Abstract
Sex of the liverwort Marchantia polymorpha is determined by the sex chromosomes Y and X, in male and female plant, respectively. Approximately half of the Y chromosome is made up of unique repeat sequences. Here, we report that part of the Y chromosome, represented by a 90-kb insert of a genomic clone pMM2D3, contains five putative genes in addition to the ORF162 gene, which is present also within the Y chromosome-specific repeat region. One of the five putative genes shows similarity to a male gamete-specific protein of lily and is expressed predominantly in male sex organs, suggesting that this gene has a male reproductive function. Furthermore, Southern blot analysis revealed that these five putative genes are amplified on the Y chromosome, but they also probably have homologs on the X chromosome and/or autosomes. These observations suggest that the Y chromosome evolved by co-amplifying protein-coding genes with unique repeat sequences.
INTRODUCTION
Unlike most animal species, which are unisexual, the majority of flowering plants such as Arabidopsis thaliana and rice (Oryza sativa) are hermaphroditic and develop bisexual flowers. Several plants, including gingko (Gingko biloba), white campion (Silene latifolia), garden sorrel (Rumex acetosa), hemp (Cannabis sativa) and the liverwort Marchantia polymorpha, are dioecious, that is, unisexual reproductive organs are formed on different individuals. In S.latifolia, the Y chromosome dominantly induces male reproductive organs by its presence in an XX/XY system similar to the mammalian pair of sex chromosomes (1). Sex determination in R.acetosa is similar to that in Drosophila, where X chromosome dosage determines sex (1).
Several attempts have been made to isolate genes carried by plant sex chromosomes. In most cases, however, the sex chromosome-derived sequences are repetitive sequences present not only on the sex chromosomes but also on autosomes (2–8). Sequences unique to the Y chromosome have been reported in R.acetosa and M.polymorpha (9,10). In terms of active genes carried by plant sex chromosomes, the S.latifolia MROS3 gene was originally identified as a gene specifically expressed in male reproductive organs (11). This gene was later shown to be X chromosome linked with a homologous sequence present on the Y chromosome (12). Recently it was reported that at least two copies of MROS3 are present in tandem on the X chromosome (13). Genes SlY1 and SlY4 are found to be Y chromosome linked and active in S.latifolia, but there are also close active homologs, SlX1 and SlX4, on the X chromosome. These sex chromosome genes are therefore not likely to have sex-specific functions such as sex determination (14,15). Genes responsible for sex determination in plants, as well as the detailed structures of the plant sex chromosomes, are still largely unresolved.
The liverwort M.polymorpha has unusually small sex chromosomes. Because of its haploidy, the Y chromosome is present only in male plants and the morphologically distinct X chromosome is found only in female plants (16). This X/Y exclusiveness strongly suggests the presence of sex determining factors on the sex chromosomes. Towards a better understanding of the sexual reproduction system in M.polymorpha, we have initiated detailed analyses of the gene structure of the Y chromosome. Genomic libraries for male and female M.polymorpha plants have enabled us to identify sequences unique to the Y chromosome (16). Here we describe five multicopy genes amplified in a highly repeated region of the Y chromosome.
MATERIALS AND METHODS
Plant materials
Male and female thalli of M.polymorpha (E lines) (17) were cultivated on M51C medium (18) at 24°C under continuous light. Sex organs were obtained from the same lines.
Sequence analyses
Shotgun sequencing of pMM2D3 was performed as described previously (10). Searches for protein coding regions were performed against the non-redundant protein sequence database at the National Center for Biotechnology Information (NCBI) using the BLASTX program (19) and against M.polymorpha ESTs (20,21) using BLASTN program (19).
The sequences reported in this paper have been deposited in the GenBank database (accession nos AF542555, AF542560, AF542556, AF542557, AF542558 and AF542559 for RS, RT, R1, R2, R3 and R4 portions in Fig. 1, respectively).
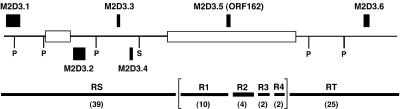
Figure 1.
