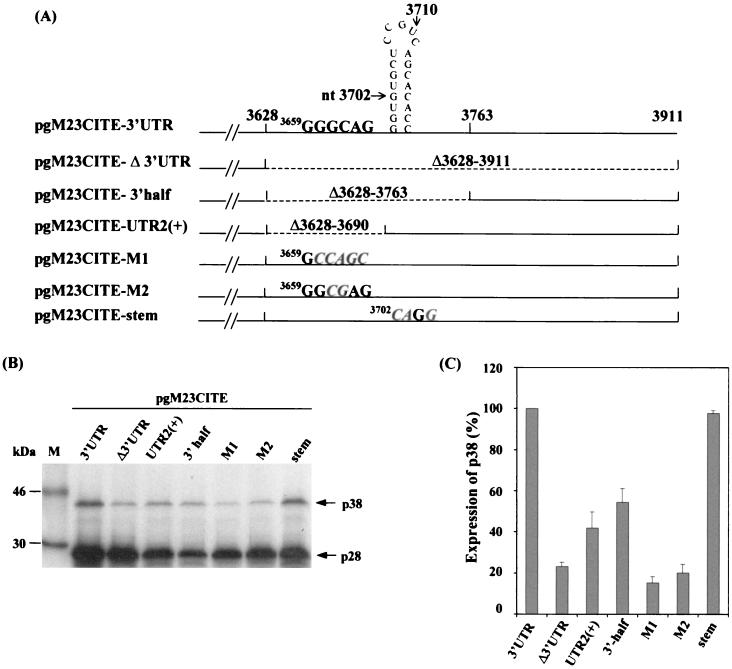
A 6-nt sequence (GGGCAG) that is complementary to the 3′ region of the 18S rRNA is essential for translational enhancement.
Deletion and mutagenesis studies were carried out to study the enhancer role of the first half of the 3′ UTR. Two deletion constructs, pgM23CITE-3′half and pgM23CITE-UTR2(+), which contain deletions of nt 3628 to 3763 and nt 3628 to 3690, respectively, were made and expressed (Fig. 6A). The results showed that p38 protein expression was reduced by twofold compared with that seen with the control, pgM23CITE-3′UTR (Fig. 6B), demonstrating that either the primary sequence or certain structural elements in this region of the 3′ UTR are essential for translational enhancement. A computer analysis revealed two interesting elements in this region of the 3′ UTR that may be relevant. The first element is a putative stem-loop structure (nt 3699 to 3719), and the second is a 6-nt element (GGGCAG) (nt 3659 to 3664) that is complementary to the 3′ region of the 18S rRNA of Nicotiana tabacum (nt 1635 to 1640).
FIG. 6.
(A) Schematic representation of pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-3′half, pgM23CITE-UTR2(+), pgM23CITE-M1, pgM23CITE-M2, and pgM23CITE-stem. (B) Analysis of the in vitro translation products of RNAs cotranscribed from nonlinearized pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-3′half, pgM23CITE-UTR2(+), pgM23CITE-M1, pgM23CITE-M2, and pgM23CITE-stem in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-15% polyacrylamide gels and visualized by autoradiography. Lane M, molecular markers. Proteins are indicated on the right. (C) Enhancement effects of the 3′ UTR on the translation of ORF p38 from pgM23CITE-3′UTR, pgM23CITE-Δ3′UTR, pgM23CITE-3′half, pgM23CITE-UTR2(+), pgM23CITE-M1, pgM23CITE-M2, and pgM23CITE-stem. Percentages are presented as the mean and standard error of the mean (n = 4). The expression of p38 from pgM23CITE-3′UTR is represented as 100%, and the expression of p38 from pgM23CITE-Δ3′UTR, pgM23CITE-3′half, pgM23CITE-UTR2(+), pgM23CITE-M1, pgM23CITE-M2, and pgM23CITE-stem is shown as a percentage of the expression of p38 from pgM23CITE-3′UTR.
The predicted stem-loop structure located at nt 3699 to 3719 was tested by using mutant pgM23CITE-stem, which was made by mutating 3 nt within the stem from 3702GTGC to 3702CAGG (Fig. 6A). The level of p38 protein expression from pgM23CITE-stem was comparable to that from the control, pgM23CITE-3′UTR (Fig. 6B and C). This observation either rules out the existence of the stem-loop structure or suggests that such a predicted secondary structure element may not be responsible for translational enhancement.
