Abstract
Reverse transcription of hepatitis B virus (HBV) pregenomic RNA is essential for virus replication. In the first step of this process, HBV reverse transcriptase binds to the highly conserved encapsidation signal, epsilon (ε), situated near the 5′ end of the pregenome. ε has been predicted to form a bulged stem–loop with the apical stem capped by a hexa– loop. After the initial binding to this apical stem– loop, the reverse transcriptase synthesizes a 4 nt primer using the bulge as a template. Here we present mutational and structural data from NMR on the apical stem–loop of ε. Application of new isotope-labeling techniques (13C/15N/2H-U-labeling) allowed resolution of many resonance overlaps and an extensive structural data set could be derived. The NMR data show that, instead of the predicted hexa–loop, the apical stem is capped by a stable UGU tri–loop closed by a C-G base pair, followed by a bulged out C. The apical stem contains therefore two unpaired pyrimidines (C1882 and U1889), rather than one as was predicted, spaced by 6 nt. C1882, the 3′ neighbour to the G of the loop-closing C-G base pair, is completely bulged out, while U1889 is at least partially intercalated into the stem. Analysis of 205 of our own HBV sequences and 1026 strains from the literature, covering all genotypes, reveals a high degree of conservation of ε. In particular, the residues essential for this fold are either totally conserved or show rare non-disruptive mutations. These data strongly indicate that this fold is essential for recognition by the reverse transcriptase.
INTRODUCTION
The hepatitis B virus (HBV) belongs to the family Hepadnaviridae, which comprises a group of highly species-specific DNA viruses (1–4). HBV is the smallest human DNA virus and has a very compact genome (Fig. 1, top). It replicates via an intermediate reverse transcription step. After transcription of HBV DNA by the host RNA polymerase II, the pregenomic RNA is transported into the cytoplasm and encapsidated into immature core particles together with HBV reverse transcriptase. Encapsidation is one of the key features in HBV replication and is triggered by the binding of HBV reverse transcriptase to the encapsidation signal, epsilon (ε), a 60-nt bulged stem–loop located at the 5′ end of the RNA pregenome (Fig. 1, bottom). The same sequence is also present at the 3′ end (Fig. 1, bottom), but this copy does not seem to be essential for viral replication (1). The HBV reverse transcriptase binds to the apical stem–loop of the 5′ ε and a 4-nt DNA primer is synthesized from the bulge within ε. The primer–reverse transcriptase complex is subsequently translocated to the primer-binding site within DR1 at the 3′ end of the pregenome, where DNA synthesis starts (2,3).
Figure 1.
HBV genome. Top, schematic diagram of the partially double-stranded genome of HBV showing + and – (coding) strands (center). Four different sizes of main RNA transcripts are shown (periphery), all of which have different initiation sites, but a common termination site (arrowheads). The pregenomic RNA (originally called 3.5 kb RNA, but really 3.3 kb large) carries a terminal redundancy of ∼130 nt, and one copy of the 60-nt ε forms at each end within the region of sequence overlap (shaded box). Bottom, a schematic linear diagram of the RNA pregenome showing the two copies of ε.
Due to the lack of proofreading during reverse transcription, the mutation rate of these viruses would be expected to be high, and similar to that of retroviruses. This tendency toward a high mutation rate is counteracted, however, by the extreme compactness of the genome, with its extensively overlapping reading frames and lack of non-coding regions (4). In fact, the apparent in vivo mutation rate of the partially double-stranded HBV genome has been found to be very low (5,6). The HBV mutations that have been described (point mutations and deletions) are often located in specific genomic regions and are believed to confer advantages on the virus through immune escape. Deletions have been suggested to arise mechanistically through faulty reverse transcription (7).
Several HBV genotypes (A–G) have been described. They have >8% sequence divergence and differ from each other in replication, clinical features and geographical origin. Genotype A differs from the other genotypes in one region especially, the precore gene, which partly overlaps with ε (Fig. 1, top, gray box). With the exception of two nucleotide positions differing in genotype A (1850 and 1858), the sequence of ε seems generally conserved between HBV genotypes. To further investigate the extent of the in vivo sequence conservation of ε, we analyzed 205 of our own HBV sequences and 1026 strains from the literature.
ε has been the subject of several secondary structure prediction studies (7–10), which all predict a stem–loop fold. Enzymatic probing studies have confirmed this conformation (9). However, no structural analysis at molecular detail has as yet been carried out and several features, such as the loop fold, remain uncertain.
Here, we present the fold of the apical loop of ε derived from NMR data on a 27-nt RNA sequence mimicking the apical stem–loop part of ε. Application of new isotope-labeling techniques (13C/15N/2H-U-labeling) made near complete assignments possible and an extensive data set of structural information was derived, including many crucial unambiguously assigned NOE (Nuclear Overhauser Effect) contacts. We also present a mutational analysis of ε in 1231 HBV strains, covering all genotypes, to firmly establish the in vivo relevance of the NMR-derived ε fold.
MATERIALS AND METHODS
Hepatitis B virus strains
The precore sequences [nucleotides 1814–1900, numbering by Okamoto et al. (11)] from 1231 HBV strains were studied. These strains had been characterized from patients with a wide variety of clinical diagnoses, ranging from acute hepatitis or healthy chronic carriership to fulminant hepatitis, cirrhosis and hepatocellular carcinoma. All seven HBV genotypes (A–G) were represented; 205 strains were characterized in our laboratory and the remaining 1026 were taken from the literature. Of these, 14 strains had been isolated from non-human primates (five chimpanzee, five gibbon, one gorilla, two orangutan and one woolly monkey).