Schematic diagram of the structure of pMM2D3. The insert of the plasmid is displayed with its SP6-end on the left. Alignments of sequence contigs are shown by the lower horizontal bars labeled as RS, RT, R1, R2, R3 and R4. Sequences of R1, R2, R3 and R4 consisted of the Y chromosome-specific repeat sequences. Approximate sizes of contigs are given in parentheses in kb. The order of contigs in brackets was not determined. Putative genes are indicated by closed boxes above (left-to-right orientation) and below (right-to-left orientation) the line. Recognition sites for PacI (P) and SfiI (S) determined by restriction digestion and electrophoresis are indicated by vertical lines. Clusters of the Y chromosome- specific repeat sequences are indicated by open boxes. The exact position of M2D3.5 within the clone is not known because of the repeat sequences, but its orientation is predicted from the orientation of the repeat sequences at the proximal ends of RS and RT.
Genomic PCR analysis
PCR for determining male specificity of putative genes was performed basically as described previously (16). Template genomic DNAs were isolated as described by Takenaka et al. (17), and 10 ng each were used as template. Sequences of primers are, 5′-CTGGACCAAGTGATTCGCTCTC and 5′-AGCCCACTGATATAACGAAGAC for M2D3.1, 5′-CCG TGACGCCGAGCGATGTGGG and 5′-CGCTCGAACGAC ACCGTATCGC for M2D3.2, 5′-GGAATGCATCCCAGT TGAGACC and 5′-AAGAGCCTCGAGCTTCTGCTTC for M2D3.3, 5′-CGGACTGGAGTACTGGAACGAT and 5′-TTCTGGTCGGAACTGCTGATCG for M2D3.4 and 5′-AAACTTTCGCTGCATCGAGCGG and 5′-TCGTCCTG TTTCTGCTTCAGCC for M2D3.6. Primers designed from ORF162 (10) and the calcium-dependent protein kinase (CDPK) gene (16) were used for quality evaluation of the genomic DNA.
Southern blot analysis
Five micrograms of total DNA were digested with BamHI and the resulting fragments were separated in a 1% agarose gel in 1× TAE buffer. After alkaline treatment and blotting onto a nylon membrane, hybridization was performed in a solution containing 5× Denhardt’s reagent, 6× SSPE pH 7.4, 0.5% SDS, 100 µg/ml denatured salmon sperm DNA and 50% formamide at 42°C. To prepare probes, DNA fragment specific to each gene was amplified by PCR with plasmid DNA of pMM2D3 as template and with the primer pairs described in ‘Genomic PCR analysis’ (Materials and Methods), and then the PCR products were labeled with [α-32P]dCTP by another round of PCR. Membranes were washed for 1 h in a solution containing 1× SSPE and 0.1% SDS at 42°C followed by two washes with 0.1× SSPE and 0.1% SDS at 65°C for 1 h. Radioactive signals were visualized with a BAS2000 Image Analyzer (Fuji Photo Film).
RT–PCR analysis
Total RNAs from male and female thalli, and from male and female sex organs, were individually prepared by the phenol/SDS method (22). Poly(A)+ RNA was prepared with the PolyATract™ System 1000 (Promega) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 5 µg of DNase-treated poly(A)+ RNA using SuperScript II™ reverse transcriptase (Gibco BRL) at 42°C with XhoSseEcoR-dT primer (5′-GAGAATTCCTGCAGGC TCGAGTTTTTTTTTTTTTTTTTT-3′) for 60 min. A 20 µl reaction mixture was diluted in 400 µl TE, and 1 µl of the diluted mixture was used as a template in a 20 µl PCR amplification mix containing 10 pmol of the same primers used for the genomic PCR. Reactions without reverse transcriptase were performed to check genomic DNA contaminations.
Northern blot analysis
Poly(A)+ RNA was prepared according to the method described above for RT–PCR. Five micrograms of poly(A)+ RNA were electrophoresed in a 0.8% denaturing agarose gel containing formaldehyde and transferred onto a nylon membrane. Hybridization was performed with ExpressHyb™ solution (CLONTECH) as described (10). Part of M2D3.4 was amplified by PCR with plasmid DNA of pMM2D3 as template and the M2D3.4 primer pair. The PCR product of M2D3.4 was labeled with [α-32P]dCTP by another round of PCR. The hybridized membrane was washed for 1 h in a solution containing 2× SSC and 0.1% SDS at 25°C followed by two washes with 0.1× SSC and 0.5% SDS at 55°C for 1 h. Radioactive signals were visualized with a BAS2000 Image Analyzer (Fuji Photo Film).
RESULTS
A male-specific clone from the Y chromosome
To identify Y chromosome-derived clones in our PAC (P1-derived artificial chromosome) library, we screened for clones containing the Y chromosome-specific repeat sequences (16). One positive clone, pMM2D3, was selected for detailed analysis, since the restriction profile of pMM2D3 is distinct from that of pMM4G7, which has been already examined (10), and there are other PAC clones that align consistently with pMM2D3 (data not shown), excluding the possibility that pMM2D3 is a chimeric clone.
