Abstract
In an attempt to identify the rotavirus receptor, we tested 46 cell lines of different species and tissue origins for susceptibility to infection by three N-acetyl-neuraminic (sialic) acid (SA)-dependent and five SA-independent rotavirus strains. Susceptibility to SA-dependent or SA-independent rotavirus infection varied depending on the cell line tested and the multiplicity of infection (MOI) used. Cells of renal or intestinal origin and transformed cell lines derived from breast, stomach, bone, or lung were all susceptible to rotavirus infection, indicating a wider host tissue range than previously appreciated. Chinese hamster ovary (CHO), baby hamster kidney (BHK-21), guinea pig colon (GPC-16), rat small intestine (Rie1), and mouse duodenum (MODE-K) cells were found to support only limited rotavirus replication even at MOIs of 100 or 500, but delivery of rotavirus particles into the cytoplasm by lipofection resulted in efficient rotavirus replication. The rotavirus cell attachment protein, the outer capsid spike protein VP4, contains the sequence GDE(A) recognized by the VLA-2 (α2β1) integrin, and to test if VLA-2 is involved in rotavirus attachment and entry, we measured infection in CHO cells that lack VLA-2 and CHO cells transfected with the human α2 subunit (CHOα2) or with both the human α2 and β1 subunits (CHOα2β1) of VLA-2. Infection by SA-dependent or SA-independent rotavirus strains was 2- to 10-fold more productive in VLA-2-expressing CHO cells than in parental CHO cells, and the increased susceptibility to infection was blocked with anti-VLA-2 antibody. However, the levels of binding of rotavirus to CHO, CHOα2, and CHOα2β1 cells were equivalent and were not increased over binding to susceptible monkey kidney (MA104) cells or human colonic adenocarcinoma (Caco-2, HT-29, and T-84) cells, and binding was not blocked by antibody to the human α2 subunit. Although the VLA-2 integrin promotes rotavirus infection in CHO cells, it is clear that the VLA-2 integrin alone is not responsible for rotavirus cell attachment and entry. Therefore, VLA-2 is not involved in the initial attachment of rotavirus to cells but may play a role at a postattachment level.
Group A rotaviruses are important viral agents of acute gastroenteritis in young children and many animal species (24). Two surface proteins, VP4 and VP7, are present on the outer capsid of rotaviruses, and trypsin cleavage of the spike protein VP4 into VP5* and VP8* enhances rotavirus infectivity and is involved in the early interaction of the virus with the cell (18, 67). Rotaviruses infect the mature enterocytes of the villus epithelium of the small intestine. This restricted tissue- and cell-type-specific tropism is mediated by one or more specific host cell surface receptors that mediate rotavirus attachment. Numerous studies have implicated glycoconjugates (glycoproteins, glycolipids, and glycosphingolipids) as the putative rotavirus receptor(s) (4, 19, 32, 34, 41, 55, 58, 59, 66). A minority of animal rotaviruses require the presence of N-acetyl-neuraminic (sialic) acid (SA) residues on the cell surface for efficient binding and infectivity, but most animal and human rotaviruses are SA independent (13). Binding of rotaviruses to SA residues has been mapped to amino acids 93 to 208 of the VP4 cleavage product VP8* (38). However, the isolation of SA-independent animal rotavirus strain variants from SA-dependent strains that are efficient in attachment and infection confirms that binding to SA is not an essential step for rotavirus infection (13, 48). Therefore, an SA-independent rotavirus receptor is likely involved in entry of both SA-dependent (after an initial interaction with SA residues) and SA-independent rotavirus strains, whose binding to cells is mediated by VP5* (67). At least three cell surface sites or molecules may be involved during early cell-rotavirus interactions, suggesting there are common steps during the entry of SA-dependent and SA-independent rotaviruses (49).
The integrins are a large family of transmembrane glycoproteins containing two large subunits (α and β) that mediate interactions with extracellular matrix components and with other cells. These heterodimeric glycoproteins act as bidirectional signaling receptors and can serve as cellular receptors or coreceptors for several viruses, including echoviruses 1 and 8 (8, 9), coxsackievirus A9 (53), foot-and-mouth disease virus (39, 40), adenoviruses (65), and papillomavirus (25). Recently, VLA-2 (α2β1), VLA-4 (α4β1), CR4 (αxβ2), and αvβ3 integrins have been implicated in rotavirus cell attachment, entry, or postattachment events in infection (17, 33, 36, 44, 68). The rotavirus outer capsid proteins VP4 and VP7 contain sequence motifs that are potentially recognized by VLA-2, VLA-4, and CR4 but not αvβ3, and antibodies to these integrins or peptides containing these recognition motifs block approximately 30 to 50% of rotavirus infectivity (17, 33, 36, 44). These studies have shown that (i) VLA-2 and VLA-4 expressed in poorly susceptible human erythroleukemic K562 cells increase binding and infectivity of the SA-dependent SA11 strain (36), (ii) the susceptibility to rotavirus infection correlates with the amount of VLA-2 expressed on the surfaces of six human cell lines exhibiting various degrees of susceptibility to rotavirus infection (44), (iii) VLA-2 is the initial receptor for a neuraminidase-resistant (SA-independent) nar3 variant but not for the parental SA-dependent RRV strain (68), and (iv) αvβ3 promotes rotavirus cell entry at a postbinding step (33).
In an attempt to identify a cell line that lacks the rotavirus cellular receptor, we tested 46 cell lines of different species and tissue origins and observed a wider host and tissue range than had been previously recognized (24). Among all the cells tested, five cell lines, including Chinese hamster ovary (CHO) cells, exhibited limited rotavirus susceptibility compared to fully permissive cells, but delivery of rotavirus particles into the cytoplasm of these cells by lipofection resulted in productive rotavirus infection, suggesting an entry level obstacle to infection. We examined whether the resistance of CHO cells to rotavirus infection was a consequence of the lack of VLA-2 expression in these cells. We found that CHO cells became 2- to 10-fold more susceptible to rotavirus infection (although a high multiplicity of infection [MOI] still had to be used) when transfected with the human α2 subunit or with both the human α2 and β1 subunits of the VLA-2 integrin. The VLA-2 integrin promoted rotavirus infection in CHO cells. However, VLA-2 did not increase binding, suggesting that VLA-2 alone is not responsible for cellular susceptibility to rotaviruses, although it may function in a postattachment event in rotavirus infection.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The characteristics, species and tissues of origin, growth media, and sources of the cell lines used in this study are listed in Table 1. SA-dependent simian SA11 Cl3 (P5B[2], G3) and RRV (P5B[3], G3), equine H-1 (P9[7], G5), bovine NCDV (P6[1], G6), and porcine OSU (P9[7], G5), SA-independent human Wa (P1A[8], G1), DS-1 (P1B[4], G2), HAL1166 (P11[14], G8), and PA169 (P11[14], G6), equine H-2 (P4[12], G3), lapine ALA (P11[14], G3), BAP-2 (P11[14], G3), and C-11 (P11[14], G3), and bovine WC3 (P7[5], G6) rotavirus strains and the SA-independent SA11Cl3 rotavirus variant 954/23R (SA11 Cl3 954/23R) strain were cultivated in rhesus monkey kidney cells (MA104) in the presence of trypsin as described previously (12).
TABLE 1.
Cell lines used in this study
| Name | Origin | Tissue | Growth mediuma | Source |
|---|---|---|---|---|
| Caco-2 | Human | Colon adenocarcinoma | DMEMb | American Type Culture Collection, Manassas, Va. |
| HT-29 | Human | Colon adenocarcinoma | DMEM | American Type Culture Collection |
| LoVo | Human | Colon adenocarcinoma | Ham’s F-12c | Tien C. Ko, University of Texas—Medical Branch, Galveston |
| T-84 | Human | Ileocecal carcinoma | DMEM/F-12d | Sheila Crowe, University of Texas—Medical Branch, Galveston |
| HCT-8 | Human | Ileocecal carcinoma | DMEM | American Type Culture Collection |
| Int 407 | Human | Embryonic small intestine | DMEM | American Type Culture Collection |
| HEK 293Te | Human | Embryonic kidney | DMEM | Richard E. Sutton, Baylor College of Medicine, Houston, Tex. |
| CCD-18 | Human | Colon | MEM-BSSf | American Type Culture Collection |
| HeLa | Human | Cervix carcinoma | DMEM | Susan J. Marriott, Baylor College of Medicine, Houston, Tex. |
| UACC-903 | Human | Breast melanoma | DMEM | Richard E. Sutton |
| HOS-TK− | Human | Osteosarcoma | DMEM | Richard E. Sutton |
| TE85 | Human | Osteosarcoma | DMEM | Ronald T. Javier, Baylor College of Medicine, Houston, Tex. |
| A549 | Human | Lung carcinoma | DMEM | Don H. Rubin, Vanderbilt University, Nashville, Tenn. |
| AGS | Human | Gastric adenocarcinoma | DMEM | American Type Culture Collection |
| HFF | Human | Foreskin fibroblasts | DMEM | Richard E. Sutton |
| Hep G2 | Human | Hepatocellular carcinoma | MEM-BSS | American Type Culture Collection |
| RD | Human | Rhabdomyosarcoma | RPMI 1640g | Martin E. Hemler, Dana-Farber Cancer Institute, Harvard Medical School, Boston, Mass. |
| RDA2h | Human | Rhabdomyosarcoma | RPMI 1640 | Martin E. Hemler |
| MA104 | Rhesus monkey | Embryonic kidney | 199i | Microbiological Associates, Bethesda, Md. |
| Vero | African green monkey | Kidney | MEM-BSS | Microbiological Associates |
| Vero C1008 | African green monkey | Kidney | MEM-BSS | American Type Culture Collection |
| CV-1 | African green monkey | Kidney | 199 | American Type Culture Collection |
| BSC-1 | African green monkey | Kidney | 199 | American Type Culture Collection |
| COS-7 | African green monkey | Kidney | DMEM | Richard E. Sutton |
| BGM | Buffalo green monkey | Kidney | 199 | American Type Culture Collection |
| LLC-MK2 | Rhesus monkey | Kidney | DMEM | American Type Culture Collection |
| LLC-RK1 | Rabbit | Kidney | DMEM | Richard E. Sutton |
| LLC-RK2 | Rabbit | Kidney | DMEM | American Type Culture Collection |
| MDBK | Bovine | Kidney | MEM-BSS | American Type Culture Collection |
| IPEC-1 | Porcine | Newborn intestinal enterocytes | DMEM/F-12j | Helen Berschneider, North Carolina State University, Raleigh, N.C. |
| MDCK-1k | Canine | Kidney | DMEM | Lennart Svensson, Karolinska Institute, Stockholm, Sweden |
| MDCK-2l | Canine | Kidney | MEM-BSS | American Type Culture Collection |
| 3T3 | Mouse | Embryo | DMEM | Ronald T.Javier |
| L-929 | Mouse | Fibroblast | IMDMm | American Type Culture Collection |
| MODE-K | Mouse | Duodenum | DMEM | John Klein, University of Texas—Dental Branch, Houston |
| MC-26 | Mouse | Colon adenocarcinoma | DMEM | Tien C. Ko |
| 148-1LMDn | Mouse | Metatastic prostate carcinoma | DMEM | Timothy C. Thompson, Baylor College of Medicine, Houston, Tex. |
| CREF | Rat | Embryonic fibroblasts | DMEM | Ronald T. Javier |
| Rielo | Rat | Small intestine | DMEM | Tien C. Ko |
| GPC-16 | Guinea pig | Colon adenocarcinoma | MEM-BSS | American Type Culture Collection |
| BHK-21 | Syrian hamster | Kidney | DMEM | Brenda G. Hogue, Baylor College of Medicine, Houston, Tex. |
| CHO-K1 | Chinese hamster | Ovary | F-12Kp | American Type Culture Collection |
| CHO-CD36q | Chinese amster | Ovary | MEM-BSS | American Type Culture Collection |
| CHO-p901r | Chinese hamster | Ovary | α-MEMs | Jeffrey M. Bergelson, The Children’s Hospital of Philadelphia, Philadelphia, Pa. |
| CHOα2t | Chinese hamster | Ovary | α-MEM | Jeffrey M. Bergelson |
| CHOα2β1u | Chinese hamster | Ovary | α-MEM | Jeffrey M. Bergelson |
Unless otherwise specified, growth media or supplements were obtained from Gibco BRL, Life Technologies, Grand Island, N.Y. and were supplemented with 10% FBS (Summit Biotechnology, Fort Collins, Colo.), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Sigma Chemical Co.).