RNA sample preparation
Unlabeled and (13C; 15N; 1′, 3′, 4′, 5′, 5″-2H)-U-labeled RNA-samples of the sequence 5′-GGCCUCCAAGCUGUGCCU UGGGUGGCC-3′ were transcribed from a synthetic DNA template using T7 RNA polymerase (12–14). (13C; 15N; 1′, 3′, 4′, 5′, 5″-2H)-labeled UTP for the U-labeled sample was synthesized by using the in vitro method as previously described (14) (for a review see 15). Large-scale transcription was carried out in 40 ml reaction mixes. Purification was carried out on 20% polyacrylamide sequencing gels containing 8 M urea. Subsequently, the RNA was electroeluted, desalted on an Econo-Pac 10 DG column (BioRad) and freeze-dried. The RNA samples were finally dissolved in 300 µl of 100 mM NaCl and 0.1 mM EDTA giving NMR samples with concentrations of 0.5 mM (unlabeled) and 0.15 mM (labeled).
NMR spectroscopy
All 1H-1H and 1H-13C NMR spectra were recorded on a Bruker DRX-600 spectrometer with a Bruker xyz-gradient TXI probe, unless stated otherwise. The 1D and 2D spectra in 90% H2O/10% D2O were recorded on the unlabeled sample at 275 K. The 2D NOESY spectra of the imino region were recorded with 100 ms mixing time. A jump-return sequence was used for water suppression (16).
Spectra of the unlabeled sample in D2O were recorded at 298 K. 2D NOESY spectra (150, 300 and 500 ms mixing times), homonuclear DIPSY-TOCSY (17) (45 and 90 ms mixing times) and DQF-COSY spectra were recorded. Natural abundance 1H-13C HMQC (18,19) and natural abundance 1H-13C HMBC spectra (20) were recorded using a Bruker z-gradient TXI cryo probe. The HMBC was recorded with selective decoupling of the H2′ resonances. The transfer delay for conversion of in-phase to multiple-quantum coherence was set to 66.76 ms, which leads to maximum transfer for JCH = 7.6 Hz. Note that, by setting the transfer delay to this value, the H5-C4/6 and H6-C4/2 cross peaks become negative due to evolution of the JH5H6 coupling.
On the (13C; 15N; 1′, 3′, 4′, 5′, 5″-2H)-U-labeled sample all spectra were recorded in D2O. 13C-decoupled 2D NOESY spectra were recorded with 80, 200 and 400 ms mixing times at 301 K. The 1H-13C HMQC spectrum was recorded at 301 K. A modified HMQC-NOESY-HMQC experiment (21) was recorded with a 200 ms mixing time at 298 K, using the cryo probe. This experiment allows selection for, or suppression of, 13C-bound protons in f1 and/or f2 (22). Four data sets were acquired in an interleaved manner with alternate phases (Ψ1,Ψ2 = x, x; x, –x; –x, x; –x, –x), where Ψ1 and Ψ2 are the phases of the second 90° carbon pulse in the 13C (f1) and 13C (f2) filter, respectively. Addition and/or subtraction of the four data sets (23) resulted in the final 13C-filtered spectra.
1H-31P HMBC spectra (24) were recorded on a Bruker AMX-500 spectrometer with a Bruker TBI probe at 297 K for both unlabeled and labeled samples. Spectra were recorded with 33 ms transfer delay with acquisition starting directly after the second 31P 90° pulse, i.e. no refocusing delay was used.
Proton and carbon shifts were calibrated against 4,4-dimethyl 4-silapentane sodium sulfonate (DSS). Phosphorus resonances were calibrated against external tri-methyl phosphate (TMP), using the IUPAC referencing number (25). Calibration against 85% phosphoric acid can be obtained by adding 3.456 p.p.m. to the TMP-calibrated phosphorus shifts (26). All spectra were processed using XWINNMR. All 2D spectra were collected in a phase-sensitive mode via the STATES-TPPI method, and processed accordingly. The 1H-31P HMBC spectra were processed in magnitude mode in f2 to remove phase distortions.
NMR assignment and analysis
To improve resonance assignment and extraction of structural parameters, the standard methods (23,27) were complemented by application of new isotope-labeling techniques involving deuteration of U residues. In addition, thanks to cryoprobe technology, through-bond H2-H8 and sugar–base correlations could be obtained via H-C correlation spectroscopy at natural abundance, instead of triple-resonance spectroscopy on 13C/15N-labeled samples.
Application of (13C; 15N; 1′, 3′, 4′, 5′, 5″-2H)-labeled U residues. The (13C; 15N; 1′, 3′, 4′, 5′, 5″-2H)-labeling of the U residues of the stem–loop sequence (14) removes all H1′, H3′-H5′/″ resonances from the spectra leading to considerable spectral simplification. Many cross peaks were assigned via combinations of spectra. Firstly, the NOESY spectra of labeled and unlabeled samples were compared. In this way H2′, H3′ and important H4′/H5′/H5″ resonances and many sequential NOE contacts could reliably be identified, e.g. in the loop region (14). The HMQC of the labeled sample was used to assign the uridine H2′ and C2′ resonances. Secondly, 13C-filtered NOESY spectra (21–23) extended/confirmed resonance assignments and extended/confirmed NOE contacts. Finally, the phosphorus resonances in the H-P HMBC spectrum that are separated from the bulk are in the standard method assigned via their through-bond connections to the 3′, 4′ and 5′/5″ protons, previously identified in the TOCSY and/or 500 ms NOESY spectra. These assignments could now be confirmed and extended to all phosphorus resonances of U residues by comparison with the H-P HMBC spectrum of the U-labeled sample.
Sugar puckering. The fraction of N-puckering, fN, was calculated from the JH1′-H2′-couplings according to the equation JH1′-H2′ = fNJN + (1 – fN)JS, where JN and JS, the J coupling values for pure N- and S-pucker sugars, respectively, were taken to be 1.2 and 8.0 Hz (28). The JH1′-H2′ couplings were measured from the splitting on the H6 (f1)-H1′(f2) in-phase cross peaks in the 500 ms NOESY spectrum.
Derivation of backbone angles. The 31P shifts were used to derive ranges on the backbone torsion angles (23,26,28). When the 31P shift (δ31P) lies in the usual range for helix residues, i.e. between –3.5 and –4.6 p.p.m. (referenced with respect to external TMP), the backbone has the regular helix conformation and the backbone torsion angles ε, ζ, α and β have regular helix values (23,26,28). Downfield shifts are often but not necessarily attributable to changes in the backbone angles next or penultimate to the phosphorus in question (ε, ζ, α and β).
