Abstract
A novel class of RNA-binding proteins, Puf, regulates translation and RNA stability by binding to specific sequences in the 3′-untranslated region of target mRNAs. Members of this protein family share a conserved Puf domain consisting of eight 36 amino acid imperfect repeats. Here we report two Puf family member genes, PfPuf1 and PfPuf2, from the human malaria parasite Plasmodium falciparum. Both genes are spliced with four and three introns clustered within or near the Puf domains, respectively. Northern and RT–PCR analysis indicated that both genes were differentially expressed in gametocytes during erythrocytic development of the parasite. Except for similarities in the Puf domain and expression profile, the deduced PfPuf1 and PfPuf2 proteins differ considerably in size and structure. PfPuf1 has 1894 amino acids and a central Puf domain, whereas PfPuf2 is much smaller with a C-terminal Puf domain. The presence of at least two Puf members in other Plasmodium species suggests that these proteins play evolutionarily similar roles during parasite development. Both in vivo studies using the yeast three-hybrid system and in vitro binding assays using the recombinant Puf domain of PfPuf1 expressed in bacteria demonstrated intrinsic binding activity of the PfPuf1 Puf domain to the NRE sequences in the hunchback RNA, the target sequence for Drosophila Pumilio protein. Altogether, these results suggest that PfPufs might function during sexual differentiation and development in Plasmodium through a conserved mechanism of translational regulation of their target mRNAs.
INTRODUCTION
Despite tremendous global efforts to control malaria, it remains a public health problem in more than 90 countries. The resurgence of malaria in the world, the result of many factors including the emergence and spread of drug-resistant parasites and insecticide-resistant vector mosquitoes, has stimulated research for novel control strategies. One strategy is to develop transmission-blocking vaccines that interrupt the malaria transmission cycle by targeting the sexual stages of the parasite (1). Sexual development is an obligate process for continued transmission of the malaria parasite by vector mosquitoes (2–4). During its development in erythrocytes, certain asexual parasites, responding to poorly understood signals, stop dividing and differentiate into gametocytes, a process known as gametocytogenesis. While the specific cues that signal this developmental transition in Plasmodium remain elusive (5), and gametocytogenesis is influenced by many environmental factors (6,7), it is clear that the capability of producing gametocytes is a genetically inherited character of the parasite (4). For example, spontaneous loss of the ability to produce gametocytes during continuous blood passage in experimental animals or blood culture of parasites without selection pressure for mosquito transmission is often associated with accumulation of chromosomal aberrations (8–11). Furthermore, mutations in certain chromosomes (e.g. Plasmodium falciparum chromosomes 9 and 12) are linked to defects in gametocytogenesis (8,12). Efforts toward transmission-blocking vaccine development and an understanding of sexual development of the malaria parasites have led to the molecular characterization of dozens of sexual stage-specific genes (reviewed in 13,14). However, the molecular mechanisms underlying this important biological process are still poorly understood.
To understand sexual development in Plasmodium, we began to investigate regulation of stage-specific gene expression in gametocytes using mRNA differential display (15). This has led us to identify a member of the newly described Puf RNA-binding protein (RBP) family. The name Puf was derived from Pumilio (Pum) protein in Drosophila melanogaster and fem-3 binding factor (FBF) protein in Caenorhabditis elegans (16,17). The Puf protein family is evolutionarily conserved in eukaryotes and widely distributed among plants, fungi, animals and protists. The signature feature of the Puf protein family is the highly conserved core RNA-binding domain, referred to as the Puf domain, which consists of eight tandem imperfect repeats of ∼36 amino acids (16,17). Despite the diversity of Puf functions, they appear to share a common, probably ancestral, role in each species, of promoting proliferation of cells and repressing their differentiation (18). Moreover, Puf proteins act through a similar molecular mechanism by repressing the translation of their respective target mRNAs and enhancing their decay (17,19–21). To date, the functions of Puf proteins have been elucidated during the developmental process of several genetically amenable organisms, including D.melanogaster, C.elegans, Dictyostelium discoideum and Saccharomyces cerevisiae. The best characterized Pum protein in Drosophila binds to the Nanos-responsive elements (NREs) in the maternal hunchback (hb) mRNA during embryogenesis. Translational repression of hb mRNA by Pum produces an antero-posterior gradient of Hunchback protein, which is essential for abdomen formation (19). Pum is also required to regulate embryonic and post-embryonic primordial germ cell development, gonadogenesis and oogenesis (22,23). In C.elegans hermaphrodites, two Puf proteins, FBF1 and FBF2, control the sperm–oocyte switch in the reproductive tract by binding to the 3′-untranslated region (3′-UTR) of fem-3 mRNA and repressing its translation (17). Additional Puf members in C.elegans appear to have an analogous function to Pum in regulating primordial germ cell development (24). In the slime mold D.discoideum, PufA represses the translation of a protein kinase (PKA) mRNA, keeping cells in vegetative division (20). Starvation relieves the repression of PKA, promoting development and fruiting body formation. In the yeast S.cerevisiae, the Puf protein Mpt5 represses expression of the HO gene, which encodes an endonuclease that stimulates mating-type switching (21). To date, the only protozoan Puf protein characterized is the TbPuf1 from Trypanosoma brucei, where it is essential for cell viability (25). Although its target gene(s) has not been isolated, it appears that TbPuf1 interacts with ESAG8, a putative regulatory protein controlling the expression site loci in T.brucei, through the most C-terminal repeats. Altogether, these studies and recent work on the crystal structures of Puf proteins have shed light on the molecular mechanism by which Puf family proteins regulate mRNA translation (26,27).
The related functions of Puf protein in germline development of Drosophila and C.elegans have prompted us to investigate the Puf homologs in the sexual development of the malaria parasite P.falciparum. Here we report the isolation and molecular characterization of two Puf members in P.falciparum that have distinct gene structures and limited sequence homology. Interestingly, both genes are differentially expressed in gametocytes, suggesting that they might play important regulatory roles during sexual differentiation and development. Moreover, using in vivo and in vitro assays we demonstrate that the PfPuf1 Puf domain has conserved binding activity for the hb NRE sequence, suggesting that the target gene(s) of PfPuf1 may share a similar consensus sequence to the hb NRE.
MATERIALS AND METHODS
Parasites and nucleic acid extraction
Plasmodium falciparum clone 3D7 was maintained as previously described (28). Mixed asexual stages were obtained at 2 days after subculturing. Gametocytes were harvested and purified at 14–16 days by Percoll gradient centrifugation (29). Synchronization of parasite culture and total RNA isolation were done as described previously (15). Parasite DNA was isolated from the parasite pellet by proteinase K digestion and phenol/chloroform extraction (30).
Isolation of the PfPuf1 gene
One 244 bp cDNA fragment, encoding a peptide with significant homology to members of the Puf RBP family, was obtained by differential display (15). To isolate the complete PfPuf1 cDNA, a gametocyte cDNA library in λgt11 was screened with the 244 bp cDNA fragment using the standard protocol (30). Inserts of the positive clones were amplified with λgt11 flanking primers and sequenced. Rapid amplification of cDNA ends (RACE) was performed to obtain 5′ and 3′ sequences of the cDNA (31). The RACE products were capture-cloned in pAMP1 using uracil DNA glycosylase (Life Technologies). To identify the genomic sequence of PfPuf1, a genomic library constructed in λZAP-Express was screened with various portions of PfPuf1 cDNA. Overlapping genomic clones in phagemid were excised from the phage in bacterial strain XLOLR according to the manufacturer’s protocol (Stratagene). The 5′ genomic sequence was obtained by inverse PCR using NdeI-digested and religated P.falciparum DNA (32).