DMEM, Dulbecco’s modified Eagle’s minimum essential medium with high glucose (4,500 mg/ml) supplemented with 1 mM sodium pyruvate, 0.2 mM nonessential amino acids (NEAA), 2 mM l-glutamine, and 10 mM HEPES (N-[2-hydroxyethyl]piperazine-N′-[2-ethane-sulfonic salt]).
Ham’s F-12 nutrient mixture supplemented with 2 mM l-glutamine.
DMEM/F-12, modified Dulbecco’s medium and Ham’s F-12 nutrient mixture supplemented with 5% newborn calf serum (Sigma Chemical Co.) and 2 mM l-glutamine.
Primary human embryonic kidney (HEK) cells immortalized with adenovirus type 5 (Ad5; strain F2853-5b) and transformed with the T antigen of simian virus 40 (21).
MEM-BSS, Eagle’s minimum essential medium with Earle’s balanced salt solution supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM NEAA, and 1.5 g of sodium bicarbonate/liter.
RPMI 1640 supplemented with 2 mM l-glutamine and 2 mg of G418 (Geneticin)/ml to maintain selection (11).
RDA2 cells are transfected with the human VLA-2 α2 subunit cDNA in the pFneo expression vector (11).
Obtained from Irvine Scientific, Santa Ana, Calif., and supplemented with 3 mM l-glutamine and 4.5 g of sodium bicarbonate/liter.
Supplemented with 5% FBS, ITS premix (insulin [5 μg/ml] transferrin [5 μg/ml], selenium [5 μg/ml]) and human epidermal growth factor (5 μg/liter) (Becton Dickinson, Sparks, Md.) (31).
MDCK-1 cells are a low-passaged highly polarized subclone of the Madin Darby canine kidney (MDCK) cell line (60).
MDCK-2 cells are a high-passaged low-resistance subclone (NBL-2) (27).
IMDM, Iscove’s modified Dulbecco’s medium (Sigma Chemical Co.) supplemented with 2 mM l-glutamine.
148-1LMD cells are androgen-insensitive 129/SV mouse metastatic prostate cancer cells (50).
DMEM supplemented with insulin (1 U/ml) (Sigma Chemical Co.).
Ham’s F-12 nutrient mixture, Kaighn’s modification (Sigma Chemical Co.), supplemented with 4 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, and 0.1 mM hypoxanthine and 0.016 mM thymidine (Sigma Chemical Co.).
CHO-CD36 cells are transfected with human CD36 and glycoprotein IV, the receptor for extracellular matrix proteins such as collagen.
CHO-p901 cells are dihydrofolate reductase-negative CHO cells (63), which express Chinese hamster integrin β1 subunit but no integrin α2 subunit, transfected with the p901 dhfr vector (9).
Nucleoside-free α-MEM (minimum essential medium) supplemented with 10% dialyzed FBS and without antibiotics.
The rabbit hyperimmune sera raised to rotavirus serotype G3 or G6 used to neutralize infectivity or to block binding of rotavirus to cells were produced and described elsewhere (12). The mouse monoclonal antibodies (MAbs) AA10 (immunoglobulin M [IgM]) and DE9 (IgG), known to recognize domain I of the α2 subunit of VLA-2 and the β1 subunit of human VLA-2, respectively, were used as ascites fluid at 1:100 dilution (8, 9). The purified MAb AK7 (IgG1) directed against the human α2 subunit (CD49b) of VLA-2, MAb MAR4 (IgG1) directed against the human β1 subunit (CD29) of VLA-2, MAb 9F10 (IgG1) directed against the human α4 subunit (CD49d) of VLA-4, MAb 23C6 (IgG1) directed against the human integrin complex αvβ3 (CD51/CD61), MAb VI-PL2 (IgG1) directed against the human β3 subunit (CD61) of αvβ3, MAb Hmα2 (hamster IgG; group 1; κ) directed against the mouse α2 subunit (CD49b) of VLA-2, MAb 9EG7 (rat IgG2a; κ) directed against the mouse β1 subunit (CD29) of VLA-2, MAb Ha1/29 (Armenian hamster IgG; group 2; λ) directed against the rat α2 subunit (CD49b) of VLA-2, and MAb Ha2/5 (Armenian hamster IgM; κ) directed against the rat β1 subunit (CD29) of VLA-2 were purchased from Pharmingen, San Diego, Calif. Additional antibodies for flow cytometry analysis, MAbs AK7, MAR4, HA1/29, and Ha2/5 conjugated to fluorescein isothiocyanate (FITC) or R-phycoerythrin (R-PE), goat polyclonal anti-mouse or anti-rat total Ig conjugated to FITC or R-PE, and a mouse anti-hamster (IgG groups 1, 2, and 3) MAb conjugated to R-PE, were also purchased from Pharmingen. The α2 subunit of VLA-2, unlike the β1 subunit, is not highly conserved across species, and MAbs directed to the α2 subunit of VLA-2 are usually species specific (7, 37). However, MAbs AK-7 and 9F10 are known to cross-react with the α2 and α4 subunits of VLA-2 and VLA-4, respectively, of baboons and rhesus and cynomolgus macaque monkeys. The purified MAb NV3901 (IgG1) directed to the recombinant Norwalk virus capsid protein (35, 64) was used to isotype match MAbs used in flow cytometry analyses.
Rotavirus infection.
Prior to infection of cells, rotaviruses were treated with 10 μg of porcine trypsin (Worthington Biochemical Corp., Lakewood, N.J.)/ml for 30 min at 37°C. Trypsin was twice crystallized, dialyzed against 1 mM HCl, and lyophilized (Worthington Biochemical Corp.). The specific activity of the trypsin preparation used was ≥180 p-toluenesulfonyl-l-arginine methyl ester units per mg of trypsin. In some cases, virus inocula were treated, following activation with trypsin, with 80 μg of soybean trypsin inhibitor type I-S (Sigma Chemical Co., St. Louis, Mo.)/ml to inactive the trypsin present in the inoculum. Cells were grown in 96-well plates, and approximately 4 × 104 cells were inoculated with 100 μl of SA-dependent or SA-independent rotavirus strains/well at an MOI of 0.1, 1, 10, 100, or 500 focus-forming units (FFU) per cell as described previously (13). Briefly, after virus adsorption for 1 to 2 h at 37°C, the inoculum was removed, and the cells were washed with medium and incubated for 18 to 24 h at 37°C with the corresponding medium with or without the presence of 1 μg of porcine trypsin/ml. The cells were fixed with cold methanol, and fluorescent foci (virus antigen) were detected by fluorescent-focus assay (FFA) (12). Viral infectivity titers and susceptibility to infection were expressed as FFU in test cell lines. In some cases, viral infectivity titers to infection were expressed as the percentage of FFU in test cell lines versus those obtained in control MA104 cells.
Treatment of MA104 cells with MAbs.
Cells were incubated with 100 μl of twofold dilutions of MAb AA10, DE9, AK7, MAR4, 9F10, 23C6, VI-PL2, or NV3901 diluted in medium, starting at 20 μg/ml (for purified MAbs) or 1/100 (for ascites), for 1 h at 37°C before virus adsorption, as described previously (17, 33). After a range of antibody concentrations was tested, a concentration of 10 μg of MAb/ml was chosen for the experiments because 10 μg/ml provided maximal inhibitory effects. Following treatment with MAbs, the cells were washed twice and infected as outlined above.
Neuraminidase treatment of cells.
In some cases, cells were treated for 1 h at 37°C with 100 μl of 20-mU/ml neuraminidase from Arthrobacter ureafaciens purified by affinity chromatography (Sigma Chemical Co.) or with 100 μl of TNC buffer (10 mM Tris [pH 7.5], 140 mM NaCl, 10 mM CaCl2) as described previously (13). Following treatment with neuraminidase, the cells were washed with TNC buffer prior to inoculation with SA-dependent or -independent rotavirus strains at an MOI of 1, 10, 100, or 500 FFU per cell. After virus adsorption was allowed for 1 h at 37°C, the inoculum was removed and the cells were washed with medium. The cells were incubated for 18 h at 37°C with the corresponding medium, washed with phosphate-buffered saline (PBS), fixed with cold methanol, and stained by FFA as described previously (12). Viral infectivity was expressed as the percentage of FFU in neuraminidase-treated cells versus that obtained in control (TNC buffer-treated) cells.
Transfection of cells with purified rotavirus double-layered particles (DLPs).
Optimal transfection conditions were determined using a plasmid encoding a red-shifted variant of wild-type green fluorescent protein (GFP) (pEGFP-N1; Clontech Laboratories, Palo Alto, Calif.) and different transfection reagents, (i) Lipofectamine plus reagent (Gibco BRL), (ii) Lipofectamine (Gibco BRL), (iii) Cellfectin (Gibco BRL), (iv) DMRIE-C (Gibco BRL), (v) Fugene6 (Boehringer-Mannheim, Indianapolis, Ind.), and (vi) Clonfectin (Clontech), according to the manufacturers’ instructions. Expression of GFP and the viabilities of pEGFP- and mock-transfected cells were assessed from 24 to 216 h posttransfection. Among all transfection reagents used, Lipofectamine plus reagent provided optimal transfection efficiencies, and by 24 to 48 h posttransfection, the transfection efficiency ranged from 60 to 70% (data not shown).
To determine if cells lacked the appropriate receptor required for efficient cell entry or were unable to synthesize rotavirus proteins, 0.5 to 2 μg of CsCl-purified and EDTA disodium salt (EDTA)-treated (50 mM; 20 min at room temperature) noninfectious rotavirus (RRV, NCDV, SA11 Cl3, OSU, H-1, Wa, WC3, H-2, ALA, C-11, or BAP-2) DLPs/ml were delivered by lipofection into the cytoplasm of cells that exhibited limited susceptibility to rotavirus infection. The concentration of each virus preparation was calculated using the formula 1 optical density unit at 260 nm ≡ 2.1 × 1012 particles ≡ 185 μg of reovirus particles, and a similar value is assumed for rotavirus particles (16). Therefore, a value of 1.1 × 1010 rotavirus particles per μg can be estimated. Lipofection was carried out in 70% confluent (≈3 × 105) cell monolayers in 6- or 24-well tissue culture plates; the cells were incubated overnight for 24 h at 37°C, washed with PBS, and fixed with cold methanol; and fluorescent foci were determined by FFA (12). The transfection efficiency of rotavirus infection was expressed as FFU per microgram of DLPs transfected or as the percentage of fluorescent cells with respect to nonfluorescent cells and was calculated from two to six replicate experiments. Control cells were inoculated with RRV or WC3 DLPs without Lipofectamine plus reagent or with Lipofectamine plus reagent alone.
Nonradioactive binding assays.