Approximately 1800 shotgun and gap-filling sequences yielded a coverage of over eight times that of the entire length of the clone (∼90 kb long) and assembly resulted in six sequence contigs. The sequences of the two ends of the insert, RS and RT, were unambiguously oriented by the SfiI and PacI recognition sites (Fig. 1). The total length of R1, R2, R3 and R4 (∼18 kb) is shorter than the actual distance between contigs RS and RT (∼26 kb) because the copy numbers of the repeat sequences are unknown. Nevertheless, any of the 1800 shotgun and gap-filling sequences aligned with one of the six contigs, indicating that the entire sequence content of pMM2D3 is represented in the six contigs.
Y chromosome-specific repeat sequences in pMM2D3
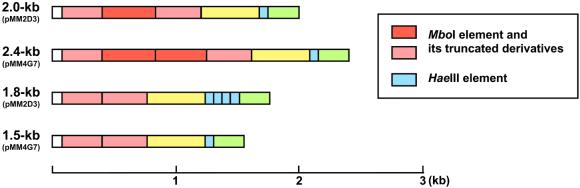
The Y chromosome-specific repeat sequences identified in clone pMM4G7 are also highly conserved in pMM2D3. In addition to the five types of Y chromosome-specific BamHI fragment (1.5, 2.4, 2.7, 5.2 and 2.2 kb) identified in clones pMM4G7 and pMM23-130F12 (10), two BamHI variants with different sizes, 2.0 and 1.8 kb, were found in pMM2D3 (Fig. 2). The 2.0-kb BamHI fragment has one MboI element while the 2.4-kb BamHI fragment has two, and the 1.8-kb BamHI fragment has three additional copies of the HaeIII element compared with the 1.5-kb BamHI fragment (Fig. 2). The 2.0- and 1.8-kb BamHI fragments found in pMM2D3 thus also consist of common sequence elements and differ only in relative copy numbers of these elements.
Figure 2.
Structures of the 2.0-kb and 1.8-kb BamHI repeat units newly identified in pMM2D3, in comparison with the 2.4-kb and 1.5-kb BamHI repeat units of pMM4G7. As in the 2.4-kb and 1.5-kb BamHI repeat units, the 2.0-kb and 1.8-kb BamHI repeat units also consist of common subrepeats, MboI (red) and HaeIII elements (blue). Other sequences are color-coded (white, yellow and green) according to their respective similarities.
Potential genes found in pMM2D3
In order to identify protein-coding genes, the sequence of pMM2D3 was searched against the 1415 M.polymorpha ESTs (20,21) and the non-redundant protein sequence database of NCBI. No M.polymorpha EST tags any portion of pMM2D3. However, five regions, M2D3.1, M2D3.2, M2D3.3, M2D3.4 and M2D3.6, of pMM2D3, show significant similarity of their deduced amino acid sequences to known proteins (Fig. 1) (Table 1). These are in addition to M2D3.5, which is a member of the Y chromosome-specific ORF162 gene family described previously (10). All these regions, with the exception of M2D3.1, align as uninterrupted open reading frames (ORFs) and thus appear to be intact protein-coding genes.
Table 1. Potential genes found in pMM2D3.
| Contig | Sequence | Length (bp) | % Identity (aa) | Similar sequences (species) | Accession no. |
|---|---|---|---|---|---|
| RS | M2D3.1 | 1173a | 67 (65/96) | Putative alliinase (A.thaliana) | T05567 |
| RS | M2D3.2 | 2073 | 68 (76/111) | Putative RAV-like B3 DNA binding protein (A.thaliana) | AAC34233 |
| RS | M2D3.3 | 492 | 35 (65/184) | Unknown protein (A.thaliana) | AAG51648 |
| RS | M2D3.4 | 588 | 33 (35/105) | LGC1: expressed exclusively in the male gametic cells (34) (Lilium logiflorum) | AAD19962 |
| R1 | M2D3.5 | 489 | 100 (162/162) | ORF162 (M.polymorpha) (10) | BAB62538 |
| RT | M2D3.6 | 367 | 47 (158/334) | Unknown protein (O.sativa) | AAG03087 |
aPutative introns were not included.