Sequence analysis
DNA was sequenced on an automated ABI 377 sequencer with the BigDye termination mix. For accuracy, the sequences were compared with the sequences from the Malaria Genome Consortium. Sequence analysis was performed using the Genetics Computer Group (GCG) software, version 10.1 (33). GenBank and P.falciparum sequence databases were searched for homologous sequences to PfPufs using various BLAST algorithms (34). Contigs with the Puf genes were retrieved from the databases for genomic organization analysis.
Phylogenetic comparisons
A total of 34 GenBank entries with complete Puf domains were retrieved for phylogenetic analysis. The Puf domains of PfPufs were trimmed and used to generate the data matrix to infer the phylogenetic relationships among Puf family members. Putative Puf members from Plasmodium yoelii and Plasmodium knowlesi were deduced based on the splicing patterns observed in the two PfPufs. Multiple alignment was performed using the CLUSTALW program (http://www. ebi.ac.uk/clustalw) and the phylogenetic tree was drawn from the alignment using the TreeView program (http://taxonomy.zoology.gla.ac.uk).
Nucleic acid hybridization
For northern analysis of PfPuf1, 10 µg of total RNA from mixed asexual stages, gradient-purified gametocytes and gametocytes 10 min after treatment with 100 µM xanthurenic acid were electrophoresed in 1% agarose/formaldehyde gels. The last sample was included to assess possible changes in PfPuf expression in gametocytes undergoing gametogenesis (35,36). For Southern analysis, 3 µg of P.falciparum genomic DNA was completely digested with HindIII, Sau3AI and XbaI and electrophoresed in a 0.7% agarose gel. Both northern and Southern blots were transferred to nylon membrane and hybridized under high stringency conditions to 32P-labeled 244 bp PfPuf1 cDNA fragment (30). Probe labeling was done with [32P]dATP using a random priming kit (Promega).
Chromosomal mapping
To determine the chromosomal locations of the PfPuf1 and PfPuf2 genes, parasite genomic DNA was resolved by pulsed field gel electrophoresis using a contour-clamped homogeneous electric fields apparatus (37). Individual chromosomal DNA bands were excised from the gel and used for PCR with a pair of PfPuf1- and PfPuf2-specific primers, respectively.
RT–PCR analysis
To study PfPuf expression in synchronized blood stages, RT–PCR was performed on cDNAs from individual asexual stages (ring, trophozoite and schizont) and gametocytes (stages I, II and IV–V) (15). PCR conditions (30 cycles) were as described previously and the actin I gene was included as a control. PfPuf1-specific primers (GTTCAGAAATGTTTAATTACC and GACATTTCTCTACAACATTAG) amplified a 337 bp fragment from cDNA and a 544 bp fragment from genomic DNA due to the presence of an intron. PfPuf2-specific primers (ACTGATGAAATTGTAAATG and GTTGTCAATAATTTCTGCATC) amplified a 209 bp fragment from cDNA and a 567 bp fragment from genomic DNA with two introns. PCR products were separated on a 1.5% agarose gel and documented using a Kodak digital camera.
Expression of the PfPuf1 Puf domain in Escherichia coli
To express the conserved RNA-binding domain of PfPuf1 in bacteria, PCR was performed with gametocyte cDNA using KlenTaq (Clontech) and two primers (CTGGAGTCCATATGAACAAACGAGGAGTAC and GAATTCCTCGAGTAAAGGATGTTTTTTAGC) designed to clone at the NdeI and XhoI sites of the expression vector pET 22b(+) (Novagen). This allows expression of the recombinant protein as a non-fusion product with six histidines at the C-terminus for purification by affinity chromatography. Protein expression in E.coli BL21 cells was induced with 1 mM IPTG for 3 h at 30°C, after which bacterial cells were harvested and resuspended in a lysis buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl). The bacteria were lysed by three or four passages through a microfluidizer. Protein purification was done under native conditions using Ni–NTA agarose resin (Qiagen). After binding of the protein to the Ni–NTA agarose, the column was washed with increasing concentrations of imidazole. The expressed protein was eluted with 200 mM imidazole and dialyzed overnight in phosphate-buffered saline at 4°C. The purified protein was analyzed by SDS–PAGE and quantified using the Bio-Rad protein assay reagents using bovine serum albumin (BSA) as the standard.
In vitro RNA binding activity of the recombinant PfPuf1 Puf domain
To test whether the Puf domain of PfPuf1 has conserved RNA binding activity to the NRE sequences of the hb mRNA, in vitro RNA binding assays were performed. For this purpose, purified native recombinant PfPuf1 protein was dialyzed extensively at 4°C in RNA binding buffer (10 mM HEPES, pH 7.4, 50 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% glycerol). To generate the DNA templates for in vitro transcription of the target RNAs, two complementary oligonucleotides corresponding to the hb NRE (38) were synthesized and annealed. The hb NRE sequence is AUUAUUUUGUUGUCGAAAAUUGUACAUAAGCC, where two motifs essential for Pum binding are underlined (Boxes A and B). Three pairs of primers with mutations in Box A or B or both were made to evaluate the binding of PfPuf1 to the mutant NREs. The Box A sequence GUUGU was mutated to CUGCU, while the Box B sequence AUUGU was mutated to AUAUG. The T7 promoter sequence was added to the end of the sense primers for in vitro transcription using T7 RNA polymerase. RNA was synthesized with an RNA transcription kit using 5 µg of template DNA following the manufacturer’s protocol (Stratagene). The RNA of the antisense and mutant NRE sequences was synthesized and used as a negative control in binding assays. Radiolabeling of RNA was done in a transcription reaction in the presence of [α-32P]UTP with a specific activity of 3000 Ci/mmol (Amersham) and labeled RNA was purified from a 6% denaturing PAGE gel. RNA binding and gel shift assays were performed essentially as described (16,39,40). The equilibrium binding constant Kd was determined using gel shift assays (41). Each binding reaction (20 µl) consisted of 10 mM HEPES–KOH, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mg/ml acetylated BSA, 0.01% Tween-20, 0.05 U/µl RNasin (Promega), 0.1 mg/ml poly(rA), 10 µg/ml yeast tRNAs, 32P-labeled RNA (∼1000 c.p.m./µl) and various amounts (0–540 nM) of purified recombinant PfPuf1 protein. For RNA competition assays, cold 10×, 50× and 100× excess RNA was included in the binding reactions. Following incubation at 20°C for 1 h, 4 µl of 50% glycerol was added and the RNA–protein complex was resolved by 6% native PAGE in 0.5× TBE buffer at 4°C, dried, and exposed to X-ray film at –80°C.
In vivo interactions of the PfPuf1 Puf domain with the NRE sequence in yeast
To investigate in vivo binding of the PfPuf1 Puf domain to the hb NRE sequence, we used the yeast three-hybrid system (42). The PfPuf1 Puf domain was cloned into the protein hybrid vector pYESTrp2, at the BamHI and XhoI sites (Invitrogen), to generate plasmid pYESTrp2-PfPuf1; PfPuf1 was expressed as a fusion protein with the B42 transactivation domain. The Drosophila NRE sequence was inserted at the SmaI and BamHI sites of the hybrid RNA vector pRH5′, to produce the bait RNA. The NRE sequence was inserted in both orientations so that either the sense or antisense NRE sequence was fused with the MS2 sequence. The yeast host strain L40uraMS2, in which Lex–MS2 fusion protein was stably integrated, was co-transformed with both pYESTrp2-PfPuf1 and one of the bait plasmids (pRH5′-antisenseNRE or pRH5′-NRE). As negative controls, yeast was co-transformed with the empty bait plasmid pRH5′ or pRH5′-IRE. Yeast co-transformed with the prey plasmid pYESTrp2-IRP and the bait plasmid carrying either the sense or antisense NRE was used as two additional negative controls. Yeast co-transformed with pYESTrp2-IRP and pRH5′-IRE was used as the positive control. Transformed yeast was plated onto a synthetic medium lacking Ura, Trp and His. Positive transformants were selected for β-galactosidase activity using both filter and liquid assays (43). For each assay, at least three yeast colonies were used.