Rotavirus binding assays were performed as described previously (36) with modifications. Briefly, confluent cell monolayers were washed twice with PBS without calcium or magnesium chloride and carefully scraped off to obtain single-cell suspensions or detached by incubation with 0.05% trypsin containing 0.53 mM EDTA (Gibco BRL) at 37°C for 5 min. The detached cells were suspended in their corresponding complete medium and incubated at 37°C for 1 h to allow reconstitution of the surface proteins. The cell suspensions were centrifuged at 500 × g at 4°C for 2 min, washed with PBS, and suspended in ice-cold culture medium without serum. For the binding assays, 250 μl of an ice-cold trypsin-activated (30 min at 37°C with 10 μg of trypsin/ml) simian SA11 Cl3 or RRV, bovine WC3, or human Wa or PA169 rotavirus strain was bound to cells (5 × 105) at an MOI of 5 for 1 h with gentle mixing at 4°C. The cell-virus complexes were subsequently washed twice with ice-cold PBS. Then, the cells were suspended in ice-cold medium containing 1 μg of trypsin (Worthington)/ml, subjected to two freeze-thaw cycles to release bound virus, and stored at −70°C. Thawed aliquots were activated with 10 μg of trypsin/ml for 30 min at 37°C, and the viral titers were determined in MA104 cell monolayers by FFA as described previously (12). Controls for binding assays included cells with medium (no virus) or equivalent amounts of virus. To measure the specificity of virus binding to cells, rotavirus strains were preincubated overnight at 4°C with a 1:100 dilution in PBS of hyperimmune sera containing neutralizing antibodies to the corresponding virus strain. Virus binding was expressed as a percentage of the titer of infectious virus bound to MA104 cells.
Alternatively, binding assays in which the amount of virus bound was quantitated by enzyme-linked immunosorbent assay (ELISA) were performed as described by Zárate et al. (67) with modifications. Single-cell suspensions of confluent cell monolayers were obtained as described above. For the binding assay, 400 μl of an ice-cold trypsin-activated simian RRV or bovine WC3 rotavirus strain, in medium containing 1% bovine serum albumin (fraction V; Calbiochem-Novabiochem Corp., La Jolla, Calif.), was bound to cells (2 × 105) at an MOI of 10 for 1 h with gentle mixing at 4°C. The cell-virus complexes were subsequently washed twice with ice-cold PBS containing 0.5% bovine serum albumin and then lysed with 100 μl of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Triton X-100). ELISA was performed essentially as described elsewhere (12). Controls for these binding assays included cells with medium (no virus) or virus (diluted in lysis buffer) with no cells.
Radioactive binding assays.
To confirm the results obtained with the nonradioactive binding assays, rotavirus radioactive binding assays were performed as described elsewhere (18) with modifications, because radioligand-binding assays are a more direct assessment of virus binding. Briefly, confluent cell monolayers in 24-well plates were washed twice and chilled to 4°C. Metabolically [35S]methionine-labeled CsCl-purified SA-dependent simian SA11 4F or SA-independent bovine WC3 rotavirus, with or without 10× unlabeled homologous virus, was added to the monolayers and incubated at 4°C with gentle rocking for 1 h. For measurement of binding, the monolayers were washed three times with ice-cold medium, lysed in 200 μl of PBS containing 1% sodium dodecyl sulfate, and counted in 5 ml of Econo-Safe (Research Products International Corp., Mount Prospect, Ill.) using an LS 3801 scintillation counter (Beckman Instruments, Inc., Fullerton, Calif.). The specific binding was calculated by subtracting the nonspecific binding from the total binding.
Flow cytometric analysis.
The cell surface expression of the α2 and/or β1 subunit of the VLA-2 integrin was determined in MA104, Caco-2, HT-29, T84, Rie1, GPC-16, BHK-21, CHO-K1, CHOdhfr, CHOα2, or CHOα2β1 cells. Confluent monolayers (4 to 6 days old) were washed twice with PBS without calcium or magnesium chloride and detached by gentle mechanical scraping of the monolayer. Monodispersed cells (105) were washed twice in PBS containing 2% (vol/vol) fetal bovine serum (PBS-FBS) and suspended in 100 μl of ice-cold PBS-FBS containing MAb AA10 (1/100), MAR4 (10 μg/ml), AK7 (10 μg/ml), Ha1/29 (10 μg/ml), Ha2/5 (10 μg/ml), or NV3901 (10 μg/ml) or no MAb for single-step (in the case of MAbs directly conjugated to FITC or PE) or two-step staining. Following an incubation period of 30 min on ice, 1 ml of PBS-FBS was added to the cells, and the cells were washed twice with PBS-FBS. When two-step staining was required, cells were incubated for 30 min on ice in the dark with 100 μl of a 1/100 dilution of FITC- or PE-conjugated goat anti-mouse Ig specific polyclonal antibody (Pharmingen) in PBS-FBS. The cells were washed and fixed with 500 μl of 4% reagent grade paraformaldehyde, pH 7.0 (Fisher Scientific, Fair Lawn, N.J.), and immunofluorescence staining of the cells was analyzed using a Profile I flow cytometer (Coulter Electronics, Hialeah, Fla.). At least 10,000 forward scatter gated events were collected per specimen. FITC fluorescence was collected using linear amplification with doublet discrimination engaged, and PE fluorescence was collected using logarithmic amplification. Controls included cells incubated in the absence of both primary and FITC- or PE-conjugated antibody to adjust for autofluorescence and cells incubated either in the absence of primary antibody or in the presence of isotype-matched antibody and stained only with FITC or PE to adjust for spectral overlap.
Statistical analysis.
Statistical analyses were performed using SPSS version 7.5 for Windows (SPSS, Inc., Chicago, Ill.). Differences in fluorescent foci (virus antigen), percent infectivity, and percent binding were compared using the Kruskal-Wallis test followed by the Mann-Whitney U test. Comparisons with a P value of <0.05 were considered statistically significant.
RESULTS
Susceptibilities of continuous human and animal cell lines of different species and tissues of origin to growth of SA-dependent and SA-independent rotaviruses.
The identification of the cellular receptor used by rotaviruses has been hampered by the inability to identify a cell line that lacks the appropriate rotavirus cellular receptor(s). It has previously been shown that cell lines of renal or intestinal origin are susceptible to rotavirus infection and that they are the most effective at propagating these viruses in the laboratory (5, 23, 24, 42). Because it will be of interest to identify cell lines that might lack the rotavirus cellular receptor(s), we tested susceptibility to SA-dependent or -independent rotavirus infection in a total of 46 human or animal cell lines derived from different tissues. We directly compared cell lines previously known to be susceptible to rotavirus infection to cell lines whose susceptibility to rotavirus infection was unknown. Unless otherwise specified, an MOI of 10 was used for all infections. As can be observed in Fig. 1, a marked difference in susceptibility to SA-dependent (RRV) or SA-independent (WC3) rotavirus infection was obtained in the cell lines tested when virus antigen was detected by FFA. The SA-dependent rotavirus strain RRV grew to higher titers (0.5 to 1 log unit) than the SA-independent rotavirus strain WC3 regardless of the cell line tested (Fig. 1). Consistent and similar results were obtained upon infection of all cell lines tested with SA-dependent (SA11 and OSU), SA-independent human (Wa, PA169, and HAL1166), or neuraminidase-resistant variant (SA11 Cl3 954/23R) strains (data not shown).
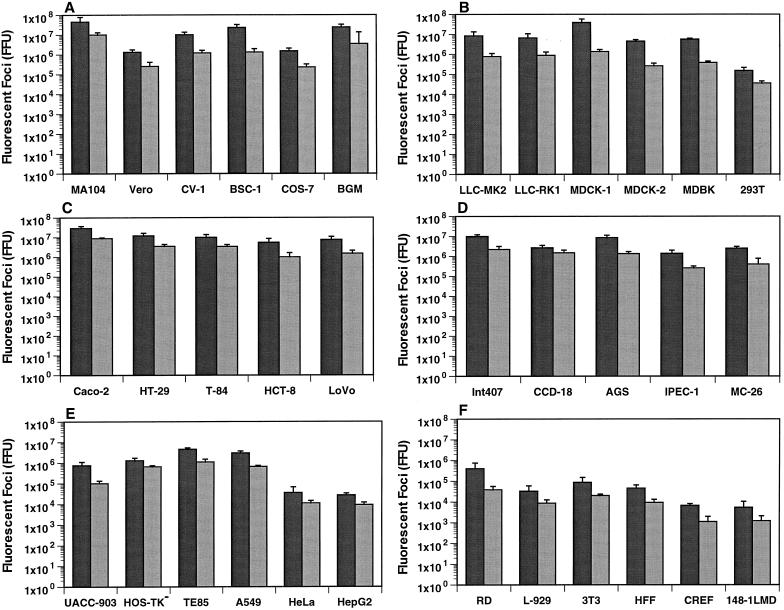
FIG. 1.
Detection of rotavirus antigen (fluorescent foci) in rhesus, African, or buffalo green monkey kidney cells (A); rhesus, rabbit, canine, bovine, or human kidney cells (B); human intestinal cells (C); human, porcine, or murine intestinal cells (D); human breast, bone, lung, cervix, or hepatic cells (E); and human rhabdomyosarcoma, murine, human, or rat fibroblasts, or murine metastatic prostate cancer cells (F). The infectivity of the SA-dependent RRV strain (solid) or the SA-independent WC3 strain (shaded) is expressed in FFU as measured by FFA. An MOI of 10 was used to infect all cells. As a control, cells incubated with medium alone were used (data not shown). The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean.
Rotavirus infection was most permissive in most, but not all, epithelial cell lines of renal and intestinal origin. A wide range of virus antigen was detected, depending on the kidney cell line infected (4.5 × 107 to 1.5 × 105 FFU [RRV] or 1 × 107 to 3.5 × 104 FFU [WC3]) (Fig. 1A and B). The kidney cell lines most susceptible to rotavirus infection were MA104, BSC-1, BGM (Fig. 1A), and MDCK-1 (Fig. 1B). CV-1, LLC-MK2, LLC-RK1, MDCK-2, and MDBK cells were 5- to 10-fold less susceptible to rotavirus infection than MA104, BSC-1, BGM, and MDCK-1 cells (Fig. 1A and B). LLC-RK2 cells were as susceptible to rotavirus infection as LLC-RK1 cells (data not shown). Cos-7, Vero (Fig. 1A), HEK 293T (Fig. 1B), and Vero C1008 cells (data not shown) were 50- to 500-fold less susceptible than MA104, BSC-1, BGM, and MDCK-1 cells. The amounts of virus antigen detected did not differ significantly (P ≥ 0.321; Mann-Whitney U test) when an MOI of 0.1 or 1 was used to infect all kidney cell lines tested (data not shown). Baby hamster BHK-21 cells were not susceptible to rotavirus infection at an MOI of 1 or 10 (data not shown).
Among intestinal cell lines tested, the amounts of virus antigen detected also exhibited a wide range (2.9 × 107 to 1.4 × 106 FFU [RRV] or 8.8 × 106 to 2.6 × 105 FFU [WC3]) (Fig. 1C and D). Caco-2, HT-29, T-84, HCT-8, LoVo, Int 407, and AGS cells (Fig. 1C and D) were as susceptible to rotavirus infection as MA104 cells (Fig. 1A). Human mesenchymal CCD-18, porcine IPEC-1, or murine MC-26 cells (Fig. 1D) showed about 10-fold less susceptibility to rotavirus infection than MA104 or Caco-2 cells (Fig. 1A and C, respectively). The amounts of virus antigen detected did not differ significantly (P ≥ 0.226; Mann-Whitney U test) when an MOI of 0.1 or 1 was used to infect all intestinal cell lines tested (data not shown), with the exception of IPEC-1 cells. IPEC-1 cells were 100- to 500-fold less susceptible to RRV or WC3 rotavirus than MA104 or Caco-2 cells when an MOI of 1 was used (P ≤ 0.034; Mann-Whitney U test) (data not shown). As observed with hamster BHK-21 kidney cells, guinea pig GPC-16, mouse MODE-K, and rat Rie1 small-intestinal or colon cells were not susceptible to rotavirus infection at an MOI of 1 or 10 (data not shown).