The segmented alignment of amino acid sequences of M2D3.1 and alliin lyase gene homologs indicates that M2D3.1 consists of six exons. The joined sequence of the six putative exons contains an uninterrupted ORF. The dinucleotides GT and AG, characteristic for 5′- and 3′-ends of eukaryotic introns, are conserved at the boundaries of the deduced introns. The alliin lyase gene of onion has four introns, all of which are located at positions identical to the putative introns in M2D3.1 (data not shown). In Chinese chive (Allium tuberosum) homolog, Lys280 has been shown to be essential for catalytic activity and to be a probable pyridoxal- phosphate-binding residue (23), the corresponding lysine residue is conserved in M2D3.1 at position 224 (Fig. 3). This suggests that the protein encoded in M2D3.1 could function as an S-alk(en)yl-l-cysteine sulfoxide lyase in the metabolism of cysteine, homocysteine and methionine or derivative compounds. Alliin lyase is thought to contribute to chemical defense in Allium plants by producing volatile sulfur-containing compounds (23). The homologous gene in M.polymorpha may have an analogous function.
Figure 3.
Multiple amino acid sequence alignment of the pyridoxal phosphate binding site in M2D3.1 and its related alliin lyase-like proteins. The numbers in parentheses indicate the positions in the respective sequence. Amino acid residues identical to those of M2D3.1 are indicated by colons. The lysine residue suggested to be the pyridoxal phosphate binding site is indicated by an arrow. AtAlh, alliin lyase homolog of A.thaliana (GenBank accession number T05567); AcAl, Allium cepa alliin lyase (AAA32639); AtuAl, A.tuberosum alliin lyase (BAA20358).
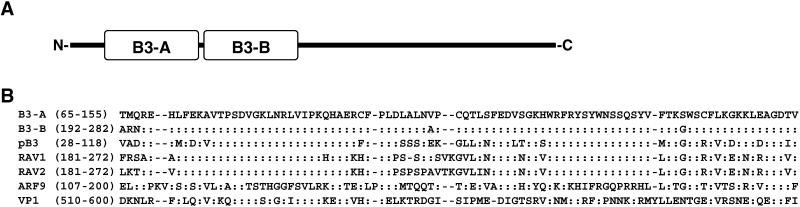
The protein encoded by M2D3.2 contains two B3 DNA-binding domains in tandem orientation. B3 DNA-binding domains are found in a variety of transcription factors such as auxin response factors (ARFs), the maize transcription factor VIVIPAROUS1 (VP1), the RAV1 DNA-binding protein of A.thaliana and their relatives (24–26). The B3 DNA-binding domains of M2D3.2 show the highest similarity to that of RAV1 (25) (Fig. 4). The B3 DNA-binding domain of RAV1 recognizes the DNA motif CACCTG (26), but its target genes have not been identified. M2D3.2 is unique in containing two B3 DNA-binding domains (Fig. 4), which are 95% identical in their amino acid sequences.
Figure 4.
Alignment of B3 DNA-binding domains found in M2D3.2. (A) Schematic illustration of M2D3.2 with two B3 DNA-binding domains, B3-A and B3-B. (B) Amino acid sequence alignment of B3 DNA-binding domains found in M2D3.2 and other proteins. The numbers in parentheses indicate the positions in the respective sequences. Amino acid residues identical to those of B3-A, are indicated by colons. Gaps are indicated by dashes. pB3, putative DNA binding protein of A.thaliana (GenBank accession no. AAC34233); RAV1, related to ABI3/VP1 1 DNA-binding protein of A.thaliana (BAA34250); RAV2, related to ABI3/VP1 2 DNA-binding protein of A.thaliana (BAA34251); ARF9, auxin response factor 9 of A.thaliana (AAD24427); VP1, VIVIPAROUS1 protein of maize (P26307).
Multiple alignment of the predicted product of M2D3.6 and the other proteins that show significant similarity to it, revealed that they contain a paired amphipathic helix repeat (PAH) domain which is important for Sin3 function as a co-repressor (Fig. 5). Similarity of M2D3.6 is limited to the PAH regions in these homologs. While Sin3 and mSin3A have four PAH domains (27), M2D3.6 has one. As PAH domains have been reported to associate with various transcription factors (27), M2D3.6 potentially interacts with a transcription factor and regulates its activity.
Figure 5.