Mutants pgM23CITE-M1 and pgM23CITE-M2, in which the nucleotide sequences are changed from 3659GGGCAG to 3659GCCAGC and from 3659GGGCAG to 3659GGCGAG, respectively, were made and expressed (Fig. 6A). The expression of the p38 protein was reduced by at least fourfold for both mutants (Fig. 6B and C). These results strongly suggest that the 6-nt sequence is crucial for the enhancer role of the 3′ UTR.
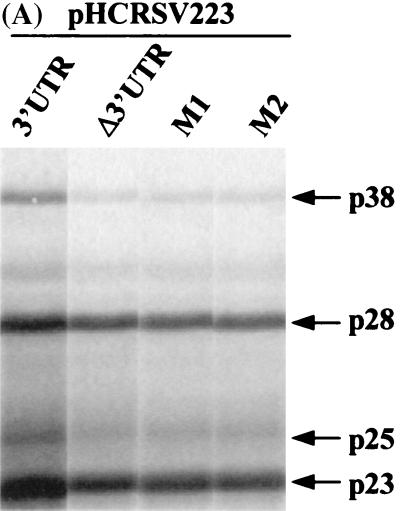
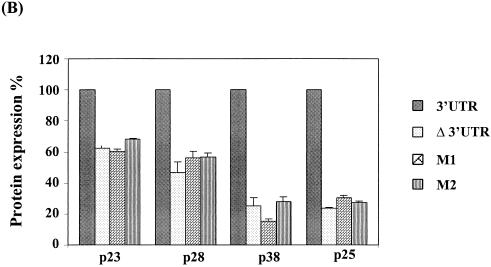
To confirm the effects of these mutations on the expression of various ORFs of gRNA, the same two sets of mutations in the 6-nt sequence were introduced into the full-length cDNA clone, generating mutants pHCRSV223-M1 and pHCRSV223-M2. The expression of pHCRSV223-M1 and pHCRSV223-M2 resulted in 1.6- to 2-fold reductions in the synthesis of the p23 and p28 proteins, whereas the expression of both p38 and p25 proteins was reduced by 4- to 6-fold, compared to that seen with the control, pHCRSV223 (Fig. 7). The expression profiles for the two mutants were similar to that for pHCRSV223-Δ3′UTR, which contains the full-length cDNA with its 3′ UTR deleted. The mutations in the 3′ UTR had a stronger influence on the expression of the p25 and p38 proteins, which were located at the 3′ portion of the genome, rather than on that of the p23 and p28 proteins, which were located at the 5′ portion. These results demonstrated that mutations of the GGGCAG sequence in both the full-length cDNA and the biscitronic constructs were sufficient to cause a drastic reduction in translational enhancement.
FIG. 7.
(A) Analysis of in vitro translation products of RNAs cotranscribed from nonlinearized pHCRSV223 (3′UTR), pHCRSV223-Δ3′UTR, pHCRSV223-M1, and pHCRSV223-M2 in wheat germ extracts with the TnT coupled translation system. [35S]methionine-labeled translation products were separated on SDS-15% polyacrylamide gels and visualized by autoradiography. Equal amounts of nonlinearized templates were used. Viral proteins are indicated on the right. (B) Enhancement effects of the 3′ UTR on the translation of ORFs p23, p28, p38, and p25. Percentages are presented as the mean and standard error of the mean (n = 4). The expression of the four proteins from pHCRSV223 is represented as 100%, and the expression of the same four proteins from pHCRSV223-Δ3′UTR, pHCRSV223-M1, and pHCRSV223-M2 is shown as a percentage of the expression from pHCRSV223.
DISCUSSION
Viruses, being obligated parasites, depend on the host for replication. They must compete with cellular mRNAs for translation and have thus evolved diverse mechanisms to redirect the translation machinery to favor viral transcripts (7). RNA viruses that lack a cap and/or a poly(A) tail have developed alternative strategies for translation regulation involving the 3′ UTR (9, 22). The roles of the 3′ UTR in translational enhancement of viral RNAs have been studied with human and animal viral systems, including HCV (13) and rotavirus (4). Translation enhancer elements within the 3′ UTRs of plant viruses that lack both the cap and the poly(A) tail have been implicated as determinants for translation efficiency in tomato bushy stunt virus (46), satellite necrosis virus (22), BYDV (1, 40), and TCV (29). In this study, we provided evidence that the expression of several viral proteins from gRNA and sgRNA of HCRSV is enhanced by at least fourfold in the presence of the 3′ UTR. Translational enhancement was also detected in a bicistronic construct, which was used to delineate the region of the 3′ UTR responsible. Deletion and mutagenesis studies showed that a 6-nt sequence (GGGCAG) in the 3′ UTR was an essential element of the translation enhancer. Furthermore, neither the terminal stem-loop nor a CU-rich region in the second half of the 3′ UTR is essential for translational enhancement, but a minimal length of 180 nt is required.