Glycosidic torsion angles. The glycosidic torsion angles were determined from a combination of three different types of data. Firstly, the H1′-H6/8 and H2′-C6/8 NOE intensities (28) provided broad ranges for the glycosidic torsion angles (for 150° < χ < 340°, 3.3 Å < dH1′H6/8 < 3.6 Å, while for 260° < χ < 340°, dH2′H6/8 < 2.5 Å, independent of sugar puckering). Secondly, the H2′ and H3′ chemical shifts provided additional information (29). Thirdly, the intensity of the H1′-C6/8 and H1′-C2/4 cross peaks in the natural abundance 1H-13C HMBC spectrum. When the H1′-C6/8 cross peak is more intense than the H1′-C2/4 cross peak, the χ angle is within anti-range (J.Schleucher, S.Flodell, B.Wu and S.Wijmenga, unpublished).
RESULTS
Analysis of HBV strains
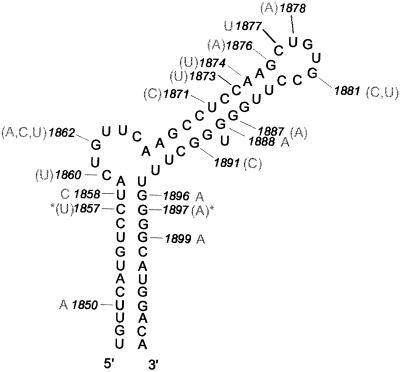
There was a large degree of homology between the ε sequences of the 1231 strains analyzed (Fig. 2). Genotypic variations without any foreseeable impact on the stability of the ε stem–loop, unless combined with additional mutations, included 1850 U→A in genotype A strains and 1858 U→C in genotype A and F strains. Two stem-stabilizing G→A mutations at positions 1896 and 1899, which have been widely described in the literature, were found in a large number of strains. The former mutation causes the well described premature translational stop in the precore gene [precore codon 28: AGG (30)]. In 11 of the 14 strains isolated from animals, G1896 was changed to U instead, thereby creating a stem disruption but not the translational stop codon often seen in human strains.
Figure 2.
The secondary structure of ε as predicted most often by secondary structure programs. The numbered positions have had at least one mutation (shown by gray lettering) described among 1025 HBV strains from the literature, and/or observed by us in 205 strains. Mutations shown in parentheses have been seen in fewer than three strains. Asterisks show a coincidental double base-pairing mutation in one strain. Numbering according to Okamoto et al. (11).
Some variability was found in the bulge region. The lower part of the stem between the unpaired pyrimidines was conserved, with the exception of a G→A mutation at position 1888, changing the predicted base pairing from U-G to U-A and thereby increasing the stability of the upper stem. In addition, a stem-disrupting change from G to C at position 1891 has been described in one patient after direct sequencing of a single PCR product (31). At position 1887, two groups have reported a stem-disrupting G→A mutation in one of 263 and one of 42 healthy carriers, respectively (32,33). The same authors also reported mutations at positions 1873 and 1874, each mutation being found in a single strain. Nucleotide 1876 G, forming part of the stabilizing base pair G-C under the apical loop predicted by secondary structure programs, has been reported to be changed to A in strains from two chronic HBV patients (31,34).
In seven strains isolated from human patients and in four primate strains, position 1877 had a U instead of C. At positions 1878–1880 (UGU) in the predicted apical loop, only one of 1231 strains appeared to have been changed (33) and nucleotides 1882 C and 1883 C were both completely conserved among all the strains analyzed.
NMR analysis
Resonance and NOE assignments. A continuous set of sequential connectivities could be seen in the NOESY spectrum (Fig. 3) between the 11 imino proton resonances, resulting in a complete sequential assignment of the imino resonances. Standard proton-based methods were employed to obtain the initial assignments of the non-exchangeable protons H1′, H6/8, H5 and H2 (Supplementary Material, Figs S1–S4 and S9). These assignments were checked via through-bond H-C correlations in natural abundance 1H-13C-HMQC (Figs S5 and S6) and 1H-13C-HMBC spectra (Figs S7 and S8) using cryo probe technology.
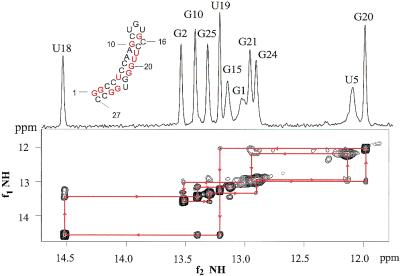
Figure 3.
1D and 2D imino spectra of the 27 nt NMR sequence of the apical loop in ε are shown. Eleven peaks are seen, indicating 11 bp in the NMR sequence. Drawn arrowed lines indicate the sequential walk between the imino protons of neighboring base pairs.