Although cell lines of renal and intestinal origin are the most susceptible to rotavirus infection, cell lines of human cervix (HeLa), liver (HepG2), myelogenous leukemic (K562), or embryonal rhabdomyosarcoma (RD) cells, mouse embryo (3T3) or fibroblast (L929) cells, and hamster ovary (CHO) cells have also been used for studies of rotavirus-cell interactions (5, 23, 33, 36, 42, 44, 56). For comparison, we retested some cell lines of nonrenal or nonintestinal origin, as well as nonrenal or nonintestinal cell lines never tested before, for susceptibility to rotavirus infection. Cell lines of human breast (UACC-903), bone (HOS-TK− and TE85), or lung (A549) origin were 5- to 10-fold less susceptible to rotavirus infection than MA104 cells (Fig. 1A and E). As previously reported, human HeLa or HepG2 (Fig. 1E), human RD (Fig. 1F), and mouse 3T3 or L929 fibroblast (Fig. 1F) cells were 100-fold less susceptible to rotavirus infection than MA104 cells (Fig. 1A). Human foreskin fibroblast (HFF) cells were 100- to 500-fold less susceptible to rotavirus infection than MA104 cells (Fig. 1A). Rat embryo (CREF) fibroblast cells and mouse prostate metastatic cancerous (148-1LMD) cells (Fig. 1F) were 5,000- to 10,000-fold less susceptible than MA104 cells (Fig. 1A). Three different Chinese hamster ovary cell lines, CHO-K1, CHO-CD36, and CHO-p901, were completely refractory to rotavirus infection when an MOI of 1 or 10 was used (data not shown).
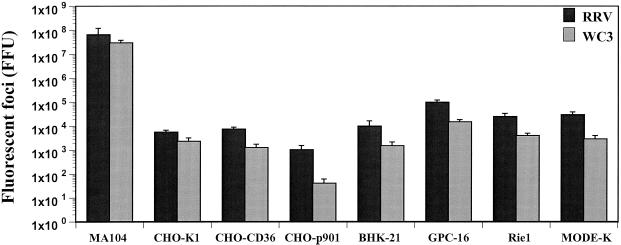
Although BHK-21, GPC-16, Rie1, MODE-K, CHO-p901, CHO-K1, and CHO-CD36 cells were resistant to rotavirus infection at an MOI of 1 or 10, infection was detected when the cells were exposed to MOIs of 500 (RRV) or 100 (WC3) (Fig. 2). However, infection of these cell lines was significantly (P ≤ 0.001; Mann-Whitney U test) less efficient than infection of MA104 cells, suggesting that these cells support only limited rotavirus infection. BHK-21, GPC-16, Rie1, and MODE-K cells were 100- to 5,000-fold less susceptible than MA104 cells, while CHO-K1 and CHO-CD36 cells were 1,000-fold less susceptible to rotavirus infection at high MOIs than MA104 cells (Fig. 2). The restricted growth of rotavirus in CHO cells was confirmed and accentuated in CHO-p901 cells, which were 10,000-fold less susceptible than MA104 cells (Fig. 2). The additional restricted growth of rotavirus in CHO-p901 cells was confirmed with at least 20 SA-dependent or SA-independent rotavirus strains (data not shown).
FIG. 2.
Detection of rotavirus antigen (fluorescent foci) in rhesus monkey kidney (MA104), Chinese hamster ovary (CHO-K-1, CHO-CD36, and CHO-p901), baby hamster kidney (BHK-21), and guinea pig (GPC-16), rat (Rie1), and murine (MODE-K) intestinal cells. The infectivity of the SA-dependent RRV strain or the SA-independent WC3 strain is expressed in FFU as measured by FFA. An MOI of 500 (RRV) or 100 (WC3) was used to infect all cells. As a control, cells incubated with medium alone were used (data not shown). The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean.
Lipofection of rotavirus DLPs into poorly susceptible cells results in rotavirus replication.
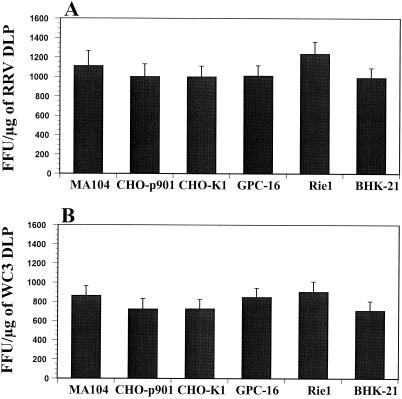
Rotavirus tissue tropism might be related to the level of expression of specific viral receptors that may alter the capacity of rotavirus to bind to or replicate in the different cell lines tested. To determine if the difference in susceptibility to rotavirus infection is at the level of attachment (binding), internalization (entry), or replication, we determined if direct delivery of transcriptionally competent rotavirus particles (DLPs) into the cytoplasm of BHK-21, GPC-16, Rie1, CHO-K1, CHO-CD36, or CHO-p901 cells would allow rotavirus replication. Noninfectious RRV or WC3 DLPs were lipofected into MA104, CHO-p901, CHO-K1, CHO-CD36, BHK-21, GPC-16, and Rie1 cells, and our results showed that the block in susceptibility to rotavirus infection was not due to the inability of any of these cells to allow synthesis of the virus proteins (Fig. 3 and data not shown). All of the cell lines were equally able to generate rotavirus antigen, although the SA-dependent RRV strain was always more efficient at replicating than the SA-independent WC3 strain (Fig. 3). No rotavirus antigen was detected in cells inoculated with RRV or WC3 DLPs without Lipofectamine plus reagent or with Lipofectamine plus reagent alone (data not shown). The transfection efficiency in CHO-CD36 cells was similar to those obtained with CHO-p901 and CHO-K1 cells (data not shown). Similar results were also obtained when noninfectious SA11, NCDV, H-1, Wa, ALA, C-11, BAP-2, or H-2 DLPs were delivered to the cytoplasm of MA104, CHO-p901, BHK-21, GPC-16, and Rie1 cells by lipofection (data not shown), and again, the ability to generate rotavirus antigen was always more efficient with SA-dependent strains (SA11, NCDV, and H-1) than with SA-independent strains (Wa, ALA, C-11, BAP-2, and H-2). These results showed that CHO-p901, CHO-K1, CHO-CD36, BHK-21, GPC-16, and Rie1 cells are permissive for rotavirus replication if normal virus entry is bypassed.
FIG. 3.
Liposome-mediated infection of cells by SA-dependent RRV strain (A) or SA-independent WC3 strain (B) with Lipofectamine plus reagent. Synthesis of RRV and WC3 rotavirus proteins was determined with 1 μg of CsCl-purified and EDTA-treated rotavirus DLPs/ml. No virus antigen was detected in cells inoculated with RRV or WC3 DLPs without Lipofectamine plus reagent or with Lipofectamine plus reagent alone (data not shown). The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean.
Effect of MAbs to different integrin subunits on the infectivity of SA-dependent and SA-independent rotaviruses.
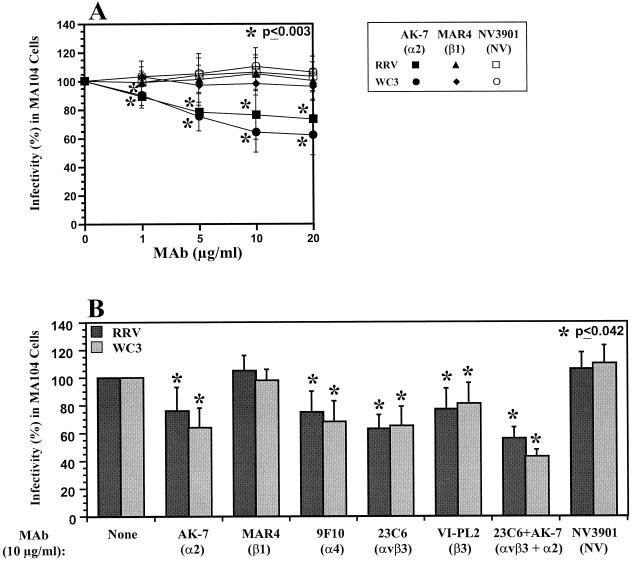
During the course of our study, several reports implicated the VLA-2 (α2β1), VLA-4 (α4β1), CR4 (αxβ2), and αvβ3 integrins in rotavirus cell attachment, entry, or other postattachment events in infection, and treatment of cells with antibodies to these integrins resulted in a reduction of rotavirus infectivity (17, 33, 36, 44, 68). Therefore, we tested the abilities of MAbs to VLA-2, VLA-4, and αvβ3 to block the infectivity of SA-dependent (RRV, SA11, and OSU) or SA-independent (WC3, Wa, PA169, and SA11 Cl3 954/23R) rotavirus strains in MA104 cells. Treatment of MA104 cells with MAbs to integrins α2, α4, β3, and αvβ3, but not β1, decreased the infectivity of RRV or WC3 rotavirus strains in a dose-dependent manner (Fig. 4A and data not shown), resulting in 25 to 40% less infectivity at 10 μg/ml (Fig. 4). The levels of inhibition of rotavirus infectivity are in accordance with those found by others for SA-dependent (RRV and SA11) or SA-independent (RV-5, Wa, and nar3) strains when MA104 cells were incubated with MAbs to the α2 (AK-7, P1E6, and RMAC11), α4 (B-5G10, P1H4, P4G9, and P4C2), or β3 (25E11) integrin subunit or to the αvβ3 (LM609) integrin (17, 33, 36, 68). The lack of inhibition by the anti-β1 MAb (MAR4) is consistent with other reports of studies that have used MAb MAR4 or other MAbs (8A2, P4C10, 4B7R, and P4G11) directed to the β1 subunit of VLA-2 or a MAb (A-1A5) directed against the common β subunit of the VLA integrin family (17, 33, 36, 68). An antibody raised against and specific to the VLA-2 heterodimer was not available and therefore was not tested. Unlike anti-α2 MAb AK-7, MAb AA10 (8) did not decrease the infectivity of RRV, SA11, Wa, or WC3 in MA104 cells (data not shown); MAb AA10 might recognize a different epitope in the α2 subunit of VLA-2. Similar results were obtained with SA11, OSU, Wa, PA169, and the variant SA11 Cl3 954/23R (data not shown). In addition, when antibodies to α2 and αvβ3 were combined to treat MA104 cells, the block in infectivity of both SA-dependent and SA-independent rotavirus strains was additive (Fig. 4B). In all cases, the infectivity of SA-independent rotaviruses was inhibited more with the MAb directed to the α2 subunit of VLA-2 than that of SA-dependent rotaviruses, although this difference was not significant (P ≥ 0.131; Mann-Whitney U test) (Fig. 4B and data not shown). Thus, rotaviruses may interact with the VLA-2 integrin.
FIG. 4.
Inhibition of RRV and WC3 rotavirus infectivity following treatment of MA104 cells with a range of concentrations (0 to 20 μg/ml) of MAb AK-7, directed to the α2 subunit of VLA-2 (A), or inhibition of RRV or WC3 rotavirus infectivity in MA104 cells by MAbs (10 μg/ml) to integrin subunits α2 (AK-7), α4 (9F10), β1 (MAR4), β3 (VI-PL2), αvβ3 integrin (23C6), and α2 (AK-7) plus αvβ3 integrin (23C6), or Norwalk virus capsid (NV3901) (B). MOI = 10. Virus infectivity is expressed as the percentage of FFU in antibody-treated cells versus those obtained in control (199 medium-treated) cells. The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean. A significant (P < 0.05; Mann-Whitney U test) decrease in rotavirus infectivity following treatment of MA104 cells with each antibody is indicated by an asterisk.