In this study, bicistronic constructs containing the EMCV IRES were used in the initial deletion and mutagenesis studies. It was noted that the EMCV IRES could function in wheat germ extracts, but with much reduced activity. As relatively efficient expression of downstream ORF p38 was observed, we do not rule out the possibility that an alternative initiation signal may exist on gRNA and sgRNA and may be responsible for the expression of the downstream ORFs (p38, p25, and others). However, the conclusions drawn from studies with bicistronic constructs, especially the role of the 6-nt sequence in translational enhancement, could apply to the full-length construct, justifying the use of these bicistronic constructs in the deletion and mutagenesis studies. The potential existence of an alternative initiation signal may also offer an explanation for the preferential regulation of the expression of different ORFs by the 3′ UTR. Experiments are being carried out to investigate and define this signal.
How does the 3′ UTR affect the translation efficiency of a given mRNA? Communication between the 5′ and 3′ termini resulting in a circularized mRNA has been demonstrated to enhance translation (14, 31). Evidence has shown that through the participation of the poly(A) tail binding protein (14), the 3′ poly(A) tail interacts synergistically with the 5′ cap to promote translation initiation and increase translation efficiency by forming a “closed-loop” initiation complex (14, 31). Circularization of mRNA is thought to enhance translation by allowing the initiation of new rounds of translation to occur rapidly (43). This activity is crucial when there is competition between viral mRNAs and cellular mRNAs for translation machinery (27) or when the availability of ribosomes or initiation factors becomes limited (28). Here we examined two such elements that could possibly bring about such an interaction.
The CU-rich element in the 3′ UTR of HCRSV is similar to the pyrimidine-rich domain in HCV that is implicated in high-affinity binding to the host PTB protein. Binding of PTB to the CU-rich region of the 3′ UTR of HCV enhances translation initiated from the 5′ IRES (13). Among carmoviruses, such host factors involved in translational enhancement have yet to be identified. It has been suggested that for two defined examples of cap-independent translational enhancement in TCV (29), BYDV (40), and satellite necrosis virus (21), PTB and other host factors may be involved. However, a recent study by Murakami et al. (24) showed that deletion of this CU-rich region in the 3′ UTR of HCV did not down-regulate translation; instead, it resulted in enhancement of translation efficiency. In our study, the deletion of a CU-rich region within the 3′ UTR did not reduce translational enhancement, indicating that the CU-rich region is not involved in enhancing translation.
Secondary structures or tertiary interactions within the viral 3′ UTR, such as the pseudoknot domain in tobacco mosaic virus (8), have been shown to enhance translation via interaction with an RNA binding protein that interacts with both the 5′ and the 3′ ends of the viral mRNA (36). Of the four predicted stem-loop structures of the 3′ UTR of HCRSV, the fourth, terminal stem-loop formed by nt 3876 to 3911 was tested, as it was the most stable predicted structure that has been shown to be conserved among all carmoviruses. This conserved structure has been shown to be functionally important for viral RNA replication as the promoter for minus-strand RNA transcription (34). Disruption of this predicted structure in HCRSV did not reduce translation efficiency. A similar observation was made for TCV when removal of the terminal hairpin structure had a relatively minor effect on translational enhancement (29).
The second possible mechanism that may facilitate the 5′-3′ communication is the existence of sequences in the 3′ UTR that may base pair to the 18S rRNA. Mauro and Edelman (20) have shown the existence of rRNA-like sequences in many eukaryotic mRNAs in both sense and antisense orientations in the UTRs. These short sequences complementary to the 3′ region of the 18S rRNA have been shown to promote translation initiation (37, 39). These include the mRNA encoding ribosomal protein S15 (38) and homeodomain Gtx (11), which have been shown to contain several sequence motifs complementary to the 3′ end of the 18S rRNA and that are able to UV cross-link with the 40S subunit. Recently, a 9-nt element in the 5′UTR of Gtx mRNA complementary to the 18S rRNA was found to contain IRES activity and to increase translation efficiency by threefold (3).