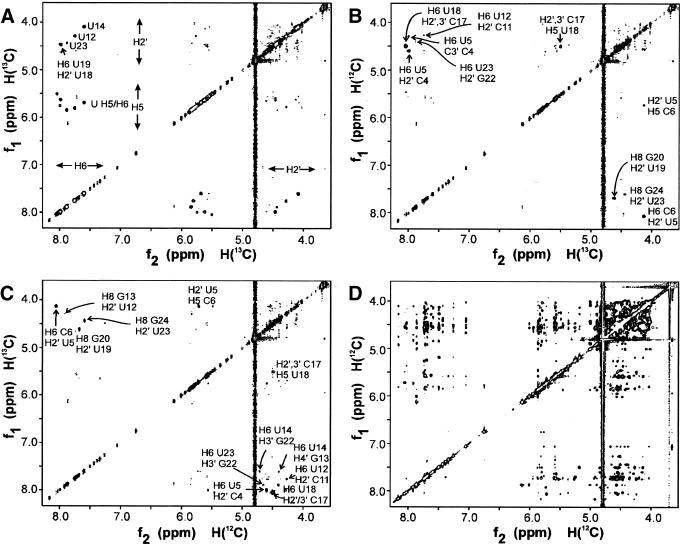
Extension of the 1H resonance assignments to H2′ to H5′/5″ is notoriously difficult due to extensive resonance overlap (27,29). Here, only some H2′ and H3′ resonances could be identified via their correlation to the H1′ in the COSY/TOCSY spectra using these standard techniques (Fig. S4). The new 2H/13C/15N-labeling of uridine residues (see Materials and Methods), located at structurally crucial positions, resolved most of the overlap problems, and assignments could reliably be extended to include many of the sugar proton resonances, as well as to carbon and phosphorus resonances. Since only the H2′ of these deuterated uridine is visible in the sugar region, many assignments could be obtained by comparison of the NOESY (14), 1H-13C-HMQC and 1H-31P-HMBC spectra (Fig. S10) of the unlabeled and labeled samples. The strength of this approach is well demonstrated by the 13C-filtered NOESY spectra (Fig. 4). These spectra established the NOE contacts between 13C-bound and 13C-bound protons (Fig. 4A), between 12C-bound (f1) and 13C-bound (f2) protons (Fig. 4B), between 13C-bound and 12C-bound protons (Fig. 4C) and between 12C-bound and 12C-bound protons (Fig. 4D). A complete list of assignments is shown in the Supplementary Material (Table S1).
Figure 4.
X-filtering. 13C-filtered NOESY spectra of the 2H/13C/15N-U-labeled sample, demonstrating the spectral simplification and improved identification of NOE contacts achieved via this new labeling method, where specific deuteration is combined with specific 13C-labeling of crucial residue types. Because in the U residues only H2′ are left on the ribose, otherwise inaccessible NOEs can now specifically be assigned. Subspectrum (A) shows the NOE contacts between 13C-bound (f1) and 13C-bound (f2) protons, e.g. between uridine H2′ and H6 protons (either intra-residue or inter-residue). The intra-residue H6/8-H2′ of U12, U14 and U23 can be seen, because they are partially or completely S-puckered, in contrast to the other U residues (Table S2). In addition, an intense sequential contact is seen between H6 of U19 and H2′ of U18. Subspectrum (B) shows cross peaks between 12C-bound protons (f1) and 13C-bound protons (f2). In the H6/8 (f1)-H2′ (f2) region (below the diagonal), two intense cross peaks are seen. They are the sequential H6/8i-H2′i–1 of C6-U5 and G20-U19. The two weaker cross peaks are the sequential H6/8i-H2′i–1 of G13-U12 and G24-U23. In the H6/8 (f2)-H2′ (f1) region (above the diagonal), sequential cross peaks from H2′i–1 (f1) to H6/8i (f2) can be seen, i.e. from C4-U5, G13-U14, C17-U18 and, weakly, G22-U23. Subspectrum (C) shows cross peaks between 13C-bound protons (f1) and 12C-bound protons (f2). Subspectra C and B are symmetry-related, i.e. each cross peak in C appears in B on the opposite side of the diagonal. Subspectrum (D) shows the 12C-bound-to-12C-bound contacts, i.e. the NOE contacts between the non-U residues. Note the crowded H2′-H5′/5″ region.
Sugar puckering, glycosidic torsion angles and backbone torsion angles. The quantitative analysis (see Materials and Methods and Table S2) shows that the helix residues have N-puckered sugars, and that deviations occur in special regions of the sequence. In the loop region, residues G13 and U14 are nearly completely S-puckered, while C11 and U12 have ∼50% N-puckering; the base-paired G15 is fully N-puckered and the bulge residue, C16, is nearly fully N-puckered. Around the unpaired residue U23, U23 itself and G22 show ∼50% N-puckering. All glycosidic torsion angles were found to lie within anti-range.
All 31P shifts for helix residues fall into the helical range (see Materials and Methods), indicating regular A-helix backbone conformations for these residues (Table S1 and Fig. S10). In the loop region encompassed by C11-P-U12-P-G13-P-U14-P-G15, only the 31P shifts of G13-P-U14 (–3.34 p.p.m.) falls just outside the regular helix range (–3.5 to –4.6 p.p.m.), while U12-P-G13 (–3.57 p.p.m.) falls just within the helical region. Thus, in the loop region, only the backbone angles ε, ζ or α between G13 and U14 (and possibly between U12 and G13) may deviate slightly from regular helix values. Around the bulged residue C16 (C1882), the phosphate C16-P-C17 below the bulge shows the largest downfield shift of all the 31P resonances. That only the 3′-phosphorus is downfield shifted is a usual observation for bulge residues (35,36). This suggests that the bulging out rotation is affected by the ε, ζ, α and β angles on the 3′ side and by γ, β on the 5′ side. Around the unpaired residue U23 (U1889), only the phosphorus on the 5′ side shows a slight downfield shift, although it still falls within the regular helix range. It is more unusual that only the 5′-phosphorus of a bulge is shifted, but it has been observed (37). This shift is smaller than for those around C16 (C1882), indicating smaller deviations from regular helix values.
Deviations from regular helix proton shifts. Proton chemical shifts are very sensitive to conformational changes and can in RNA be back-calculated with good accuracy, i.e. with an r.m.s.d. 0.08 p.p.m. (29). They are therefore good indicators for deviations from regular helix conformation. If no shift deviations >∼0.1 p.p.m. are present, the residue in question has a standard helix conformation (29). The average deviations of the observed H6/8, H5, H2, H1′ and H2′ shifts from standard helix values are shown in Figure S11.