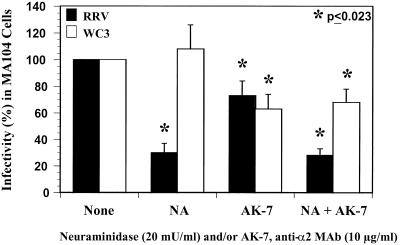
Effect of neuraminidase treatment of cells and anti-α2 MAb on the infectivity of SA-dependent and SA-independent rotaviruses.
To determine the roles of SA residues and the VLA-2 integrin as functional receptors for rotavirus, we treated MA104 cells with neuraminidase and/or α2 MAb AK-7 and infected the cells with SA-dependent (RRV and SA11) and SA-independent (WC3, SA11 Cl3 954/23R, PA169, and Wa) rotaviruses. Infection of MA104 with RRV, but not WC3, was significantly inhibited (P = 0.001; Mann-Whitney U test) by pretreatment of the cells with neuraminidase (Fig. 5). Furthermore, the combination of neuraminidase and α2 MAb had no additional inhibitory effect on the infectivity of RRV compared to that of neuraminidase alone (P = 0.871; Mann-Whitney U test). Similar results were obtained with the SA-dependent rotavirus SA11 strain (data not shown). However, it is important to keep in mind that SA-dependent rotavirus strains require SA residues for efficient binding and infectivity (2, 13, 15, 48, 49), and treatment of MA104 cells with neuraminidase does not specifically address the capacity of SA-dependent RRV or SA11 rotavirus strains to engage VLA-2 because integrins are glycosylated and sialylated proteins (7, 37). As expected, pretreatment of cells with neuraminidase had no effect on the infectivity of WC3, and the decrease of WC3 infectivity was due only to the α2 MAb (Fig. 5). Similar results were obtained with the SA11 Cl3 954/23R, PA169, and Wa SA-independent rotavirus strains (data not shown). These data suggest that SA-dependent rotaviruses utilize SA residues on the cell surface, not VLA-2 integrin, for efficient infectivity of susceptible MA104 cells.
FIG. 5.
Inhibition of RRV or WC3 rotavirus infectivity in MA104 cells treated with 20 mU of neuraminidase (NA) (A. ureafaciens)/ml and/or MAb (10 μg/ml) to the α2 integrin subunit (AK-7). MOI = 10. Virus infectivity is expressed as the percentage of FFU in neuraminidase-treated and/or antibody-treated cells versus those obtained in control (TNC buffer- and 199 medium-treated) cells. The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean. A significant (P < 0.05; Mann-Whitney U test) decrease in rotavirus infectivity following treatment of MA104 cells with neuraminidase (NA) and/or AK-7 MAb is indicated by an asterisk.
Cellular expression of VLA-2 integrin in cell lines exhibiting poor susceptibility to rotavirus infection.
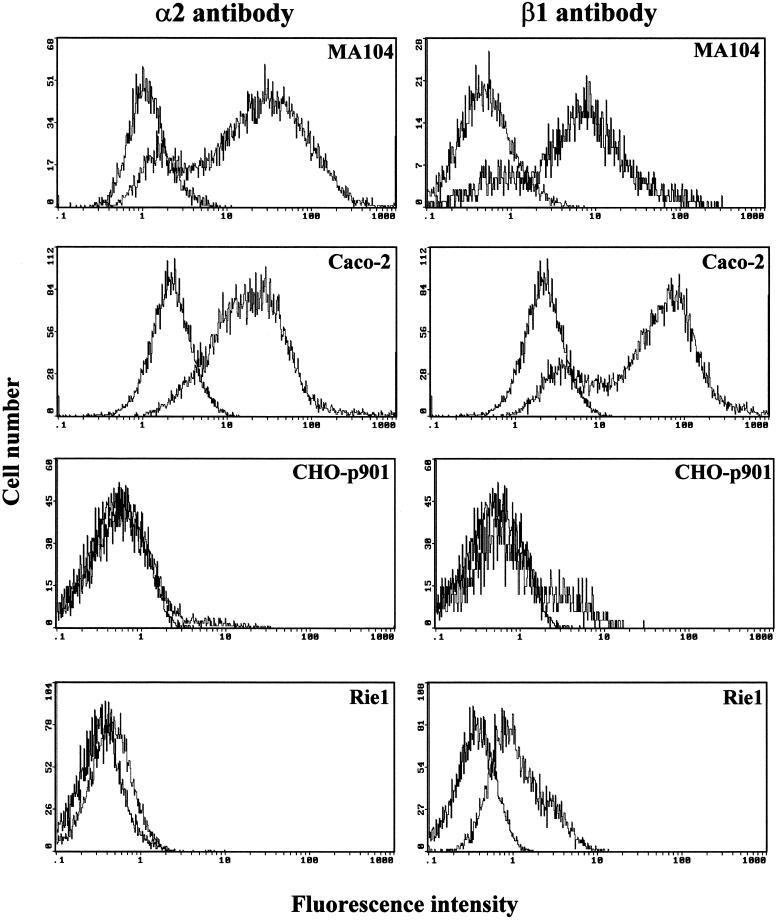
To determine if the restricted susceptibility of CHO-K1, CHO-p901, Rie1, MODE-K, GPC-16, or BHK-21 cells to rotavirus infection was due to low surface expression of integrin subunits α2 and β1, we performed flow cytometry. Of these cell lines, only CHO and BHK-21 cells are known not to express endogenous VLA-2 (1, 29). The expression of endogenous VLA-2 integrin on the surfaces of human Caco-2, HT-29, and HeLa and rhesus monkey MA104 cells was analyzed with a cross-reactive antibody to the human α2 subunit of VLA-2, while that of murine 3T3, L929, and MC-26 cells was analyzed with an antibody to the mouse α2 subunit of VLA-2. The α2 integrin subunit was not detected in CHO-K1, CHO-p901, or BHK-21 cells using antibody to the human, mouse, or rat α2 subunit of VLA-2 (Fig. 6 and data not shown). The α2 integrin subunit was abundantly detected in MA104, Caco-2, HT-29, HeLa, 3T3, L929, and MC-26 cells, while it was detected in small amounts in mouse MODE-K and rat Rie1 cells when antibody to the mouse or rat α2 subunit of VLA-2, respectively, was used (Fig. 6 and data not shown). The α2 integrin subunit was not detected in guinea pig GPC-16 cells when antibody to the human α2 subunit of VLA-2 was used, but it was detected in small amounts when antibody to the mouse or rat α2 subunit of VLA-2 was used (data not shown). Since an antibody to the guinea pig α2 subunit of VLA-2 is not available, it is possible that the limited detection of guinea pig VLA-2 is due to not having the proper species-specific antibody to detect it, because the α2 subunit of VLA-2, unlike the β1 subunit, is not highly conserved across species (1, 22). Except for CHO-K1 and CHO-p901, all cells were shown to express the β1 integrin subunit (Fig. 6 and data not shown). The endogenous expression of the hamster β1 subunit is very low (10 to 16%) in CHO-K1 and CHO-p901 cells (Fig. 6 and data not shown).
FIG. 6.
Flow cytometry histograms of the cell surface expression of integrin subunits α2 and β1 of VLA-2. The cell line analyzed is indicated in each histogram. Expression of α2 or β1 was determined by using 10 μg of MAb AK-7 (MA104, Caco-2, and CHO-p901) or MAb Ha1/29 (Rie1) or of MAb MAR4 (MA104, Caco-2, and CHO-p901) or MAb Ha2/5 (Rie1)/ml, respectively, in a single- or two-step stain. MAb NV3901, directed against the recombinant Norwalk virus capsid (35, 64), was used to match the isotype (IgG1) in a two-step stain.
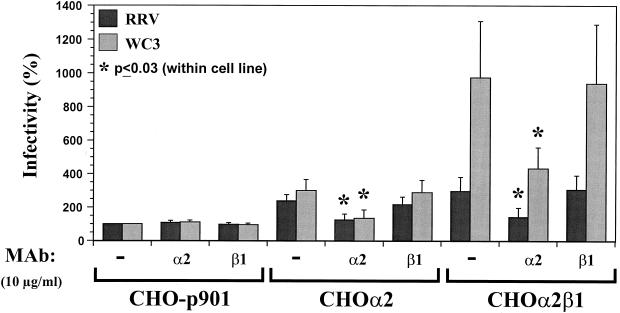
Infection of CHO cells transfected with the human α2 subunit of VLA-2 (CHOα2) or both the α2 and β1 subunits of VLA-2 (CHOα2β1) with SA-dependent and SA-independent rotaviruses.
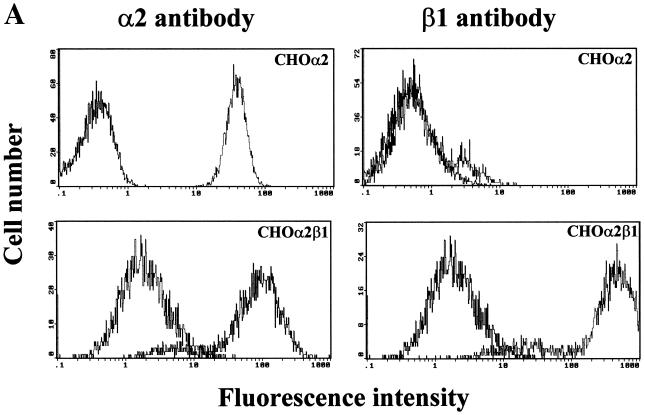
To determine if the α2 integrin subunit is essential and sufficient for rotavirus infectivity, we examined rotavirus infectivity in stable CHO-p901 cells transfected with the human α2 subunit of VLA-2 (CHOα2) or cotransfected with both the α2 and the β1 subunit of VLA-2 (CHOα2β1). All transfectant CHO cells were maintained in nucleoside-free medium because the parental CHO dihydrofolate reductase-negative cells do not grow in that medium. Transfectant CHOα2 and CHOα2β1 cells were sorted for homogenous expression (≥99.5% positive) of human α2 or both human α2 and β1 integrin subunits, respectively, by fluorescence-activated cell sorting (data not shown). Testing of CHOα2 and CHOα2β1 transfectant cells during the course of the experiments (including freezing and thawing of the cells) showed that the levels of expression of the integrins were stable (data not shown). Flow cytometry analysis of human α2 subunit or α2β1 integrin surface expression in control CHO-p901, CHOα2, and CHOα2β1 revealed that CHOα2 and CHOα2β1 (Fig. 7A), but not CHO-p901 (Fig. 6), expressed high levels of integrin subunit α2 and that only CHOα2β1 expressed high levels of VLA-2.
FIG. 7.
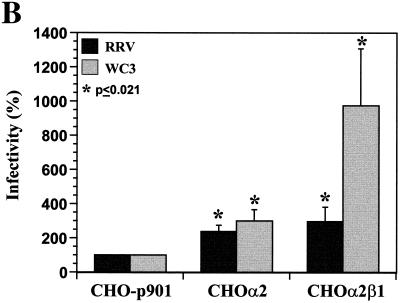
(A) Flow cytometry histograms of the surface expression of human integrin subunits α2 and β1 of VLA-2 in CHOα2 and CHOα2β1 cells. The cell line analyzed is indicated in each histogram. Expression of α2 or β1 was determined by using 10 μg of MAb AK-7 or MAb MAR4/ml, respectively, in a single-step stain. MAb NV3901, directed against the recombinant Norwalk virus capsid (35, 64), was used to match the isotype (IgG1) in a two-step stain. (B) VLA-2 integrin promotes CHO cell infection. CHO-p901 or stably transfectant CHOα2 and CHOα2β1 cells were infected with SA-dependent RRV or SA-independent WC3 rotaviruses (MOI = 500 or 100, respectively). Virus infectivity is expressed as the percentage of FFU in CHOα2 and CHOα2β1 cells versus those obtained in CHO-p901 cells. The values shown are the arithmetic means of at least five replicate experiments. The error bars represent one standard error of the mean. A significant (P < 0.05; Mann-Whitney U test) increase in rotavirus infectivity of CHOα2 and CHOα2β1 cells with respect to infectivity of CHO-p901 cells is indicated by an asterisk.