In this study, we identified a 6-nt sequence (GGGCAG) within the HCRSV 3′ UTR that could possibly base pair directly to the 3′ region of 18S rRNA. Mutations of this primary sequence in both the full-length cDNA and the bicistronic constructs resulted in a drastic reduction in translation efficiency. The mutations abolished the translational enhancement effect exerted by the 3′ UTR on the expression of viral products. The influence of the 3′ UTR was more pronounced on the expression of the ORFs located at the 3′ region of the genome than on the expression of those located at the 5′ region. These results strongly indicate the involvement of the 6-nt sequence (GGGCAG) in the translational enhancement of ORFs located at the 3′ portion of gRNA. It has been suggested that such complementary sequences allow the binding of the 40S ribosomal subunit to the 5′UTR of viral RNAs in a cap-independent manner (17, 25, 26). As the influence of this 6-nt segment was stronger on the 3′ ORFs, it is likely that the translation initiation of ORFs located in the 3′ portion of gRNA is brought about by interaction of the 3′ UTR with a possible IRES that could involve RNA-RNA base pairing with 18S rRNA.
This primary sequence was conserved in carmoviruses such as cardamine chlorotic fleck virus (33), cowpea mottle virus (47), melon necrotic spot virus (30), carnation mottle virus (10), saguaro cactus virus (44), TCV (2), and other plant viruses lacking a poly(A) tail. The potential binding of the 3′ UTR of HCRSV to the 18S rRNA resembles the Shine-Dalgarno mRNA-ribosome interactions in the prokaryotic system (32), where translation initiation occurs via interactions between the 16S rRNA and the polycistronic mRNA. Such interactions, shown to involve initiation factors and to promote the binding of mRNA to the 30S ribosomal subunit in prokaryotes (35), would probably bring both the 5′ and the 3′ ends together for translation to be initiated at the 5′ end.
Acknowledgments
This research was supported by the National Science and Technology Board, Singapore, and The National University of Singapore (research grant R-154-000-111-112).
REFERENCES
- 1.Allen, E. S., S. Wang, and W. A. Miller. 1999. Barley yellow dwarf mosaic virus RNA requires cap-independent translation sequence because it lacks a 5′ cap. Virology 253:139–144. [DOI] [PubMed] [Google Scholar]
- 2.Carrington, J. C., L. A. Heaton, D. Zuidema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 139:22–31. [DOI] [PubMed] [Google Scholar]
- 3.Chappell, S. A., G. M. Edelman, and V. P. Mauro. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosomal entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 97:1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chizhikov, V., and J. T. Patton. 2000. A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151–174. [DOI] [PubMed] [Google Scholar]
- 6.Dreher, T. W., C. H. Tsai, C. Florentz, and R. Giege. 1992. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry 31:9183–9189. [DOI] [PubMed] [Google Scholar]
- 7.Gale, M., Jr., S. L. Tan, and M. G. Katz. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108–2116. [DOI] [PubMed] [Google Scholar]
- 9.Gallie, D. R. 1998. The tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1–11. [DOI] [PubMed] [Google Scholar]
- 10.Guilley, H., J. C. Carrington, E. Balaz, G. Jonard, K. Richards, and T. J. Morris. 1985. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 13:6663–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, M. C., P. Tranque, G. M. Edelman, and V. P. Mauro. 1999. rRNA-complementarity in the 5′ untranslated region of mRNA specifying the Gtx homeodomain protein: evidence that base-pairing to 18S rRNA affects translational efficiency. Proc. Natl. Acad. Sci. USA 96:1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, M., D. C. Y. Koh, L. J. Weng, M. L. Chang, L. Zhang, and S. M. Wong. 2000. Complete nucleotide sequence and genome organization of hibiscus chlorotic ringspot virus, a new member of the genus Carmovirus: evidence for the presence and expression of two novel open reading frames. J. Virol. 74:3149–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., S. M. Tahara, and M. M. C. Lai. 1998. The 3′ untranslated region of the hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson, A. 1996. Poly(A) metabolism and translation: the closed loop model, p.451–480. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 15.Jang, S. K., H. G. Krausslic, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, D. R., and G. M. Behncken. 1980. Hibiscus chlorotic ringspot, a widespread virus disease in the ornamental hibiscus rosa-sinensis. Aust. J. Plant Pathol. 9:4. [Google Scholar]