Three groups of residues show deviations >0.1 p.p.m. First, the deviations seen for 5′-end G1 and G2 and 3′-end C27 are probably caused by end-fraying effects. Secondly, U5, G21 and U23 show large deviations. U23 is the unpaired residue and deviations are expected here. Its H5 proton has a shift of 5.85 p.p.m., slightly up field from the random coil shift [6.04 p.p.m. (29)]. Thus, the base of U23 still experiences some ring current effects from neighboring bases (usually on the 5′ side), suggesting that U23 may be partially intercalated in the stem. U5 is located cross-strand from U23, and G21 is penultimate from U23. Thirdly, all residues in the loop region (C10–C16) show deviations from regular helix values, with the proposed bulge residue C16 showing the largest deviations. The H5 resonance of C16 has near random coil shift (29). This strongly suggests that the C16 bulge lies outside of the helix, as it does not experience the ring currents from the bases of the flanking residues.
Folding of the apical stem–loop
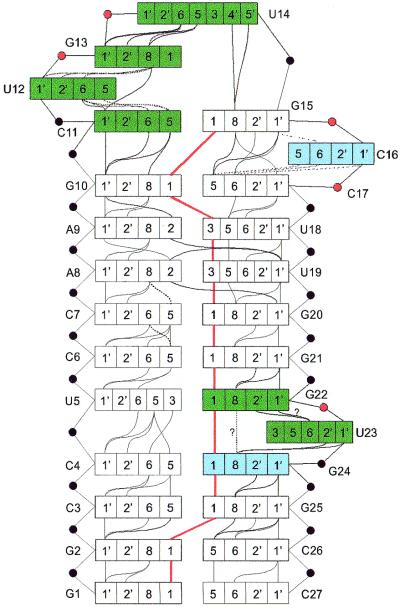
This extensive and reliable set of structural parameters, NOE contacts, torsion angles and chemical shifts was used to derive a schematic model for the folding of the apical stem–loop of HBV ε (Fig. 5).
Figure 5.
Schematic fold of the apical stem–loop of ε. In the loop and bulges, important structural features are indicated, such as sugar puckering (green box, >40% S; blue box, <40% S; white box, N), non-helical phosphorus shifts (red circles) and NOE contacts (solid lines). Dotted lines indicate weaker contacts. Thick red lines indicate imino–imino contacts. The contact between H8 G22 and H8 G24 is very weak, making it uncertain whether it is a true contact or a noise peak. The H1′ G22 to H6 U23 could not be seen due to overlap between H1′ G22 and H5 U23. However, the H2′ G22 to H6 U23 NOE is clearly visible (data not shown). The 31P with resonances slightly shifted from the bulk (Materials and Methods) are indicated in red.
Epsilon is capped by a UGU tri–loop with a CG closing base pair. The 11 peaks in the imino spectra show that 11 base pairs are formed in the stem (Fig. 3). Together with the sequential NOE contacts between the imino protons (Fig. 3), this shows that a UGU tri–loop is formed with a CG closing base pair.
The apical tri–loop shows 5′-3′ helical sequential NOE contacts. The following NOE contacts are observed. (i) Sequential 5′-3′ H1′-H6/8 NOE contacts from C11 (stem) up to U14 (Fig. 5) and a NOE from H1′ C11 to H5 U12. (ii) Sequential 5′-3′ H2′-H6/8 contacts observed in the NOESY spectra of the labeled sample (Fig. 4) (14). (iii) Sequential base-to-base contacts (Fig. S3). Together, these NOE contacts suggest that the residues C11, U12, G13 and U14 stack upon each other. Although these observations are typical for a loop where the base stacking is continuous in the 5′-3′ direction going from the stem into the loop, structure calculations have to show the exact nature of the base conformations in the tri–loop.
No sequential NOEs are seen between U14 and G15, except for H8 G15 to U14 H4′ and U14 H5′/5″, observed in the 500 ms NOESY. Furthermore, the H4′ and H5′/5″ resonances of U14 are shifted to values lower than 4.0 p.p.m. This shows that the sugar of U14 is located over the base of G15, so that the sugar resonances experience strong ring current effects (38), creating a turn in the backbone. Usually this turn is brought about by a trans γ angle of the G15 residue. The overlap in the NOESY or COSY spectra makes it impossible to directly determine the G15 γ angle from JH4′H5′ couplings. However, no deviating phosphorus resonance is present in the backbone between U14 and G15, indicating that the angles ε, ζ and α have normal values. This leaves only the γ angle to affect the turn.
The bulged residue is C16 and it is completely bulged out. The remaining ambiguity, after assignment of the imino resonances, was whether the bulge was C16 (C1882) or C17 (C1883). The uncertainty could be resolved, after assignment of the non-exchangeable protons, via the 1H-31P HMBC spectrum, because the H3′ of G15 was found to be J-coupled to the same phosphorus as H5′/5″ of residue C16 (Fig. S10). This assignment was also found to be consistent with the sequential NOEs between non-exchangeable protons.
NOE contacts between H8 of G15-H1′ and H5 of C17 are visible. No H6/8-H6/8 NOE contacts are seen between G15 and C16 or between C16 and C17. H1′ and H5 of C16 possess random coil values (29) (Table S1). In contrast, H5 of C17, which is influenced by ring currents from the 5′ neighbor (G15), has a helix shift value, indicating a position for the base nearly as in a regular helix. Taken together these data show a completely bulged out C16 and that the base pair C11-G15 completely stacks through the bulge onto the base pair G10-C17.
The unpaired uridine is partially intercalated in the stem. The conformation around the unpaired residue U23 (U1889) follows from the observed NOE contacts. An NOE contact is seen between the imino protons of G24 and U5, which would not be possible with U23 completely intercalated between G22 and G22. On the other hand, there are regular 5′-3′ sequential contacts between H1′/H2′ G22 to H6 U23 and H1′/H2′ U23 to G24 H8. In addition, H6 U23 contacts H8 G24 and H8 G22 (Fig. S3). These latter data indicate at least partial stacking and partial intercalation. The near random coil shift of the H5 of U23 indicates that the U23 base is rotated away from its 5′ neighbour G22. Furthermore, there is a weak NOE contact between H8 G22 and H8 G24. This suggests that U23 is intercalated, but rotated away from a regular stack, allow ing the NOE contact between G22-G24. Consistently, the G22-P-U23 and U23-P-G24 phosphorus shifts are inside or on the edge of the normal helix region, suggesting no dramatic changes in the backbone torsion angles.