Expression of the α2 and β1 subunits of human VLA-2 rendered CHO-p901 cells 2 to 10 times more susceptible to rotavirus infection than the parental cells (P ≤ 0.030; Mann-Whitney U test). CHOα2 cells were two- to threefold more susceptible to RRV or WC3 infection, respectively (Fig. 7B). Because the α2 subunit is expressed on the cell surface mainly in a VLA-2 heterodimer and the α2 subunit is not known to interact with subunits other than β1 (7, 37), the α2 subunit in CHOα2 cells was expressed in association with the endogenous hamster β1 subunit (8, 9). CHOα2β1 cells, which expressed human VLA-2, were 3-fold more susceptible to RRV infection and 10-fold more susceptible to WC3 infection (Fig. 7B). This difference in SA-dependent and SA-independent rotavirus infectivity in CHOα2β1 cells was also observed with additional SA-dependent (SA11 and OSU) and SA-independent (Wa, ALA, HAL1166, and PA169) rotavirus strains (data not shown). The block in susceptibility to rotavirus infection of CHOα2, CHOα2β1, CHO-K1, CHO-CD36, BHK-21, GPC-16, and Rie1 cells was not due to the inability of any of these cells to support synthesis of the virus proteins (Fig. 3 and data not shown). All of the cell lines were equally able to express rotavirus antigen following lipofection, although the SA-dependent RRV strain (Fig. 3A) was always more efficient at replicating than the SA-independent WC3 strain (Fig. 3B). The transfection efficiencies of noninfectious RRV or WC3 DLPs lipofected into CHOα2 and CHOα2β1 were similar to that in CHO-p901 (Fig. 3 and data not shown).
Introduction of the full-length cDNA clone of the α2 subunit of VLA-2 into the human RD tumor cell line resulted in the expression of VLA-2 in transfected RD cells (RDA2) (11). Parental RD cells were 100-fold less susceptible than MA104 cells (Fig. 1F), and similar to results obtained in CHOα2β1 cells (Fig. 7B), RDA2 cells were 2- to 3-fold more susceptible to RRV infection and 10-fold more susceptible to WC3 infection than the parental RD cells (P ≤ 0.023; Mann-Whitney U test) (data not shown). The same pattern of increased susceptibility to RRV or WC3 rotavirus infection was observed with additional SA-dependent (SA11, NCDV, and OSU) and SA-independent (Wa, DS-1, ALA, C-11, H-2, HAL1166, and PA169) rotavirus strains (data not shown).
The increase of rotavirus infectivity in CHOα2, CHOα2β1, and RDA2 cells was blocked by preincubation of the cells with a MAb to the α2, but not the β1, subunit of VLA-2 (Fig. 8 and data not shown); this result provided further evidence that the increase in rotavirus infectivity observed in these cells was due to the expression of VLA-2 integrin. Although VLA-2 integrin promotes rotavirus infectivity in CHO cells, CHOα2β1 cells were still 1,000-fold less susceptible to rotavirus infection than MA104 cells, indicating that lack of VLA-2 integrin is not solely responsible for the decreased amount of rotavirus infection of these cells. Whether the enhanced infection of CHOα2β1 observed with SA-independent rotavirus strains over that of SA-dependent rotavirus strains (Fig. 7B and data not shown) depends on the use of VLA-2 as an entry factor (or coreceptor) remains to be determined.
FIG. 8.
Inhibition of RRV or WC3 rotavirus infectivity in stably transfectant CHOα2 and CHOα2β1 cells by MAb (10 μg/ml) to integrin subunit α2 (AK-7). MOI = 500 (RRV) or 100 (WC3). The data are expressed as the percentage of the virus infectivity obtained when the CHO-p901 cells were preincubated with α-MEM as a control. The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean. An asterisk indicates a significant (P < 0.05; Mann-Whitney U test) decrease in rotavirus infectivity of CHO-p901, CHOα2, or CHOα2β1 cells following treatment of the cells with antibody to the α2 or β1 subunit of VLA-2 integrin with respect to the infectivity obtained in corresponding untreated CHO-p901, CHOα2, or CHOα2β1 cells, respectively.
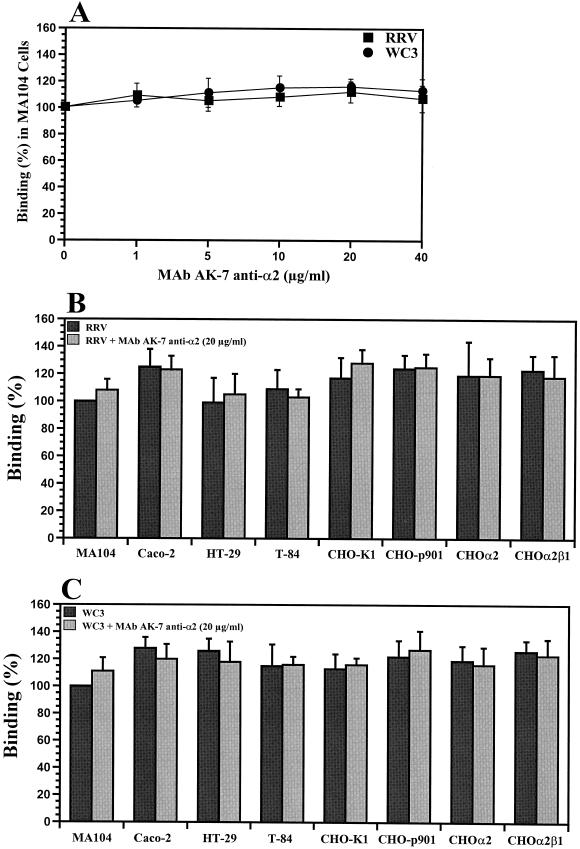
Binding of SA-dependent or SA-independent rotaviruses to continuous human and animal cell lines of different species and tissue origins.
To determine if the increase in susceptibility to rotavirus infection of CHOα2 and CHOα2β1 resulted from an increased capacity to bind virus, we measured the abilities of SA-dependent and SA-independent rotaviruses to bind to CHO-K1, CHO-p901, CHOα2, CHOα2β1, BHK-21, GPC-16, Rie1, MA104, Caco-2, HT-29, and T-84 cells by using FFA, ELISA, or radiolabeled virus binding assays. Testing of the capacity of different concentrations of MAb AK-7 (directed to the α2 subunit of VLA-2) to inhibit infectious RRV or WC3 virus binding in MA104 cells revealed that MAb AK-7 was not able to inhibit binding of either RRV or WC3 to these cells (Fig. 9A). Binding of infectious RRV or WC3 virus to the CHOα2 and CHOα2β1 cell lines, as measured by FFA, was not increased (P ≥ 0.640; Mann-Whitney U test) over binding to poorly susceptible CHO-p901 or CHO-K1 or susceptible MA104, Caco-2, HT-29, or T-84 cell lines (Fig. 9B and C). Levels of binding of additional SA-dependent (SA11, OSU, and H-1) and SA-independent (Wa, ALA, H-2, HAL1166, PA169, and SA11 Cl3 954/23R) rotaviruses to CHOα2, CHOα2β1, CHO-p901, CHO-K1, MA104, Caco-2, HT-29, or T-84 cells were also equivalent (P > 0.523; Mann-Whitney U test) (data not shown). Binding results were confirmed with a more direct radioactive assay that employed purified radiolabeled and nonradiolabeled virus (data not shown). When binding was analyzed by ELISA, similar results were also obtained (data not shown). Also, binding of infectious RRV or WC3 virus to poorly susceptible BHK-21, GPC-16, or Rie1 cells was not increased over binding to highly susceptible MA104, Caco-2, HT-29, or T-84 cell lines (data not shown). However, binding of SA-dependent RRV or SA-independent WC3 to cells was specifically blocked if the viruses were preincubated with hyperimmune sera containing neutralizing antibodies to the viruses tested (data not shown). Binding of RRV, but not of WC3, was decreased when cells were pretreated with 20 mU of neuraminidase/ml, suggesting SA residues on the surfaces of cells play a role in the initial attachment of RRV to these cells (data not shown).
FIG. 9.
Capacities of different concentrations (0 to 40 μg/ml) of MAb AK-7, directed to the α2 subunit of the VLA-2 integrin, to inhibit binding of RRV or WC3 rotavirus strains in MA104 cells (A) and binding of infectious SA-dependent rotavirus (RRV) in the absence or presence of MAb AK-7 to intestinal (Caco-2, HT-29, and T-84), kidney (MA104), or ovary (CHO-K1, CHO-p901, CHOα2, and CHOα2β1) cells (B) or SA-independent rotavirus (WC3) in the absence or presence of MAb AK-7 to intestinal (Caco-2, HT-29, and T-84), kidney (MA104), or ovary (CHO-K1, CHO-p901, CHOα2, and CHOα2β1) cells (C). Binding of infectious rotavirus was blocked (90 to 100%) when the virus was preincubated with rabbit hyperimmune serum containing neutralizing antibodies to RRV or WC3 (data not shown). The data are expressed as the percentage of infectious virus binding when MA104 cells were preincubated with 199 medium as a control. The values shown are the arithmetic means of at least three replicate experiments. The error bars represent one standard error of the mean.
Since CHOα2 and CHOα2β1 cells bound equal amounts of SA-dependent and SA-independent rotavirus, VLA-2 by itself cannot be responsible for rotavirus binding to cells. Furthermore, binding of the infectious RRV (Fig. 9B) or WC3 (Fig. 9C) rotavirus strain to CHOα2 and CHOα2β1 cells was not blocked by MAb AK-7 to α2 at a concentration of 20 μg/ml. Thus, binding of infectious rotavirus to the surface of CHO cells did not appear to be a critical step in determining the differential susceptibilities exhibited by MA104 and CHO cells.
DISCUSSION
Cellular receptor-virus associations are the initial step in a viral infection, and many different cell surface molecules can serve as receptors for the attachment or entry of viruses. The specificity of viruses for their cellular receptors has implications for host and tissue range, interspecies barriers, and viral pathogenesis. Cellular receptors used by viruses belong to different families of proteins, carbohydrates, or lipids, often embedded in complex cell surface matrices that are involved in immune modulation, signaling pathways, cell adhesion, or some other unknown function. The identity of the cellular receptor(s) for rotavirus has been controversial. Several molecules, including SA residues and VLA-2 and αvβ3 integrins, have been proposed as cellular receptors for rotaviruses, but the initial steps of rotavirus binding to and entry into cells are complex and involve a multistep process (2, 4, 13, 15, 17, 19, 32–34, 36, 41, 46, 55, 58, 59, 66–68).