RESULTS
Identification and cloning of PfPuf1 and PfPuf2 cDNA
Preliminary analysis of a 244 bp differential display product obtained from mature gametocytes revealed significant similarity to the Puf domain of the Puf RBP family (15). Screening a gametocyte cDNA library in λgt11 (∼106 plaques) with the 244 bp cDNA isolated 15 clones, all with inserts of ∼600 bp. Sequencing of three cDNA clones showed identical inserts, partially overlapping with the 244 bp cDNA fragment, plus a short 3′ end extension of the sequence. 5′ and 3′ sequences of the cDNA were procured using 5′ and 3′ RACEs. The incorporation of long-range PCR in two 5′ RACEs extended the 5′ PfPuf1 sequence to ∼170 bp beyond a putative translation initiation codon. Further analysis by RT–PCR and S1 nuclease mapping indicated that the PfPuf1 transcription initiation site(s) was ∼1 kb upstream of the first ATG codon (data not shown). Results from 3′ RACE showed that PfPuf1 mRNA terminated within a region with a homopolymer of 22 T residues, ∼2.6 kb downstream from the Puf domain. A poly(A) tail was located ∼110 bases downstream from the putative stop codon. Two putative polyadenylation signals (AATAAA) were located at ∼80 and ∼90 bases from the stop codon. The estimated size of the PfPuf1 cDNA was ∼7 kb.
Since most of the organisms in which Puf proteins have been characterized have multiple Puf members, we tried to determine if this was true for P.falciparum. When the deduced amino acid sequence of the putative Puf domain of PfPuf1 was used in a TBLASTN search of P.falciparum sequence databases, a sequence with significant homology to Puf domains was identified from the chromosome 4 assembly. Accordingly, we refer to these two P.falciparum Puf genes as PfPuf1 and PfPuf2, respectively.
Mapping and genomic organizations of PfPuf1 and PfPuf2
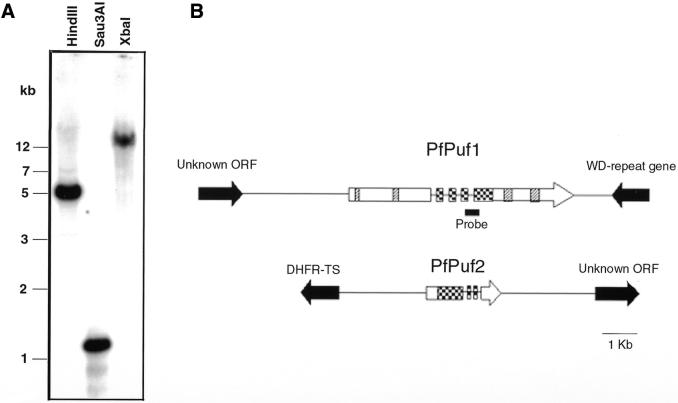
The genomic Southern blot of PfPuf1 detected a single restriction fragment for each enzyme digestion, suggesting that this is a single copy gene (Fig. 1A). To determine the chromosomal locations of the two PfPuf genes, PCRs were performed with individual chromosomal DNAs separated by pulse field gel electrophoresis. The results showed that PfPuf1 and PfPuf2 were amplified from chromosomes 5 and 4, respectively (data not shown). Identification of PfPuf1 and PfPuf2 sequences from specific chromosome assemblies from the Malaria Genome Project further confirmed their chromosomal locations.
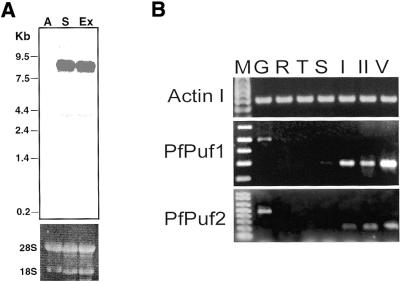
Figure 1.
Genomic organization of PfPuf genes. (A) Genomic Southern of PfPuf1. In each lane, 3 µg of P.falciparum DNA was digested with HindIII, Sau3AI and XbaI and electrophoresed in a 0.7% agarose gel. The blot was probed with the 32P-labeled 244 bp PfPuf1 cDNA fragment. DNA markers are in kb. The location of the probe for genomic Southern and northern analyses is indicated in (B). (B) A schematic representation of the genomic structure of PfPuf loci. Exons are indicated as boxes and introns and intergenic regions as solid lines. The conserved RNA-binding domain is shown as checkered boxes. Other repetitive sequences in PfPuf1 (Fig. 2A) are indicated as hatched boxes. The orientations of the genes are indicated by arrows. Genes flanking the Puf genes are shown as solid arrows.
Four overlapping PfPuf1 genomic clones were isolated by screening a P.falciparum genomic DNA library with labeled 5′ and 3′ RACE products. These clones were sequenced and compared with the cDNA sequence to identify introns. Both the PfPuf1 and PfPuf2 genes and their flanking regions were retrieved from GenBank for detailed sequence analysis using the GCG program. The results showed that PfPuf1 contained four introns (129, 147, 196 and 207 bp), clustered within or immediately upstream of the Puf domain (Fig. 1B). Sequencing of two RT–PCR fragments of PfPuf2 demonstrated the presence of three introns (233, 125 and 124 bp), located within or downstream of the Puf domain (Fig. 1B). The splice donor/acceptor signals (GT/AG) of all introns conform to the consensus for eukaryotic genes. Similar to other P.falciparum genes, the intron sequences (86% AT) are more AT rich than the coding sequences (70%). It is noteworthy that the locations and sizes of the introns are not conserved between PfPuf1 and PfPuf2. Analysis of retrieved genomic sequences showed that PfPuf1 is located within a 10 kb region of chromosome 5, flanked by an upstream gene encoding a hypothetical protein and a downstream gene with homology to the yeast WD repeat gene. PfPuf2 is located within an 8 kb region of chromosome 4, flanked by DHFR-TS and an unknown ORF. All the putative intergenic regions (3 kb 5′ and 1 kb 3′ of the PfPuf1 ORF; 2 kb 5′ and 3.5 kb 3′ of the PfPuf2 ORF) are highly AT rich (86–88%), containing homopolymer runs of A or T.
Predicted PfPuf proteins and the Puf domains
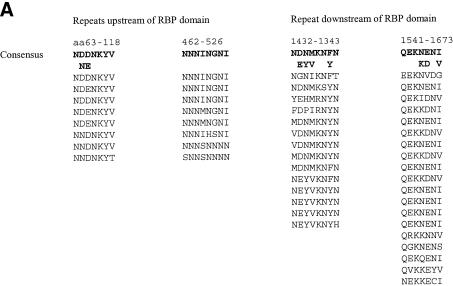
Except for the conserved Puf domains, the deduced PfPuf proteins differ considerably. PfPuf1 is a relatively large protein with 1894 amino acids and a predicted molecular weight of 224 kDa, whereas PfPuf2 has 514 amino acids and a predicted molecular weight of 61.4 kDa. The deduced PfPuf1 is slightly basic with a pI of 8.2, whereas PfPuf2 is more acidic with a pI of 6.2. In addition, the Puf domain of PfPuf1 is located at the center of the polypeptide (823–1109), whereas that of PfPuf2 is at the C-terminus (184–476). Unlike other members of the Puf family, both predicted PfPuf proteins lack Glu/Ala-rich and Ser-rich regions upstream of the Puf domain, two features that were recently identified for most Puf members (44). As is true for many P.falciparum proteins predicted from chromosome 2 and 3 sequences (45,46), PfPuf1 also contains regions of low complexity, including tandem arrays of repeated peptide motifs and homopolymer runs of a single residue (Fig. 2A). For example, PfPuf1 is rich in the amino acid Asn (27%) and a sequence of 33 asparagines (613–645) is located upstream of the Puf domain. The entire deduced PfPuf1 protein contains many repetitive sequences, including the eight imperfect repeats of the Puf domain and four types of short peptide repeats (Fig. 2A). These short repetitive sequences, with the repeat units being comprised of seven or eight amino acids, range from 56 to 133 amino acids. In contrast, the deduced PfPuf2 is not as complex as PfPuf1 and it lacks other repeat motifs except the Puf repeats.