DISCUSSION
HBV replicates via reverse transcription of its pregenomic RNA. A highly conserved bulged stem–loop (ε) at the 5′ end of the pregenomic RNA is essential for encapsidation and replication. Here we studied the fold of the apical stem–loop of ε by NMR using new isotope labeling techniques and through-bond correlation at natural abundance using cryo probe technology. The 2H/13C/15N-U-labeling in the study of ε demonstrates the spectral simplification, confirmed the assignments, resolved ambiguities in crowded regions and provided many NOE contacts. This type of labeling is crucial for NMR studies of larger RNA systems.
The NMR data show that the loop region, which caps the stem, folds into a UGU tri–loop with a CG closing base pair (Figs 5 and 6). In the stem underlying the tri–loop, the unpaired residue C16 (C1882) is completely bulged out, while the unpaired residue U23 (1889) is partly intercalated. C16 and U23 are spaced 6 bp apart. A large number of NOE contacts (Fig. 5) and narrow resonances were observed throughout the loop region, which are all accounted for (e.g. Fig. 4). This indicates no conformational exchange at the ms time-scale region or at slower time-scales, suggesting a stable and well defined loop. In agreement with this, thermodynamic studies on a series of tri–loops have shown that U-rich tri–loops with a CG closing base pair are highly stable (39).
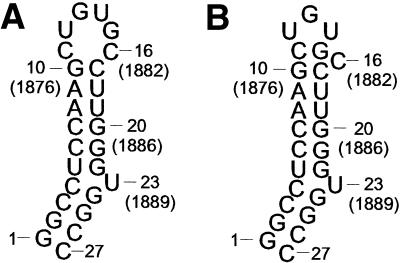
Figure 6.
Comparison between the predicted (A) and the new proposed fold (B) of the HBV ε apical stem–loop is shown. Instead of the previously described hexa–loop, our data support a UGU tri–loop with a CG closing base pair and a C-bulge.
In view of this we hypothesize that the loop fold is essential for the recognition by the reverse transcriptase. If so, mutations in the HBV strains should be structurally silent. In the next section we consider the mutations in the 1231 analyzed HBV strains in the light of this hypothesis.
Mutations in the apical stem–loop region of ε
Both the primary sequence and secondary structure of ε are apparently very well conserved (Fig. 2). However, some important mutations have been observed in the loop region [e.g. 1878 U→A, (33)], and in the closing base pair (1877 C→U; 1881 G→C/U). The 1878 U→A mutation does not affect the loop stability significantly, since both a U and an A residue at the 5′ end of the tri–loop result in stable tri–loops (39). The 1877 C→U mutation, which creates a UG closing base pair, has been seen in several strains (32,33; K. Kidd-Ljunggren, unpublished data). Although a UG base pair is not as stable as a CG base pair in the context of a regular A-helix, it still forms a stable closing base pair of the tri–loop (39). It might be sufficiently stable under intracellular conditions to keep the bulged C in the correct position. Furthermore, there is structural evidence for the formation of such a tri–loop. A tri–loop with a UG trans-wobble closing base pair and a bulged U has been described for one of the loops in the internal ribosomal entry site (IRES) in the hepatitis C virus genome (40,41). However, the folding patterns of the IRES loop seem to differ considerably from the folding of the ε apical loop.
Two rare mutations (1881 G→C/U and 1876 G→A) have been observed (<3 out of 1231 strains) that could potentially disrupt the stable UGU-tri–loop fold with bulged out C observed here. However, observation of these rare mutations may be an experimental artifact of the PCR method used. Amplifying a region of viral DNA by PCR may introduce mutations, a rare occurrence, but more frequent if polymerases without proofreading capacity are used. This risk is increased if nested PCR techniques are used in order to detect very low levels of viral DNA. Samples containing low levels of DNA showed a greater variability in sequences when they were reamplified and sequenced, than samples with high levels of DNA (42).
Alternatively, rare mutations could occur during the reverse transcription of a viable virus, creating non-viable virus particles. In addition, a ‘non-viable’ mutant strain might still be able to replicate if complemented by a wild-type strain. It is not known what proportion of wild-type strains is necessary to complement mutated strains in order to ensure viable viral replication. These can then only exist in the presence of wild-type strains. Direct sequencing of PCR products from such a sample without an intermediate cloning step is often used. The resulting sequence is generally believed to represent the most prevalent clone present in the sample, but it does not necessarily have to be the case.
Although these mutations create unusual and rather unstable base pairs, their formation cannot be excluded. The 1881 G→C/U mutations (31) create a CC or CU closing base pair in the proposed apical loop fold. Unusual pyrimidine– pyrimidine closing base pairs have been observed (43,44). In addition, the C-UGU-G loop is highly stable [–2.2 kcal/mol (39)] and the free energy penalty incurred by formation of CC or CU instead of a CG closing base pair might be insufficient to disrupt the tri–loop. The second rare mutation from G to A at position 1876 creates an AC base pairing in the stem directly below the closing base pair of the proposed apical loop. Whereas the stability of the stem may be affected by this mutation in both of these strains, the stable CG closing base pair remains intact (31,34). In any case, whatever the actual events during replication might be, the fact that the mutations are very rare makes them likely to hamper, if not abolish, recognition by the reverse transcriptase.