The present study demonstrates that SA-dependent and SA-independent rotaviruses are able to infect a wider and more varied range of host and target cells than had been previously recognized. Cultured epithelial cells of renal or intestinal origin have been thought to be the most susceptible to rotavirus infection (5, 23, 24, 42, 46), but cell lines of bone, breast, stomach, and lung origin are as susceptible to rotavirus infection as are cells of renal and intestinal origin. In nature, rotavirus is known to infect many avian and mammalian species, including humans, monkeys, rabbits, cows, pigs, dogs, and mice (all species represented in the cell lines tested), and although we recently reported that rats are susceptible to rotavirus infection (14), guinea pigs and hamsters are not known to serve as natural hosts for rotaviruses. Cells derived from several human and animal tissues, including kidney, intestine, cervix, breast, bone, lung, stomach, skin, prostate, ovary, and liver, can be classified into cells that are highly, moderately, or poorly susceptible to rotavirus infection, but out of the 46 cell lines tested, none was completely refractory to infection. The majority of the cell lines tested were highly susceptible to rotavirus infection, some cell lines were moderately susceptible (HEK 293T, HeLa, HepG2, RD, 148-1LMD, L-929, 3T3, HFF, and CREF cells), and several cell lines were poorly susceptible to rotavirus infection. Rotavirus infection of CHO (including three different cell lines, CHO-K1, CHO-CD36 and CHO-p901), Rie1, GPC-16, MODE-K, and BHK-21 cells was significantly (P ≤ 0.001; Mann-Whitney U test) less efficient (1,000- to 10,000-fold difference) than rotavirus infection of most cells examined, and infection could be detected only when the cells were exposed to MOIs of 100 (WC3) or 500 (RRV). Nevertheless, all of these poorly susceptible cell lines become permissive for rotavirus replication, similar to the level of susceptible cells, if entry is bypassed. Our ability to infect the majority of the cell lines tested suggests that rotaviruses have a broad tissue tropism, reflecting a single widely expressed cellular receptor or more than one receptor for rotavirus. Further studies are required to understand the biological importance of the wide tropism exhibited by rotaviruses in cell culture and the limited cell tropism during infection in an animal or human host. Under normal circumstances, rotavirus infection is restricted to the mature enterocytes of the villi of the small intestine; however, in immunocompromised hosts, extraintestinal rotavirus infection, mainly in the liver, has been reported (26, 30, 42, 52, 62).
Most integrins are expressed on a variety of cells; most cells express several, but not all, integrins (7, 22, 37); and several viruses utilize integrins as cellular receptors or coreceptors (8, 9, 25, 39, 40, 53, 65). Recently, several reports have implicated VLA-2 integrin in rotavirus cell attachment or entry (17, 36, 44, 68). In this study, we demonstrate that rotavirus infection of CHO or RD cells, which normally do not express VLA-2 and are 1,000- to 10,000- or 100-fold less susceptible than fully permissive MA104 cells, respectively, increases 2- to 10-fold when VLA-2 is introduced into CHO cells via stable transfection. Moreover, enhanced infection of SA-dependent rotaviruses in CHO cells is determined by the human α2 subunit and does not require expression of human β1 (2-fold increase), but infection of SA-independent rotaviruses is further enhanced by expression of both human α2 and β1 (10-fold increase). The level of enhanced susceptibility obtained in CHO cells is similar to that obtained in poorly susceptible K562 cells expressing VLA-2 with the SA-dependent SA11 rotavirus strain (36). Since the level of permissivity of CHO cells transfected with VLA-2 does not approach that of fully permissive cell lines, VLA-2 is not likely the primary rotavirus receptor. In addition, we show for the first time that SA-dependent and SA-independent rotavirus strains bind efficiently, and at equivalent levels as measured by three distinct methods, to all highly and poorly susceptible cell lines tested, regardless of their VLA-2 expression phenotype. Therefore, CHO and the other poorly susceptible cell lines tested bear the receptor(s) for virus binding but lack at least one of the cell surface proteins (or lipids or carbohydrates) that form part of the rotavirus-receptor complex required for rotavirus entry.
The complex nature of the interactions of rotaviruses with gastrointestinal cells is highlighted by the fact that upon binding or entry, rotavirus or its gene products stimulate cytokine and chemokine responses (10, 54, 57), an enterotoxin (3), the enteric nervous system (45), and cell signaling pathways (43). Our data and previous findings (68) suggest that VLA-2 may function in postattachment events or as a coreceptor for virus entry. Alternatively, the interaction of rotavirus with VLA-2, or other integrins, could induce signaling pathways. Although the rotavirus VP4 protein, which contains the sequence motifs DGE and IDA potentially recognized by VLA-2 and VLA-4, respectively, enhances NF-κB activation through the tumor receptor-associated factor 2 (TRAF2) that inhibits Jun N-terminal kinase signaling (43), it is unclear if this enhancement is an early or late event during infection. The region of VP4 that mediates this effect remains uncharacterized. It will be of interest to determine if rotaviruses induce such signaling pathways via initial interaction with integrins.
If integrins play a role in rotavirus attachment or entry, they must be accessible to the virus in natural infections. Although integrins are found associated with the basolateral cell surfaces of enterocytes (6, 7, 37, 47, 61), recent data (15, 20, 51, 60) indicate that rotavirus infection of polarized intestinal epithelial Caco-2 cells causes disruption of tight junctions and loss of transepithelial electrical resistance in the absence of cell death; thus, once tight junctions are disrupted, virus would have access to the basolateral surface. In this regard, we recently found that SA-independent rotaviruses, unlike SA-dependent rotaviruses, infect polarized cells efficiently through the basolateral surface (15), which may explain why expression of VLA-2 promotes SA-independent rotavirus infection eight times more than that of SA-dependent rotaviruses.
Based on analysis of the surface expression of VLA-2 of six human cell lines (MA104, Caco-2, COS-7, HepG2, RD, and K562), Londrigan et al. (44) reported that cell surface expression of VLA-2 correlates with susceptibility to rotavirus infection. Our data suggest that VLA-2 cannot be the sole determinant because cells that completely lack VLA-2 (i.e., CHO-p901, CHO-K1, and BHK-21 [references 1 and 29 and this report]) are as poorly susceptible as cells with a small amount of VLA-2 (i.e., Rie1, GPC-16, and MODE-K). Although the absence or reduced amount of VLA-2 in these cell lines could explain their low susceptibilities to rotavirus infection, human HeLa and murine L929 and 3T3 cells express levels of VLA-2 similar to those on MA104 cells (references 9 and 22 and this report) but their susceptibility to rotavirus infection is 100-fold less than that of MA104 cells.
Combined treatment with neuraminidase and α2 MAb show that SA-dependent rotaviruses utilize SA residues, not VLA-2, as functional receptors, suggesting that antibodies to VLA-2 integrin may block rotavirus infection due to steric hindrance. Also, rotavirus may interact with the αvβ3 integrin (although none of the rotavirus proteins have the consensus RGD sequence recognized by the αvβ3 integrin) through a site proximal to the RGD recognition domain in a fashion similar to that of hantaviruses (28, 33). Because antibodies to VLA-2 and αvβ3 have an additive effect on blocking rotavirus infectivity in MA104 cells (reference 33 and this report), coexpression of both α2β1 and αvβ3 in CHO cells may result in further enhancement of rotavirus infectivity in these cells.
In conclusion, this study furthers our understanding of rotavirus tropism and contributes to the identification of the primary rotavirus cellular receptor. Receptor binding is just the first step in infection. Several cell molecules may be required for efficient rotavirus infectivity, and different rotavirus strains may bind to distinct molecules in various cell lines or distinct molecules in the same cells. Although our data indicate that VLA-2 is not a functional attachment receptor for rotavirus, it appears to possibly function as an internalization receptor or coreceptor required to establish a productive infection. Recently, five individual proteins of 30, 45, 57, 75, and 110 kDa which inhibited rotavirus infectivity in MA104 cells were identified (2, 32). Several of these molecules may serve as receptors for rotavirus. Also a MAb to an as-yet-unidentified MA104 surface protein inhibits rotavirus infectivity in a fashion similar to that of MAbs to the α2 subunit of VLA-2, and the protein recognized by this MAb is present in poorly susceptible cells, such as CHO or L929 cells (2, 46, 68). Therefore, the classical concept of the rotavirus cellular receptor as a single entity that participates in the recognition process has been superseded by new data indicating that binding and entry is a multistep process, often involving different virus attachment proteins and more than one host cell receptor most likely immersed in glycosphingolipid-enriched plasma membrane microdomains (2, 49). Identification of such receptors may allow a rational approach to designing drugs to inhibit the process of rotavirus infection.
Acknowledgments
We thank Martin E. Hemler (Dana-Farber Cancer Institute, Harvard Medical School, Boston, Mass.), Lennart Svensson (Karolinska Institute, Stockholm, Sweden), Tien C. Ko and Sheila Crowe (University of Texas Medical Branch, Galveston), John Klein (University of Texas Dental Branch, Houston), Helen Berschneider (North Carolina State University, Raleigh), Don H. Rubin (Vanderbilt University, Nashville, Tenn.), and Richard E. Sutton, Marc-Henri van Maanen, Ronald T. Javier, Isabel Latorre, Susan J. Marriott, and Brenda G. Hogue (Baylor College of Medicine, Houston, Tex.) for supplying cell lines. We also thank Jeffrey M. Scott (Center for AIDS Research [CFAR] Immunology Core, Department of Immunology, Baylor College of Medicine) for help in flow cytometric analyses.
This work was supported by National Institute of Diabetes and Digestive and Kidney Disease grants DK30144 and DK56338 to the Texas Gulf Coast Digestive Diseases Center, NRSA NIH F32-AI10604 (S.E.B. and E.C.), and Pediatric Gastroenterology Training Grant DK07664.