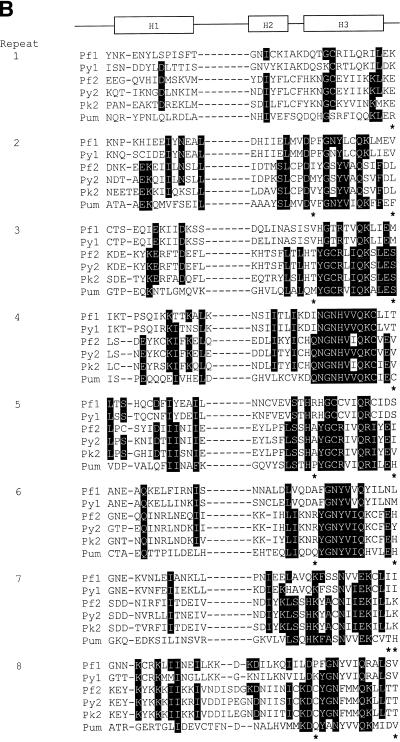
Figure 2.
RNA-binding domains of the deduced PfPuf proteins and other short repeats of PfPuf1. PfPuf1 and PfPuf2 sequences were deposited in GenBank with accession nos AY098937 and AY099486. (A) Short repetitive sequences of PfPuf1. Four types of short repeats are aligned and their locations in the protein and consensus sequences indicated above the sequences. (B) Alignment of Puf domains (eight imperfect repeats) of five Plasmodium Pufs and Drosophila Pum. Plasmodium Puf genes are PfPuf1 (Pf1), PfPuf2 (Pf2), P.yoelii Puf1 (Py1), P.yoelii Puf2 (Py2) and P.knowlesi Puf2 (Pk2). Matching amino acids (at least 4 of 6) are shaded and gaps (–) are introduced to optimize alignment. Asterisks indicate amino acids that are likely to confer RNA binding specificity. Three boxes, labeled H1, H2 and H3, respectively, indicate regions that were determined to form three α-helices in Pum and human Pumilio1.
The Puf RBP family is defined by the presence of eight copies of an imperfect repeat of ∼36 amino acids (16,17). As revealed by a BLASTX search, the repeats of PfPuf proteins share striking homology to those of other Puf family members (Fig. 2B). When the eight repeats of PfPuf1 and PfPuf2 were aligned with those of the Pum protein, the most conserved sequences are in the regions corresponding to the core consensus sequences (47,48). These core sequences have been determined in crystal structures of the Pum and human Pumilio1 RNA-binding domains to form the α-helices that most likely interact with the target RNA (26,27). The sequences forming the RNA-binding helices are 41% identical between PfPuf1 and Pum and 58% identical between PfPuf2 and Pum. The conservation of the repeats of the Puf domain suggests that PfPufs may form a similar 3-dimensional structure and possess similar RNA binding activity, albeit the divergence between the Puf domains of PfPuf1 and PfPuf2 (27% identity) may imply distinct mRNA targets and different roles during parasite development.
The Puf RBP family
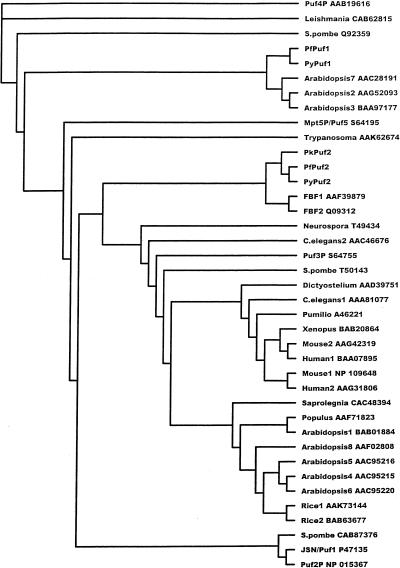
Puf members constitute a large protein family found in evolutionarily diverse eukaryotic organisms such as animals, plants, fungi and protists. A recent BLASTX search of GenBank revealed more than 60 sequences with significant homology to the Puf domains of PfPuf proteins. In the databases of the Malaria Genome Consortium, we have identified two Puf homologs from the rodent malaria parasite P.yoelii (PyPuf1 and PyPuf2) and one from the primate malaria parasite P.knowlesi (PkPuf2) (Fig. 2B). A phylogenetic tree was constructed based on CLUSTALW alignment of 34 GenBank sequences with complete Puf domains and five Plasmodium Puf sequences (Fig. 3). Overall, most Puf genes from plants (Arabidopsis, rice and Populus) and animals (human, mouse, Xenopus, Drosophila and nematode) clustered into two separate groups. Interestingly, the two Plasmodium Puf1 genes are more related to three Arabidopsis Puf genes, whereas the three Plasmodium Puf2 genes are more related to the FBF genes from C.elegans (Fig. 3). Sequence analysis also revealed that Plasmodium Puf genes form two groups (Puf1 and Puf2). Within the Puf1 group, PfPuf1 and PyPuf1 are 78% identical in amino acid sequence of their Puf domains. Members of the Puf2 group (PfPuf2, PyPuf2 and PkPuf2) are 61–70% homologous. However, between the two groups, there is only 27% identity in amino acid sequences. Moreover, Plasmodium Puf genes within the same group are not only conserved in their Puf domains, but also in their genomic organization (in both flanking sequences and introns), conforming to the synteny among Plasmodium species (49). For instance, the three Plasmodium Puf2 sequences are all flanked by an upstream DHFR-TS gene and a downstream unknown ORF, and the flanking sequences are highly homologous. We speculate that all the malaria parasite species may have at least the homologs corresponding to PfPuf1 and PfPuf2.
Figure 3.
A phylogenetic tree showing the relationship between the amino acid sequences of Puf members. GenBank entries with significant homology to the Puf domain of PfPuf1 were retrieved for detailed analysis. Among these, 34 members with complete Puf domains and five Plasmodium Pufs were used for phylogenetic analysis. Only the Puf domains were used for alignment. Each entry is identified by its GenBank accession no.
The expression of PfPuf genes in parasites during erythrocytic development
The expression of the Puf family members, as shown for Pum and FBF, is developmentally regulated. To confirm the results of differential display, northern analysis was performed using RNA from asexual and gametocyte stages. The labeled PfPuf1 cDNA fragment hybridized to an 8 kb mRNA only from gametocytes and gametocytes induced to undergo exflagellation, but not to RNA from asexual stages (Fig. 4A). The discrepancy between the size of the predicted PfPuf1 cDNA (7 kb) and that observed in the northern analysis may be due to the presence of an additional 5′ sequence and a long poly(A) tail. For RT–PCR analysis of PfPuf expression in blood stage parasites, cDNAs were synthesized using total RNA isolated from synchronized parasite stages. The results showed that expression of the actin I gene of the parasite was relatively constitutive during parasite development in erythrocytes, whereas both PfPuf1 and PfPuf2 showed preferential expression in gametocyte stages. The expression patterns of the two PfPuf genes resembled that of the sexual stage-specific gene pfs16 (50).