In conclusion, we observe that the structure of the upper stem–loop of ε is conserved among all known human HBV strains. Thus, evolutionary pressure maintains this structure in viable human HBV, strongly suggesting that it is critical for recognition. Beyond the scope of the present study, it is of interest to consider some aspects of the subsequent binding process. The reverse transcriptase recognizes and binds to ε (1–3,45) and subsequently, encapsidation, primer synthesis, translocation and then DNA synthesis take place (1–3,45). This process also requires cellular chaperones. For human HBV, as yet no in vitro studies can be carried out on the reverse transcriptase binding to ε. Such studies have been done using an in vitro reconstituted duck reverse transcriptase HBV (45). They show, via chemical probing, that in the primer competent complex, the upper stem–loop of ε has melted, at least in part, in both duck and heron HBV. In light of the above and the function of a polymerase, it seems likely that for human HBV after initial recognition of the conserved structure of free ε, also subsequently structural changes occur in ε. Interestingly, duck reverse transcriptase recognizes both duck ε, with a well defined upper stem–loop structure, though different from human HBV, as well as heron ε, where several of the base pairs in the upper stem are non-canonical and may be absent. Thus, in the in vitro system the exact structure of the upper-stem loop of ε does not seem critical (45). This raises interesting questions about potential differences in the recognition and binding process. More studies are needed to define the exact nature of the binding process. We finally note that irrespective of the exact nature of binding process the conservation of the structure of the upper stem–loop of free ε in human HBV makes it an interesting target for potential anti-viral drugs.
CONCLUSION
The NMR data show that, instead of the predicted hexa–loop, a stable well defined UGU tri–loop is formed with a CG closing base pair (Figs 5 and 6). In the stem underlying the tri–loop, the unpaired residue C16 (C1882) is completely bulged out, while the unpaired residue U23 (1889) is partly intercalated. C16 and U23 are spaced 6 bp apart. Analysis of mutations in 1231 human HBV strains, covering all human HBV genotypes, strongly suggests that this fold is conserved in vivo. We therefore propose that the fold of the apical stem–loop is essential for recognition of the HBV reverse transcriptase. The structural information on ε from this study complemented with residual dipolar couplings will be used to derive a detailed three-dimensional structure of ε. Knowing the three-dimensional structure will help in the screening for small molecules that can bind to ε, thereby preventing viral replication. This might lead to the discovery of new, more efficient antiviral agents against hepatitis B.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank J. Williamson, for providing the overproducing strains for the enzymes needed in the synthesis described, and J. J. Dunn for providing the strain for producing T7 RNA polymerase. A. H. Kidd and F. Girard are gratefully acknowledged for valuable discussions. This work was supported by grants from the Swedish Natural Science Research Council (K5104-1655/2001), Swedish Medical Research Council (Proj. Nos K1999-16X-011592-04B and K2001-16GX-14075-01), Biotechnology Fund Umeå University, Foundation for Strategic Research / I&V (S.W.) and Kempe Foundation (S.F. and J.C.).
REFERENCES
- 1.Rieger A. and Nassal,M. (1996) Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J. Virol., 70, 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang G. and Seeger,C. (1993) Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol., 67, 6507–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem D., Pollack,J. and Tavis,J. (1994) Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication. Infect. Agents Dis., 3, 85–93. [PubMed] [Google Scholar]
- 4.Ganem D. and Varmus,H. (1987) Molecular biology of the hepatitis B virus. Annu. Rev. Biochem., 56, 651–693. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto H., Imai,M., Kametani,M., Nakamura,T. and Mayumi,M. (1987) Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through materno-fetal transmission. Jpn. J. Exp. Med., 57, 231–236. [PubMed] [Google Scholar]
- 6.Bläckberg J. and Kidd-Ljunggren,K. (2000) Occult hepatitis B virus after acute self-limited infection persisting for 30 years without sequence variation. J. Hepatol., 33, 992–997. [DOI] [PubMed] [Google Scholar]
- 7.Kidd A.H. and Kidd-Ljunggren,K. (1996) A revised secondary structure model for the 3′-end of hepatitis B virus pregenomic RNA. Nucleic Acids Res., 24, 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskus T., Rakela,J. and Persing,D. (1994) The stem–loop structure of the cis-encapsidation signal is highly conserved in naturally occurring hepatitis B virus variants. Virology, 200, 809–812. [DOI] [PubMed] [Google Scholar]
- 9.Knaus T. and Nassal,M. (1993) The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem–loop structure that is critical for its function. Nucleic Acids Res., 21, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollack J. and Ganem,D. (1993) An RNA stem–loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol., 67, 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto H., Tsuda,F., Sakugawa,H., Sastroewignjo,R.I., Imai,M., Miyakawa,Y. and Mayumi,M. (1988) Typing hepatitis B virus by homology in nucleotide sequence comparison of surface antigen subtypes. J. Gen. Virol., 69, 2575–2583. [DOI] [PubMed] [Google Scholar]
- 12.Milligan J. and Uhlenbeck,O. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 13.Nikonowicz E., Sirr,A., Legault,P., Jucker,F., Baer,L. and Pardi,A. (1992) Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res., 20, 4507–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flodell S., Cromsigt,J., Schleucher,J., Kidd-Ljunggren,K. and Wijmenga,S. (2002) Structure elucidation of the hepatitis B virus encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn., 19, 627–636. [DOI] [PubMed] [Google Scholar]
- 15.Scott L., Tolbert,T. and Williamson,J. (2000) Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol., 317, 18–38. [DOI] [PubMed] [Google Scholar]