- 17.Le, S. Y., J. H. Chen, N. Sonenberg, and J. V. Maizel. 1992. Conserved tertiary structure elements in the 5′ untranslated region of human enteroviruses and rhinoviruses. Virology 191:858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, D. X., D. Cavanagh, P. Green, and S. C. Inglis. 1991. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology 184:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, D. X., H. Y. Xu, and T. D. K. Brown. 1997. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 71:1814–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mauro, V. P., and G. M. Edelman. 1997. rRNA-like sequences occur in diverse primary transcripts: implications for control of gene expression. Proc. Natl. Acad. Sci. USA 94:422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meulewaeter, F., X. Danthinne, M. V. Montagu, and M. Cornelissen. 1998. 5′ and 3′ sequences of the satellite necrosis virus RNA promoting translation in tobacco. Plant J. 14:169–176. [DOI] [PubMed] [Google Scholar]
- 22.Meulewaeter, F., Van Montagu, M., and M. Cornelissen. 1998. Features of autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA 4:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537–546. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, K., M. Abe, T. Kageyama, N. Kamoshita, and A. Nomoto. 2001. Down regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729–741. [DOI] [PubMed] [Google Scholar]
- 25.Pestova, T. V., C. U. T. Hellen, and E. Wimmer. 1991. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 65:6194–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, Y. V. Svitkin, A. N. Sinyakov, and V. I. Agol. 1992. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68:119–131. [DOI] [PubMed] [Google Scholar]
- 27.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail promoted translation in yeast. Nature 392:516–520. [DOI] [PubMed] [Google Scholar]
- 28.Proweller, A., and S. Butler. 1997. Ribosome concentration contributes to discrimination against poly(A)− mRNA during translation initiation in Saccharomyces cerevisiae. J. Biol. Chem. 272:6004–6010. [DOI] [PubMed] [Google Scholar]
- 29.Qu, F., and T. J. Morris. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 74:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riviere, C. J., and D. M. Rochon. 1990. Nucleotide sequence and genome organization of melon necrotic spot virus. J. Gen. Virol. 71:1887–1896. [DOI] [PubMed] [Google Scholar]
- 31.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting from the beginning, middle and end: translation initiation in eukaryotes. Cell 89:831–838. [DOI] [PubMed] [Google Scholar]
- 32.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribisomal RNA: complementarity to nonsense triplets bad ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skotnicki, M. L., A. M. Mackenzie, M. Torronen, and A. J. Gibbs. 1993. The genome sequence of cardamine chlorotic fleck carmovirus. J. Gen. Virol. 74:1933–1937. [DOI] [PubMed] [Google Scholar]
- 34.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6–14. [DOI] [PubMed] [Google Scholar]
- 35.Steinz, J. A. 1975. Ribosome recognition of initiator regions in the RNA bacteriophage genome, p.319–352. In N. D. Zinder (ed.), RNA phages. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Tanguay, R. L., and D. R. Gallie. 1996. Isolation of characterization of the 102-kilodalton RNA-binding protein that binds to the 5′ and 3′ translational enhancers of tobacco mosaic virus RNA. J. Biol. Chem. 271:14316–14322. [DOI] [PubMed] [Google Scholar]
- 37.Teerink, H., H. O. Voorma, and A. A. Thomas. 1995. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim. Biophys. Acta 1264:403–408. [DOI] [PubMed] [Google Scholar]
- 38.Tranque, P., M., M. C. Hu, G. M. Edelman, V. P. Mauro. 1998. rRNA complementarity within mRNAs: a possible basis for mRNA-ribosome interactions and translational control. Proc. Natl. Acad. Sci. USA 95:12238–12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagner, S., M. C. Gensac, A. Maret, F. Bayard, F. Amalric, H. Prats, and A. C. Prats. 1995. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 15:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S., K. S. Browning, and W. A. Miller. 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 16:4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterworth, H. E. 1980. Hibiscus chlorotic ringspot virus. CMI/AAB descriptions of plant viruses no. 227. Association of Applied Biologists, Warwick, United Kingdom.
- 42.Waterworth, H. E., R. H. Lawson, and R. L. Monroe. 1976. Purification and properties of hibiscus chlorotic ringspot virus. Phytopathology 64:570–575. [Google Scholar]
- 43.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135–140. [DOI] [PubMed] [Google Scholar]
- 44.Weng, Z., and Z. Xiong. 1997. Genome organization and gene expression of saguaro cactus carmovirus. J. Gen. Virol. 78:525–534. [DOI] [PubMed] [Google Scholar]
- 45.Wong, S. M., and C. G. Chng. 1992. Occurrence of hibiscus chlorotic ringspot virus in Singapore. Phytopathology 82:722.
- 46.Wu, B., and K. A. White. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 73:8982–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You, X. J., J. W. Kim, G. W. Stuart, and R. F. Borzarth. 1995. The nucleotide sequence of cowpea mottle virus and its assignment to the genus Caromovirus. J. Gen. Virol. 76:2841–2845. [DOI] [PubMed] [Google Scholar]