Figure 4.
Expression of PfPuf genes in blood stage parasites. (A) Northern analysis of PfPuf1 expression. Aliquots of 10 µg of total RNA from P.falciparum asexual stages (A), purified stage IV–V gametocytes (S) and gametocytes 10 min after stimulation with 100 µM xanthurenic acid (Ex) were electrophoresed in 1% agarose/formaldehyde gels and transferred to nylon membranes for hybridization to 32P-labeled 244 bp PfPuf1 cDNA fragment. The upper panel shows the autoradiograph of the hybridization and the lower panel shows the rRNAs in the ethidium bromide stained gel as loading controls. RNA sizes (in kb) are indicated. The faint bands of ∼4 kb might be due to cross-hybridization to the PfPuf2 mRNA. (B) RT–PCR analysis of PfPuf expression during erythrocytic development of the parasite. RT–PCR was performed on total RNA isolated from synchronized asexual parasites as rings (R), trophozoites (T), schizonts (S) and gametocytes at stage I (I), stage II (II) and mixed stages IV and V (V). G indicates PCR amplification from P.falciparum genomic DNA. The actin I gene was used as an internal control, which showed a relatively constitutive expression in erythrocytic stages.
Expression of the PfPuf1 Puf domain in E.coli
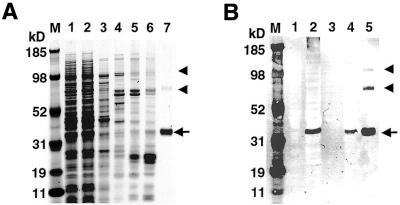
For Pum protein, the eight 36 amino acid repeats plus short sequences on each side (334 amino acids) are essential for RNA binding activity (16). To study PfPuf1 RNA binding activity, the putative Puf domain of PfPuf1 (344 amino acids) corresponding to the Pum RNA-binding domain was expressed in a bacterial expression system. Protein expressed in E.coli was purified using Ni–NTA agarose resin under native conditions since preliminary studies showed that the recombinant protein was soluble (Fig. 5A). Protein expression was confirmed by immunoblotting using a monoclonal anti-His tag antibody, which detected a 40 kDa protein in induced, but not in uninduced, cells (Fig. 5B). The result was consistent with the predicted molecular size of the PfPuf1 Puf domain. The expressed protein was purified to almost homogeneity (Fig. 5A). Two more bands with molecular sizes two and three times that of the expressed monomeric protein were also observed in the eluted proteins. These proteins were likely dimers and trimers of the recombinant protein because they also reacted with the monoclonal anti-His tag antibody (Fig. 5B). The yield of the recombinant protein was 0.5– 0.8 mg/l of bacterial culture.
Figure 5.
Expression and purification of the PfPuf1 RNA-binding domain in a bacterial expression system. (A) SDS–PAGE analysis of protein samples. Lane 1, lysate of induced bacteria; lane 2, lysate passed through a Ni–NTA agarose column; lane 3, 15 mM imidazole wash; lane 4, 30 mM imidazole wash; lane 5, 50 mM imidazole wash; lane 6, 80 mM imidazole wash; lane 7, 200 mM imidazole elution. Proteins were electrophoresed in a 4–12% NuPAGE gradient gel and visualized by Coomassie blue staining. (B) An immunoblot of PfPuf1 RNA-binding domain expression. The samples were separated in a 4–12% SDS–PAGE gel and transferred to nitrocellulose membrane for immunoblotting with monoclonal anti-His tag antibody. Lane 1, lysate of uninduced bacteria; lane 2, lysate of induced bacteria; lane 3, 30 mM imidazole wash; lane 4, 80 mM imidazole wash; lane 5, 200 mM imidazole elution. The 40 kDa PfPuf polypeptide eluted in 200 mM imidazole is indicated by an arrow. Two bands corresponding to the dimers and trimers of the 40 kDa protein are indicated with arrowheads. The MultiMark multi-colored standard (Invitrogen) is indicated in kDa (M).
In vitro binding activity of the recombinant Puf domain of PfPuf1
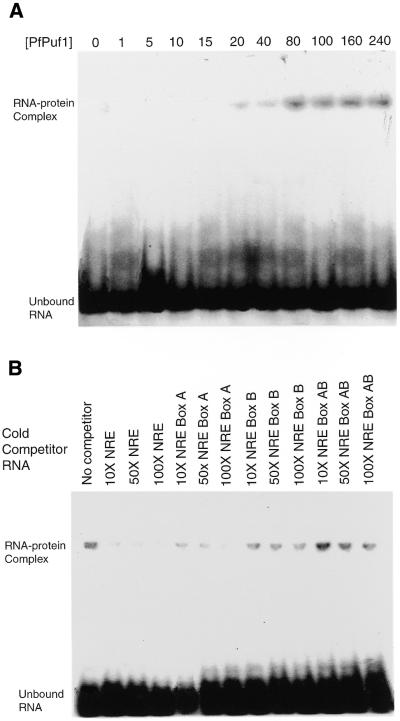
Homology among the repeat sequences forming the H3 helices in Puf proteins suggests similar binding activity to the target sequences. Both the human and mouse recombinant Puf proteins produced in bacteria bind to the Drosophila NRE sequence in vitro (16,44), which suggests that hb NREs may be used as artificial targets to study the binding activity of other Puf family proteins, especially when their authentic target mRNAs are unknown. To determine if the recombinant Puf domain of PfPuf1 has the conserved RNA binding activity, in vitro binding assays were performed using hb NRE as a target RNA. Our preliminary results showed that the recombinant PfPuf1 Puf domain bound to the hb NRE RNA but not the antisense RNA (data not shown). No mobility shift of the labeled RNA was observed when BSA was the only protein in the binding reaction. Using various concentrations (1–240 nM) of recombinant PfPuf1, we further determined the Kd for NRE to be <20 nM (Fig. 6A), comparable to the binding of Pum to NRE. In contrast, mutations at either Box A, B or both, of the NRE sequence impaired the binding (Kd >40 nM for Box A mutation, >60 nM for Box B mutation and >120 nM for simultaneous mutations of Boxes A and B) (data not shown). In addition, excess cold NRE was able to efficiently compete for binding (Fig. 6B). Although NRE with the Box A mutation was still able to compete for binding, this ability was significantly reduced for NREs with mutations at Box B or at both sites. This further suggested that the Box B sequence (UUGU) was more essential for PfPuf1 binding. The NRE sequence may not be an optimal target sequence for PfPuf1, but PfPuf1–NRE interactions demonstrated the RNA binding activity of the recombinant PfPuf1, and the NRE sequence may be useful for the identification of authentic target sequence(s) for PfPuf1.
Figure 6.
RNA binding analysis of recombinant PfPuf1 RNA-binding domain. (A) A gel retardation assay using ∼30 fmol of 32P-labeled NRE RNA and various concentrations (0–240 nM) of recombinant PfPuf1. The Kd was estimated to be <20 nM. (B) A gel retardation assay using ∼30 fmol of 32P-labeled NRE RNA and 60 nM recombinant PfPuf1 in the presence of a 10–100× excess of cold competitor RNAs. NRE and three mutants (Box A, Box B and both Boxes A and B) were included as competitors.