REFERENCES
- 1.Amadori, M., G. Volpe, P. Defilippi, and C. Berneri. 1997. Phenotypic features of BHK-21 cells used for production of foot-and-mouth disease vaccine. Biologicals 25:65–73. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C. F., C. Guerrero, E. Méndez, S. Zárate, P. Iša, R. Espinosa, P. Romero, and S. López. 2001. Early events of rotavirus infection: the search for the receptor(s). Novartis Found. Symp. 238:47–63. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. M., P. Tian, C. Q.-Y. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104. [DOI] [PubMed] [Google Scholar]
- 4.Bass, D., E. Mackow, and H. B. Greenberg. 1991. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology 183:602–610. [DOI] [PubMed] [Google Scholar]
- 5.Bass, D., M. Baylor, C. Chen, E. M. Mackow, M. Bremont, and H. B. Greenberg. 1992. Liposome-mediated transfection of intact particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J. Clin. Investig. 90:2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basson, M., I. M. Modlin, and J. A. Madri. 1992. Human enterocyte (Caco-2) migration is modulated in vitro by extracellular matrix composition and epidermal growth factor. J. Clin. Investig. 90:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaulieu, J. 1999. Integrins and the human intestinal cell functions. Front. Biosci. 4:D310–D321. [DOI] [PubMed] [Google Scholar]
- 8.Bergelson, J. M., M. P. Shepley, B. M. C. Chan, M. E. Hemler, and R. W. Finberg. 1992. Identification of the integrin VLA-2 as a receptor for echovirus 1. Science 255:1718–1720. [DOI] [PubMed] [Google Scholar]
- 9.Bergelson, J. M., N. St. John, S. Kawaguchi, M. Chan, H. Stubdal, J. Modlin, and R. W. Finberg. 1993. Infection by echoviruses 1 and 8 depends on the α2 subunit of human VLA-2. J. Virol. 67:6847–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casola, A., M. K. Estes, S. E. Crawford, P. L. Ogra, P. B. Ernst, R. Garofalo, and S. E. Crowe. 1998. Rotavirus infection of cultured intestinal epithelial cells induces secretion of CXC and CC chemokines. Gastroenterology 114:947–955. [DOI] [PubMed] [Google Scholar]
- 11.Chan, B., N. Matsuura, Y. Takada, B. Zetter, and M. E. Hemler. 1991. In vitro and in vivo consequences of VLA-2 expression on rhabdomyosarcoma cells. Science 251:1600–1602. [DOI] [PubMed] [Google Scholar]
- 12.Ciarlet, M., M. K. Estes, C. Barone, R. F. Ramig, and M. E. Conner. 1998. Analysis of host range restriction determinants in the rabbit model: comparison of homologous and heterologous rotavirus infections. J. Virol. 72:2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen Virol. 80:943–948. [DOI] [PubMed] [Google Scholar]
- 14.Ciarlet, M., M. E. Conner, M. J. Finegold, and M. K. Estes. 2002. Group A rotavirus infection and age-dependent diarrheal disease in rats: a new animal model to study the pathophysiology of rotavirus infection. J. Virol. 76:41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciarlet, M., S. E. Crawford, and M. K. Estes. 2001. Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J. Virol. 75:11834–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombs, K. M., B. N. Fields, and S. C. Harrison. 1990. Crystallization of the reovirus type 3 Dearing core. Crystal packing is determined by the λ2 protein. J. Mol. Biol. 215:1–5. [DOI] [PubMed] [Google Scholar]
- 17.Coulson, B. S., S. Londrigan, and D. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford, S. E., M. Labbé, J. Cohen, M. Burroghs, J.-Y. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delorme, C., H. Brüssow, J. Sidoti, N. Roche, K.-A. Karlsson, J.-R. Neeser, and S. Teneberg. 2001. Glycosphingolipid binding specificities of rotaviruses: identification of a sialic acid-binding epitope. J. Virol. 75:2276–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickman, K., S. Hempson, J. Anderson, S. Lippe, L. Zhao, R. Burakoff, and R. Shaw. 2000. Rotavirus alters paracellular permeability and energy metabolism in caco-2 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G757–G766. [DOI] [PubMed] [Google Scholar]
- 21.DuBridge, R. B., and M. P. Calos. 1988. Recombinant shuttle vectors for the study of mutation in mammalian cells. Mutagenesis 3:1–9. [DOI] [PubMed] [Google Scholar]
- 22.Edelman, J., B. Chan, S. Uniyal, H. Onodera, D. Wang, N. St John, L. Damjanovich, D. Latzer, R. Finberg, and J. M. Bergelson. 1994. The mouse VLA-2 homologue supports collagen and laminin adhesion but not virus binding. Cell Adhes. Commun. 2:131–143. [DOI] [PubMed] [Google Scholar]
- 23.Estes, M. K., D. Y. Graham, C. Gerba, and E. Smith. 1979. Simian rotavirus SA11 replication in cell cultures. J. Virol. 31:810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estes, M. K. 2001. Rotaviruses and their replication, p.1747–1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Evander, M., I. Frazer, E. Payne, Y. Qi, K. Hengst, and N. McMillan. 1997. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J. Virol. 71:2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitts, S., M. Green, J. Reyes, B. Nour, A. Tzakis, and S. Kocoshis. 1995. Clinical features of nosocomial rotavirus infection in pediatric liver transplant recipients. Clin. Transplant. 9:201–204. [PubMed] [Google Scholar]
- 27.Gaush, C. R., W. L. Hard, and T. F. Smith. 1966. Characterization of an established line of canine kidney cells (MDCK). Proc. Soc. Exp. Biol. Med. 122:931–935. [DOI] [PubMed] [Google Scholar]
- 28.Gavrilovskaya, I., M. Shepley, R. Shaw, M. Ginsberg, and E. R. Mackow. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giancotti, F., and E. Ruoslahti. 1990. Elevated levels of the α5β1 fibronectin receptor suppress the transformed phenotype of Chinese hamster ovary cells. Cell 60:849–859. [DOI] [PubMed] [Google Scholar]
- 30.Gilger, M. A., D. Matson, M. E. Conner, H. Rosenblatt, M. J. Finegold, and M. K. Estes. 1992. Extraintestinal rotavirus infections in children with immunodeficiency. J. Pediatr. 12:912–917. [DOI] [PubMed] [Google Scholar]
- 31.González-Vallina, R., H. Wang, R. Zhan, H. M. Berschneider, R. M. Lee, N. O. Davidson, and D. D. Black. 1996. Lipoprotein and apolipoprotein secretion by a newborn piglet intestinal cell line (IPEC-1). Am. J. Physiol. 271:G249–G259. [DOI] [PubMed] [Google Scholar]
- 32.Guerrero, C. A., S. Zárate, G. Corkidi, S. López, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerrero, C. A., E. Méndez, S. Zárate, P. Iša, S. López, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644–14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, C.-T., O. Nakagomi, M. Mochizuchi, H. Ishida, M. Kiso, Y. Ohta, T. Suzuki, D. Miyamoto, K. Hidari, and Y. Suzuki. 1999. Ganglioside GM1a on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem. 126:683–688. [DOI] [PubMed] [Google Scholar]
- 35.Hardy, M. E., T. N. Tanaka, N. Kitamoto, L. J. White, J. M. Ball, and M. K. Estes. 1996. Antigenic mapping of the recombinant Norwalk virus capsid protein using monoclonal antibodies. Virology 217:252–261. [DOI] [PubMed] [Google Scholar]
- 36.Hewish, M., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hynes, R. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11–25. [DOI] [PubMed] [Google Scholar]
- 38.Iša, P., S. López, L. Segovia, and C. F. Arias. 1997. Functional and structural analysis of the sialic acid-binding domain of rotavirus. J. Virol. 71:6749–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson, T., A. Sharma, R. Ghazaleh, W. Blakemore, F. Ellard, D. Simmons, J. Newman, D. I. Stuart, and A. M. Q. King. 1997. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth-disease viruses to the purified integrin αvβ3 in vitro. J. Virol. 71:8357–8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. King. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jolly, C., B. Beisner, and I. H. Holmes. 2000. Rotavirus infection of MA104 cells is inhibited by Ricinus lectin and separately expressed single binding domains. Virology 275:89–97. [DOI] [PubMed] [Google Scholar]
- 42.Kitamoto, N., R. F. Ramig, D. Matson, and M. K. Estes. 1991. Comparative growth of different rotavirus strains in differentiated cells (MA104, HepG2, and CaCo-2). Virology 184:729–737. [DOI] [PubMed] [Google Scholar]
- 43.LaMonica, R., S. S. Kocer, J. Nazarova, W. Dowlong, E. Geimonen, R. D. Shaw, and E. R. Mackow. 2001. VP4 differentially regulates TRAF2 signaling, disengaging JNK activation while directing NF-κB to effect rotavirus-specific cellular responses. J. Biol. Chem. 276:19889–19896. [DOI] [PubMed] [Google Scholar]
- 44.Londrigan, S., M. Hewish, M. Thompson, G. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81:2203–2213. [DOI] [PubMed] [Google Scholar]
- 45.Lundgren, O., A. Peregrin, K. Persson, S. Kordasti, I. Uhnoo, and L. Svensson. 2000. Role of the enteric nervous system in the fluid and electrolyte secretion of rotavirus diarrhea. Science 287:491–495. [DOI] [PubMed] [Google Scholar]
- 46.López, S., R. Espinosa, P. Iša, M. T. Merchant, S. Zárate, E. Méndez, and C. F. Arias. 2000. Characterization of a monoclonal antibody directed to the surface of MA104 cells that blocks the infectivity of rotaviruses. Virology 273:160–168. [DOI] [PubMed] [Google Scholar]
- 47.McCormick, B. A., A. Nusrat, C. Parkos, L. D’Andrea, P. Hofman, D. Carnes, T. W. Liang, and J. L. Madara. 1997. Unmasking of intestinal epithelial lateral membrane β1 integrin consequent to transepithelial neutrophil migration in vitro facilitates inv-mediated invasion by Yersinia pseudotuberculosis. Infect. Immun. 65:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Méndez, E., C. F. Arias, and S. López. 1993. Binding to sialic acid is not an essential step for the entry of animal rotavirus to epithelial cells in culture. J. Virol. 67:5253–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Méndez, E., S. López, M. Cuadras, P. Romero, and C. F. Arias. 1999. Entry of rotaviruses is a multistep process. Virology 263:450–459. [DOI] [PubMed] [Google Scholar]
- 50.Nasu, Y., T. Timme, G. Yang, C. Bangma, L. Li, C. Ren, S. Park, M. DeLeon, J. Wang, and T. C. Thompson. 1998. Suppression of caveolin expression induces androgen sensitivity in metastatic androgen-insensitive mouse prostate cancer cells. Nat. Med. 4:1062–1064. [DOI] [PubMed] [Google Scholar]
- 51.Obert, G., I. Peiffer, and A. Servin. 2000. Rotavirus-induced structural and functional alterations in tight junctions of polarized intestinal Caco-2 cell monolayers. J. Virol. 74:4645–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riepenhoff-Talty, M., K. Schaekel, H. F. Clark, W. Mueller, I. Uhnoo, T. Rossi, J. Fisher, and P. L. Ogra. 1993. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr. Res. 33:394–399. [DOI] [PubMed] [Google Scholar]
- 53.Roivainen, M., L. Piirainen, T. Hovi, I. Virtanen, T. Riikonen, J. Heino, and T. Hyypia. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology 203:357–365. [DOI] [PubMed] [Google Scholar]
- 54.Rollo, E., K. Kumar, N. Reich, J. Cohen, J. Angel, H. B. Greenberg, R. Sheth, J. Anderson, B. Oh, S. Hempson, E. R. Mackow, and R. D. Shaw. 1999. The epithelial cell response to rotavirus infection. J. Immunol. 163:4442–4452. [PubMed] [Google Scholar]
- 55.Rolsma, M. D., T. Kuhlenschmidt, H. Gelberg, and M. S. Kuhlenschmidt. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruiz, M. C., M. J. Abad, A. Charpilienne, J. Cohen, and F. Michelangeli. 1997. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J. Gen. Virol. 78:2883–2893. [DOI] [PubMed] [Google Scholar]
- 57.Sheth, R., J. Anderson, T. Sato, B. Oh, S. Hempson, E. Rollo, E. R. Mackow, and R. D. Shaw. 1996. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology 221:251–259. [DOI] [PubMed] [Google Scholar]
- 58.Srnka, C., M. Tiemeyer, J. Gilbert, M. Moreland, H. Schweingruber, B. de Lappe, P. James, T. Grant, R. Willoughby, R. Yolken, M. Nashed, S. Abbas, and R. Laine. 1992. Cell surface ligands for rotavirus: mouse intestinal glycolipids and synthetic carbohydrate analogs. Virology 190:794–805. [DOI] [PubMed] [Google Scholar]
- 59.Superti, F., and G. Donelli. 1991. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J. Gen. Virol. 72:2467–2474. [DOI] [PubMed] [Google Scholar]
- 60.Svensson, L., B. Finlay, D. Bass, C.-H. von Bonsdorff, and H. B. Greenberg. 1991. Symmetric infection of rotavirus on polarized human intestinal epithelial (Caco-2) cells. J. Virol. 65:4190–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tafazoli, F., A. Holmstrom, A. Forsberg, and K. E. Magnusson. 2000. Apically exposed, tight junction-associated β1-integrins allow binding and YopE-mediated perturbation of epithelial barriers by wild-type Yersinia bacteria. Infect. Immun. 68:5335–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhnoo, I., M. Riepenhoff-Talty, T. Dharakul, P. Chegas, J. Fisher, H. B. Greenberg, and P. Ogra. 1990. Extramucosal spread and development of hepatitis in immunodeficient and normal mice infected with rhesus rotavirus. J. Virol. 64:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urlaub, G., and L. A. Chasin. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. USA 77:4216–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickham, T., P. Mathias, D. Cheresh, and G. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309–319. [DOI] [PubMed] [Google Scholar]
- 66.Willoughby, R. E., R. H. Yolken, and R. L. Schnaar. 1990. Rotaviruses specifically bind to neutral glycosphingolipid asialo-GM1. J. Virol. 64:4830–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zárate, S., R. Espinosa, P. Romero, E. Méndez, C. F. Arias, and S. López. 2000. The VP5 domain of VP4 can mediate attachment of rotaviruses to cells. J. Virol. 74:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zárate, S., R. Espinosa, P. Romero, C. Guerrero, C. F. Arias, and S. López. 2000. Integrin α2β1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50–54. [DOI] [PubMed] [Google Scholar]