- 16.Plateau P. and Gueron,M. (1982) Exchangeable proton NMR without base-line distortion using new strong-pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 17.Shaka J., Lee,C. and Pines,A. (1988) Iterative schemes for bilinear operations—application to spin decoupling. J. Magn. Reson., 77, 274–293. [Google Scholar]
- 18.Bax A., Griffey,R. and Hawkins,B. (1983) Correlation of proton and nitrogen-15 chemical shifts by multiple quantum NMR. J. Magn. Reson., 55, 301–315. [Google Scholar]
- 19.Bendall M., Pegg,D. and Doddrell,D. (1983) Pulse sequence utilizing the correlated motion of coupled heteronuclei in the transverse plane of the doubly rotating frame. J. Magn. Reson., 52, 81–117. [Google Scholar]
- 20.Van Dongen M., Heus,H., Wijmenga,S., Eritja,R., Azorín,F. and Hilbers,C. (1996) Through-bond correlation of adenine H2 and H8 protons in unlabeled DNA fragments by HMBC spectroscopy. J. Biomol. NMR, 8, 207–212. [DOI] [PubMed] [Google Scholar]
- 21.Kay L., Clore,M., Bax,A. and Gronenborn,A. (1990) Four-dimensional heteronuclear triple-resonance NMR spectroscopy of interleukin-1β in solution. Science, 249, 411–414. [DOI] [PubMed] [Google Scholar]
- 22.Otting G., Senn,H., Wagner,G. and Wuthrich,K. (1986) Editing of 2D H-1-NMR spectra using X half-filters—combined use with residue-selective N-15 labeling of proteins. J. Magn. Reson., 70, 500–505. [Google Scholar]
- 23.Wijmenga S. and van Buuren,B. (1998) The use of NMR methods for conformational studies of nucleic acids. Prog. in NMR Spectrosc., 32, 287–387. [Google Scholar]
- 24.Hurd R. and John,B. (1991) Gradient-enhanced proton-detected heteronuclear multiple-quantum coherence spectroscopy. J. Magn. Reson., 91, 648–653. [Google Scholar]
- 25.Markley J., Bax,A., Arata,J., Hilbers,C., Kaptein,R., Sykes,B., Wright,P. and Wütrich,K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl. Chem., 70, 117–142. [DOI] [PubMed] [Google Scholar]
- 26.Roongata A., Jones,C. and Gorenstein,D. (1990) Effect of distortions in the deoxyribose phosphate backbone conformation of duplex oligodeoxyribo-nucleotide dodecamers containing GT, GG, GA, AC and GU base-pair mismatches on 31P NMR spectra. Biochemistry, 29, 5245–5258. [DOI] [PubMed] [Google Scholar]
- 27.Cromsigt J., van Buuren,B., Schleucher,J. and Wijmenga,S. (2001) Resonance assignment and structure determination for RNA. Methods Enzymol., 318, 371–399. [DOI] [PubMed] [Google Scholar]
- 28.Wijmenga S., Mooren,M. and Hilbers,C. (1993) NMR of Macromolecules—A Practical Approach. Oxford University Press, New York, pp. 217–288.
- 29.Cromsigt J., Hilbers,C. and Wijmenga,S. (2001) Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR, 21, 11–29. [DOI] [PubMed] [Google Scholar]
- 30.Carman W.F., Jacyna,M.R., Hadziyannis,S., Karayiannis,P., McGarvey,M.J., Makris,A. and Thomas,H.C. (1989) Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet, 2, 588–591. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues-Frias F., Buti,M., Jardi,R., Cotrin,R.E. and Guardia,J. (1995) Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology, 22, 1641–1647. [DOI] [PubMed] [Google Scholar]
- 32.Lok A.S.F., Akarca,U. and Greene,S. (1994) Mutations in the precore region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl Acad. Sci. USA, 91, 4077–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramvis A., Bukofzer,S., Kew,M.C. and Song,E. (1997) Nucleic acid sequence analysis of the precore region of hepatitis B virus from sera of Southern African black adult carriers of the virus. Hepatology, 25, 235–240. [DOI] [PubMed] [Google Scholar]
- 34.Hannoun C., Norder,H. and Lindh,M. (2000) An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol., 81, 2267–2274. [DOI] [PubMed] [Google Scholar]
- 35.Niconowicz E., Roongata,V., Jones,C. and Gorenstein,D. (1989) Sequence-dependent variations in the 31P NMR spectra and backbone torsional angles of wild-type and mutant Lac operator fragments. Biochemistry, 28, 8714–8725. [DOI] [PubMed] [Google Scholar]
- 36.Rosen M., Live,D. and Patel D. (1992) Comparative NMR study of A(n)-bulge loops in DNA duplexes: intrahelical stacking of A, A-A and A-A-A bulge loops. Biochemistry, 31, 4004–4014. [DOI] [PubMed] [Google Scholar]
- 37.Kalnik M., Norman,D., Li,B., Swann,P. and Patel,D. (1990) Conformational transitions in thymidine bulge-containing deoxytridecanucleotide duplexes. Role of flanking sequence and temperature in modulating the equilibrium between looped out and stacked thymidine bulge states. J. Biol. Chem., 28, 294–303. [PubMed] [Google Scholar]
- 38.Wijmenga S., Kruithof,M. and Hilbers,C. (1997) Analysis of 1H chemical shifts in DNA: assessment of the reliability of 1H chemical shifts calculations for use in structure refinement. J. Biomol. NMR, 10, 337–350. [DOI] [PubMed] [Google Scholar]
- 39.Shu Z. and Bevilaqcua,P. (1999) Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry, 38, 15369–15397. [DOI] [PubMed] [Google Scholar]
- 40.Lukavsky P., Otto,G., Lancaster,A., Sarnow,P. and Puglisi,J. (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site. Nature Struct. Biol., 7, 1105–1110. [DOI] [PubMed] [Google Scholar]
- 41.Klinck R., Westhof,E., Walker,S., Afshar,M., Collier,A. and Aboul-ela,F. (2000) A potential RNA drug target in the hepatitis C virus internal entry site. RNA, 6, 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunther S., Meisel,H., Reip,A., Miska,S., Kruger,D. and Will,H. (1992) Frequent and rapid emergence of mutated pre-C sequences in HBV from e-antigen positive carriers who seroconvert to anti-HBe during interferon treatment. Virology, 187, 271–279. [DOI] [PubMed] [Google Scholar]
- 43.Hilbers C., Heus,H., van Dongen,M. and Wijmenga,S. (1994) The hairpin elements of nucleic acid structure: DNA and RNA folding. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, New York, NY, Vol. 8, pp. 56–104.
- 44.Leeper T., Martin,M., Kim,H., Cox,S., Semenchenko,V., Schmidt,F. and van Dooren,S. (2002) Structure of the UGAGAU hexaloop that braces Bacillus RNase P for action. Nature, 9, 397–403. [DOI] [PubMed] [Google Scholar]
- 45.Beck J. and Nassal,M. (1998) Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol., 18, 6265–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.