In vivo interactions between the PfPuf1 Puf domain and the NRE sequence in yeast
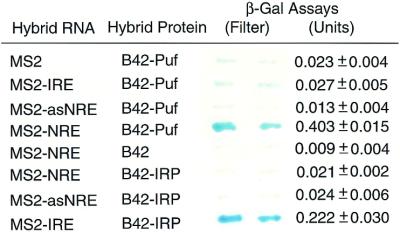
To further confirm the results of the in vitro binding assays, we studied the binding of the PfPuf1 Puf domain to NRE using the yeast three-hybrid system (42). Yeast transformed with the plasmid pYESTrp2-PfPuf1 grew well on the selective medium and produced the recombinant BS42–PfPuf1 protein as detected by PfPuf1-specific antibodies (data not shown). When this yeast strain was transformed with different bait RNA constructs, only the one with sense NRE sequence grew vigorously on the selective medium with 5 mM 3-aminotriazole, whereas those transformed with the vectors pRH5′, pRH5′-IRE or pRH5′-antisenseNRE did not grow well. In addition, yeast transformed with pRH5′-NRE produced more β-galactosidase than the positive control (MS2–IRE + B42–IRP) (Fig. 7). In addition, neither the sense NRE nor the antisense NRE interacted with iron regulatory protein (IRP), an RBP. These results further demonstrated that the binding of the PfPuf1 Puf domain to the NRE RNA was specific.
Figure 7.
In vivo interactions of the PfPuf1 Puf domain with the Drosophila NRE sequence in yeast. Yeast strain L40uraMS2 was transformed with the plasmid pYESTrp2-PfPuf1 to express the hybrid protein of the B42 transactivation domain and the PfPuf1 Puf domain. One of the bait plasmids expressing MS2, MS2–NRE, MS2–IRE or MS2–antisense NRE (MS2– asNRE) was co-transformed with the hybrid protein plasmid. The MS2–NRE and MS2–asNRE were co-transformed with pYESTrp2-IRP as additional negative controls. The yeast expressing the hybrid protein B42–IRP and bait RNA MS2–IRE was included as the positive control. Both filter and liquid assays for β-galactosidase are shown. For liquid assay, the number represents the mean ± SD using at least three yeast colonies.
DISCUSSION
The Puf protein family
Translational regulation of gene expression plays an essential role in the development of a wide variety of organisms (51). In many cases, translational regulation is mediated by cis-acting elements residing in the 3′-UTR of cytoplasmic mRNAs (52). Trans-acting factors, including the new family of RBPs, Puf, which interact specifically with the 3′-UTR sequences, have been identified in diverse organisms. Puf proteins are evolutionarily conserved and widely distributed among eukaryotes. We have shown here that the malaria parasite P.falciparum possesses two distinct Puf members with similar expression patterns during gametocytogenesis. Furthermore, these genes are conserved among the Plasmodium species and their homologs are present in the P.yoelii and P.knowlesi genomes. The fact that these genes form two quite distinct groups strongly suggests that duplication and divergence of the Puf genes occurred prior to the speciation events in the genus Plasmodium.
Translational regulation by Puf proteins: a conserved molecular mechanism
Conservation of the Puf domain sequences suggests that this novel RBP family may act through a similar molecular mechanism by binding to the 3′-UTR regulatory sequences in their target mRNAs. So far, studies using various Puf/mRNA systems, including Pum/hb, FBF/fem-3, PufA/PKA and Mpt5/HO, all support this assumption (17,19–21). The only exception is Puf3p in yeast, which does not appear to act through translational repression. Puf3p binds to the 3′-UTR of COX17 mRNA and promotes its deadenylation and degradation, a phenomenon similar to one aspect of Pum/hb mRNA interactions (53,54). Nevertheless, translational repression and acceleration of mRNA decay could be the result of the same pathway (18). In the best-characterized systems, translational repression of target mRNAs not only requires Puf, but also other proteins that interact with Puf. Surprisingly, even the Puf-interacting proteins may be evolutionarily conserved, as Puf has been found to interact with Nanos (NOS) in Drosophila and NOS homologs in C.elegans and Xenopus (55–57). Particularly, studies in Drosophila and C.elegans show that evolutionarily distinct organisms share the Pum/NOS-mediated mechanism for germline development, a role most likely reflecting their ancestral function (58). Recently, it has been shown that Pum is able to recruit NOS to hb mRNA and translational repression of hb requires the formation of a quaternary complex including Pum, NOS, Brat and NRE-containing hb RNA (55,59). In Plasmodium, even though the exact biological functions of PfPuf proteins are unknown at present, it is likely that PfPufs act by a similar mechanism of translational regulation. This suggestion is based on sequence conservation of the Puf domain and the innate, specific binding activity of the PfPuf1 Puf domain to the Drosophila NRE sequence. The differential expression of PfPuf genes during gametocytogenesis, similar to that of pfs16, further suggests that PfPufs play essential roles during sexual development of the parasite. Moreover, the presence of Puf genes in P.yoelli and P.knowlesi indicates evolutionary conservation and implies functional importance of Puf proteins in the genus Plasmodium.
Multiple Puf members in one organism: redundant or distinct in functions?
Many eukaryotic organisms contain more than one Puf homolog. For example, there are at least 10 Puf members in C.elegans and five Puf homologs in S.cerevisiae (17,21). In Drosophila, even a single Pum gene encodes two functional protein isoforms of 156 and 130 kDa (23). Our analysis of the five Puf genes from three Plasmodium species indicates the presence of two Puf homologs in each Plasmodium species. The presence of more than one Puf gene or isoform in a given organism raises an interesting question about their biological functions. It is plausible that some are functionally redundant, whereas others may act in a tissue- and/or time-specific manner to regulate the translation of different mRNAs, and thus have distinct biological functions. In C.elegans, FBF1 and FBF2 seem redundant in regulating gametogenesis (17), whereas other Puf homologs appear to be required for primordial germ cell development (24). In baking yeast, however, two of the five Puf members, Puf3p and Mpt5, appear to regulate the expression of two different genes, Cox17 and HO, respectively (21,54). Furthermore, Mpt5 seems to be a global regulator with multiple targets, since it regulates aging, mating-type switching and pheromone response (21,60,61). Even the single pum gene in Drosophila, where pum mRNA and Pum protein are widely distributed at all stages of development, participates in many developmental processes (46,47). Originally identified as a maternal effect gene required for posterior patterning in early Drosophila embryos (62), Pum is also directly involved in germline stem cell maintenance and division (23,63,64). In addition, Pum may also have other functions in somatic cells. The achievement of such diverse functions during Drosophila development may require that Pum interact with different proteins and bind different RNA targets (23,64). In fact, there are at least three mRNAs (hb, bicoid and cyclin B) that bear the conserved NRE sequences and may interact with Pum (38,59). In P.falciparum, although the two PfPuf genes share similar expression patterns, it is very likely that they play different roles in parasite development instead of being temporally redundant. First, PfPuf1 and PfPuf2 are structurally divergent and only share limited sequence identity in the Puf domain (∼25%). Second, the 14 residues presumed to confer RNA binding specificity (Fig. 2B) are all different between the two PfPuf proteins and therefore may bind different target RNA sequences. Finally, the eighth Puf repeats of the two PfPuf proteins, which correspond to the region that interacts with NOS in Pum, are also divergent, implying that they interact with different proteins.
The RNA-binding domains and terminal extensions
Two recent studies of the crystal structures of the Puf domains showed that the eight Pum repeats, corresponding to eight copies of a single structural motif, are packed together to form a rainbow-shaped molecule (26,27). The single structural motif consists of one short H2 α-helix making up the ridge and two longer helices, H1 and H3, covering the outer and inner surfaces, respectively. The asymmetric charge distribution on the Puf structure renders the concave surface highly basic and the convex surface acidic. It is speculated that the concave surface binds RNA, while the convex surface mediates interactions with NOS and Brat. Alignment of the repeats demonstrates that such a structure with a basic concave face is conserved among all Puf-like proteins (27). A close comparison of the Plasmodium Puf proteins also revealed that the most homologous regions of the Puf repeats correspond to the sequences forming the RNA-binding helix, H3 (Fig. 2B). Furthermore, the H3 consensus F/Y-G-X4-Q-K/R-X2-E is also conserved in most of the Plasmodium Puf repeats. Such a striking similarity in the H3 regions between the PfPuf proteins and Pum strongly suggests that the Puf domain of PfPuf1 may also recognize the NRE sequence. This assumption was verified in our experiments using in vitro binding assays and the yeast three-hybrid analysis (Figs 6 and 7). It is plausible that the conserved UUGU motif in hb, fem-3, Cox17 and HO mRNAs may be the general Puf recognition sequence motif, while flanking nucleotide sequences confer more binding specificity for their respective Puf proteins and proteins that interact with the Puf–RNA complex. For instance, the inability of a ternary structure consisting of Drosophila Pum, Nos and cyclin B mRNA to recruit Brat probably lies in the sequences that flank the NRE-like elements in cyclin B mRNA (59).
The Puf domain seems to contain all necessary elements that are required for Pum’s proper function in posterior patterning, as it is sufficient to rescue the abdominal segmentation defects in pum mutant embryos (19). Yet, in addition to the Puf domains, most Puf proteins have long N-terminal sequences, whose functions have not been elucidated so far. Seemingly divergent, the N-terminal extensions of many Puf proteins share an S-rich and Q/A-rich motif (43). In Plasmodium, however, the two PfPufs lack these two common features, and PfPuf1 is even more unusual in having equally long N- and C-terminal sequences. Uniquely, the deduced PfPuf1 protein contains four additional repetitive motifs that might be important for PfPuf1 interactions with other proteins or provide more specificity to target RNA selection. Therefore, further analysis of PfPufs may provide new insights into their functions in sexual development of all Plasmodium species.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Xinzhuan Su for the determination of the chromosomal locations of PfPuf genes. Some sequence data were obtained from the Malaria Genome Consortium: The Sanger Centre, The Institute for Genomic Research and The Stanford DNA Sequencing and Technology Center. This work was supported by a National Institutes of Health grant (AI46472) to L.C.
DDBJ/EMBL/GenBank accession nos AY098937, AY099486
REFERENCES
- 1.Kaslow D.C. (1993) Transmission-blocking immunity against malaria and other vector-borne diseases. Curr. Opin. Immunol., 5, 557–565. [DOI] [PubMed] [Google Scholar]
- 2.Garnham P.C.C. (1988) Malaria parasites of man: life-cycles and morphology. In Wernsdorfer,W.H. and McGregor,I. (eds), Malaria: Principles and Practice of Malariology. Churchill Livingstone, London, UK, Vol. 1, pp. 61–96.
- 3.Alano P. and Carter,R. (1990) Sexual differentiation in malaria parasites. Annu. Rev. Microbiol., 44, 429–449. [DOI] [PubMed] [Google Scholar]
- 4.Sinden R.E., Butcher,G.A., Billker,O. and Fleck,S.L. (1996) Regulation of infectivity of Plasmodium to the mosquito vector. Adv. Parasitol., 38, 53–117. [DOI] [PubMed] [Google Scholar]
- 5.Williams J.L. (1999) Stimulation of Plasmodium falciparum gametocytogenesis by conditioned medium from parasite cultures. Am. J. Trop. Med. Hyg., 60, 7–13. [DOI] [PubMed] [Google Scholar]
- 6.Carter R. and Miller,L.H. (1979) Evidence for environmental modulation of gametocytogenesis in Plasmodium falciparum in continuous culture. Bull. World Health Organ., 57 (suppl. 1), 37–52. [PMC free article] [PubMed] [Google Scholar]
- 7.Carter R. and Graves,P.M. (1988) Gametocytes. In Wernsdorfer,W.H. and McGregor,I. (eds), Malaria: Principles and Practice of Malariology. Churchill Livingstone, London, UK, Vol. 1, pp. 253–305.
- 8.Day K.P., Karamalis,F., Thompson,J., Barnes,D.A., Peterson,C., Brown,G.V. and Kemp,D.J. (1993) Genes necessary for expression of a virulence determinant and for transmission of Plasmodium falciparum are located on a 0.3-megabase region of chromosome 9. Proc. Natl Acad. Sci. USA, 90, 8292–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pologe L.G. (1994) Aberrant transcription and the failure of Plasmodium falciparum to differentiate into gametocytes. Mol. Biochem. Parasitol., 68, 35–43. [DOI] [PubMed] [Google Scholar]
- 10.Alano P., Roca,L., Smith,D., Read,D., Carter,R. and Day,K. (1995) Plasmodium falciparum: parasites defective in early stages of gametocytogenesis. Exp. Parasitol., 81, 227–235. [DOI] [PubMed] [Google Scholar]
- 11.Guinet F., Dvorak,J.A., Fujioka,H., Keister,D.B., Muratova,O., Kaslow,D.C., Aikawa,M., Vaidya,A.B. and Wellems,T.E. (1996) A developmental defect in Plasmodium falciparum male gametogenesis. J. Cell Biol., 135, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaidya A.B., Muratova,O., Guinet,F., Keister,D., Wellems,T.E. and Kaslow,D. (1995) A genetic locus on Plasmodium falciparum chromosome 12 linked to a defect in mosquito-infectivity and male gametogenesis. Mol. Biochem. Parasitol., 69, 65–71. [DOI] [PubMed] [Google Scholar]
- 13.Alano P. (1991) Plasmodium sexual stage antigens. Parasitol. Today, 7, 199–203. [DOI] [PubMed] [Google Scholar]
- 14.Lobo C.A. and Kumar,N. (1998) Sexual differentiation and development in the malaria parasite. Parasitol. Today, 14, 146–150. [DOI] [PubMed] [Google Scholar]
- 15.Cui L., Rzomp,K.A., Fan,Q., Martin,S.K. and Williams,J. (2001) Plasmodium falciparum: differential display analysis of gene expression during sexual development of the parasite. Exp. Parasitol., 99, 244–254. [DOI] [PubMed] [Google Scholar]
- 16.Zamore P.D., Williamson,J.R. and Lehmann,R. (1997) The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA, 3, 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B., Gallegos,M., Puoti,A., Durkin,E., Fields,S., Kimble,J. and Wickens,M.P. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature, 390, 477–484. [DOI] [PubMed] [Google Scholar]
- 18.Wickens M., Bernstein,D.S., Kimble,J. and Parker,R. (2002) A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet., 18, 150–157. [DOI] [PubMed] [Google Scholar]
- 19.Wharton R.P., Sonoda,J., Lee,T., Patterson,M. and Murata,Y. (1998) The pumilio RNA-binding domain is also a translational regulator. Mol. Cell, 1, 863–872. [DOI] [PubMed] [Google Scholar]
- 20.Souza G.M., da Silva,A.M. and Kuspa,A. (1999) Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development, 126, 3263–3274. [DOI] [PubMed] [Google Scholar]
- 21.Tadauchi T., Matsumoto,K., Herskowitz,R. and Irie,K. (2001) Post-transcriptional regulation through the HO-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J., 20, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asaoka-Taguchi M., Yamada,M., Nakamura,A., Hanyu,K. and Kobayashi,S. (1999) Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nature Cell Biol., 1, 431–437. [DOI] [PubMed] [Google Scholar]
- 23.Parisi M. and Lin,H. (1999) The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics, 153, 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramaniam K. and Seydoux,G. (1999) nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development, 126, 4861–4871. [DOI] [PubMed] [Google Scholar]
- 25.Hoek M., Zanders,T. and Cross,G.A.M. (2002) Trypanosoma brucei expression-site-8 protein interacts with a Pumilio family protein. Mol. Biochem. Parasitol., 120, 269–283. [DOI] [PubMed] [Google Scholar]
- 26.Edwards T.A., Pyle,S.E., Wharton,R.P. and Aggarwal,A.K. (2001) Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell, 105, 281–289. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Zamore,P.D. and Hall,T.M.T. (2001) Crystal structure of a Pumilio homology domain. Mol. Cell, 7, 855–865. [DOI] [PubMed] [Google Scholar]
- 28.Trager W. and Jensen,J.B. (1976) Human malaria parasites in continuous culture. Science, 193, 673–675. [DOI] [PubMed] [Google Scholar]
- 29.Kariuki M.M., Kiaira,J.K., Mulaa,F.K., Mwangi,J.K., Wasunna,M.K. and Martin,S.K. (1998) Plasmodium falciparum: purification of the various gametocyte developmental stages from in vitro-cultivated parasites. Am. J. Trop. Med. Hyg., 59, 505–508. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Frohman M.A. (1990) RACE: rapid amplification of cDNA ends. In Innis,M.A., Gelfand,D.H., Sninsky,J.J. and White,T.J., (eds), PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, pp. 28–38.
- 32.Ochman H., Medhora,M.M., Garza,D. and Hartl,D.L. (1990) Amplification of flanking sequences by inverse PCR. In Innis,M.A., Gelfand,D.H., Sninsky,J.J. and White,T.J. (eds), PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, pp. 219–227.
- 33.Devereaux J., Haeberli,P. and Smithies,O. (1984) A comprehensive set of sequence analysis programs for the VAX. Gene, 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Billker O., Lindo,V., Panico,M., Etienne,A.E., Paxton,T., Dell,A., Rogers,M., Sinden,R.E. and Morris,H.R. (1998) Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature, 392, 289–292. [DOI] [PubMed] [Google Scholar]
- 36.Garcia G.E., Wirtz,R.A., Barr,J.R., Woolfitt,A. and Rosenberg,R. (1998) Xanthurenic acid induces gametogenesis in Plasmodium, the malaria parasite. J. Biol. Chem., 273, 12003–12005. [DOI] [PubMed] [Google Scholar]
- 37.Dolan S.A., Adam,R.D. and Wellems,T.E. (1993) Chromosome mapping methods for parasitic protozoa. Methods Mol. Biol., 21, 319–332. [DOI] [PubMed] [Google Scholar]
- 38.Wharton R.P. and Struhl,G. (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell, 67, 955–967. [DOI] [PubMed] [Google Scholar]
- 39.Murata Y. and Wharton,R.P. (1995) Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell, 80, 747–756. [DOI] [PubMed] [Google Scholar]
- 40.Curtis D., Treiber,D.K., Tao,F., Zamore,P.D., Williamson,J.R. and Lehmann,R. (1997) A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J., 16, 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setzer D.R. (2000) Measuring equilibrium and kinetic constants using gel retardation assays. In Haynes,S.R. (ed.), RNA-Protein Interaction Protocols. Humana Press, Totowa, NJ, pp. 115–128. [DOI] [PubMed]
- 42.SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Te Heesen S. and Stagljar,I. (2000) Two-hybrid system applicable to membrane protein. In Zhu,L. and Hannon,G.J. (eds), Yeast Hybrid Technologies. Eaton Publishing, Natick, MA, pp. 117–127.
- 44.White E.K., Moore-Jarrett,T. and Ruley,H.E. (2001) PUM2, a novel murine Puf protein and its consensus RNA-binding site. RNA, 7, 1855–1866. [PMC free article] [PubMed] [Google Scholar]
- 45.Gardner M.J., Tettelin,H., Carucci,D.J., Cummings,L.M., Aravind,L., Koonin,E.V., Shallom,S., Mason,T., Yu,K., Fujii,C. et al. (1998) Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum.Science, 282, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 46.Bowman S., Lawson,D., Basham,D., Brown,D., Chillingworth,T., Churcher,A.M., Craig,A., Davies,R.M., Devlin,K., Feltwell,T. et al. (1999) The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature, 400, 532–538. [DOI] [PubMed] [Google Scholar]
- 47.Barker D.D., Wang,C., Moore,J., Dickinson,L.K. and Lehmann,R. (1992) Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev., 6, 2312–2326. [DOI] [PubMed] [Google Scholar]
- 48.McDonald P.M. (1992) The Drosophila pumilio gene: an unusually long transcription unit and unusual protein. Development, 114, 221–232. [DOI] [PubMed] [Google Scholar]
- 49.Carlton J.M.-R., Vinkenoog,R., Waters,A.P. and Walliker,D. (1998) Gene synteny in species of Plasmodium.Mol. Biochem. Parasitol., 93, 285–294. [DOI] [PubMed] [Google Scholar]
- 50.Dechering K.J., Kaan,A.M., Mbacham,W., Wirth,D.F., Eling,W., Konings,R.N. and Stunnenberg,H.G. (1999) Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite Plasmodium falciparum. Mol. Cell. Biol., 19, 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickens M., Kimble,J. and Strickland,S. (1996) Translational control of developmental decisions. In Hershey,J., Mathews,M. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 411–450.
- 52.Wickens M., Anderson,P. and Jackson,R.J. (1997) Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev., 7, 220–232. [DOI] [PubMed] [Google Scholar]
- 53.Wreden C., Werrotti,A.C., Schisa,J.A., Lieberfarb,M.E. and Strickland,S. (1997) Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development, 124, 3015–3023. [DOI] [PubMed] [Google Scholar]
- 54.Olivas W. and Parker,R. (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J., 19, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonoda J. and Wharton,R.P. (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev., 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraemer B., Crittenden,S., Gallegos,M., Moulder,G., Barstead,R., Kimble,J. and Wichens,M. (1999) NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol., 9, 1009–1018. [DOI] [PubMed] [Google Scholar]
- 57.Nakahata S., Katsu,Y., Mita,K., Inoue,K., Nagahamas,Y. and Yamashita,M. (2001) Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with Nanos homolog (Xcat-2) and a cytoplasmic polyadenylation element-protein (CPEB). J. Biol. Chem., 276, 20945–20953. [DOI] [PubMed] [Google Scholar]
- 58.Parisi M. and Lin,H. (2000) Translational repression: a duet of Nanos and Pumilio. Curr. Biol., 10, R81–R83. [DOI] [PubMed] [Google Scholar]
- 59.Sonoda J. and Wharton,R.P. (2001) Drosophila Brain Tumor is a translational repressor. Genes Dev., 15, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen T. and Kurjan,J. (1997) Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol. Cell. Biol., 17, 3429–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy B.K., Gotta,M., Sinclair,D.A., Mills,K., McNabb,D.S., Murthy,M., Pak,S.M., Laroche,T., Gasser,S.M. and Guarente,L. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell, 89, 381–391. [DOI] [PubMed] [Google Scholar]
- 62.Lehmann R. and Nüsslein-Volhard,C. (1987) Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature, 329, 167–170. [Google Scholar]
- 63.Lin H. and Spradling,A.C. (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development, 124, 2463–2476. [DOI] [PubMed] [Google Scholar]
- 64.Forbes A. and Lehmann,R. (1998) Nanos and pumilio have critical roles in the development and function of Drosophila germline stem cells. Development, 125, 679–690. [DOI] [PubMed] [Google Scholar]