Amino acid sequence alignment of the PAH domains found in M2D3.6 and other proteins. The numbers in parentheses indicate the positions in the respective sequence. Amino acid residues identical to those of the PAH domain of M2D3.6 are indicated by colons. Gaps are indicated by dashes. OsUP, unknown protein of rice (GenBank accession no. AAG03087); AtTRl, transcription regulator-like protein of A.thaliana (T51447); MmSin3A, Sin3 transcription regulator homologous protein of Mus musculus (AAA89119); ScSin3, Sin3 transcription regulator protein of Saccharomyces cerevisiae (RGBYS3).
Analysis of sex specificity of the five putative genes found in pMM2D3
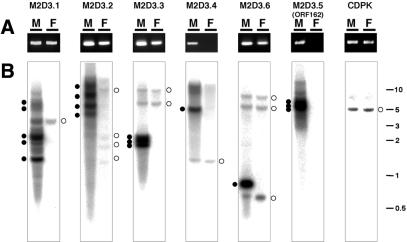
To investigate their sex specificities, we performed diagnostic genomic PCR of the five putative genes (Fig. 6A). A primer pair was designed for the conserved region of each of the putative genes, M2D3.1, M2D3.2, M2D3.3, M2D3.4 and M2D3.6. M2D3.4 did not yield a PCR product for female DNA. Three additional different primer sets for M2D3.4 again yielded no PCR products (data not shown). Primer sets of the other four genes readily detected homologous sequences in the female DNA. This result indicates that at least one homologous sequence for each of the putative genes, M2D3.1, M2D3.2, M2D3.3 and M2D3.6, is present on the X chromosome and/or autosomes.
Figure 6.
The sex specificity and copy number of the five putative genes. (A) Sex specificity of the five putative genes was examined by genomic PCR with primer pairs designed for the conserved regions of the respective five genes. CDPK served as a quality control for the genomic DNAs. (B) The copy numbers of the five putative genes were examined by genomic Southern blot analyses. Genomic DNAs of male (M) and female (F) plants were digested with BamHI, and were probed with DNA fragments of the respective ORFs and CDPK. Closed circles indicate intense signals detected specifically in male DNAs. Open circles indicate signals detected in both male and female DNAs. Sizes of signals are given in kb on the right.
The five putative genes are present in multicopy on the Y chromosome
In order to investigate the copy number of the five putative genes, genomic Southern blot analysis was performed. The five putative genes showed at least one intense signal in the male but not the female DNA, revealing that the male genome carries numerous copies of these putative genes on the Y chromosome (Fig. 6B, indicated by closed circles). In contrast to M2D3.5 (ORF162), which as expected shows no signal in the female DNA (10), the probes for the five putative genes gave several weakly hybridizing fragments in the female DNA (Fig. 6B, indicated by open circles), indicating the existence of homologous sequences on the X chromosome and/or autosomes. Since all the DNA fragments detected in the female DNA are also found in the male DNA (Fig. 6B, indicated by open circles), these homologous sequences are likely to be located on autosomes. Arabidopsis thaliana genes, similar to M2D3.2 and M2D3.6, have a few diverged homologs on different A.thaliana chromosomes but do not appear to be multicopy genes like the liverwort genes. Arabidopsis thaliana genes similar to the other liverwort genes, M2D3.1, M2D3.3, M2D3.4 and M2D3.5, are single-copy genes.
Transcription analysis of the novel putative genes
Expression analyses of the five putative genes were carried out by diagnostic RT–PCR. Except for M2D3.3, four of the putative genes, M2D3.1, M2D3.2, M2D3.4 and M2D3.6, were detected to be expressed in male thalli as well as in mature male sex organs (Fig. 7). The size discrepancy of 103 bp for M2D3.1 between the 225-bp PCR products from cDNA (lanes + in Fig. 7) and the 328-bp product from the genomic clone pMM2D3 (lane P in Fig. 7) matches well with the size of a predicted intron of 103 bp. The RT–PCR assay also demonstrated that homologous sequences of the three putative genes, M2D3.1, M2D3.2 and M2D3.6, are expressed in female thalli and mature sex organs, suggesting that their respective X chromosomal and/or autosomal copies are functional.
Figure 7.
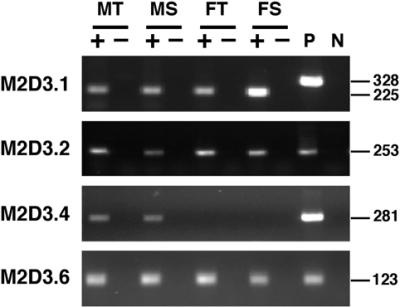
RT–PCR analyses of the putative genes. Poly(A)+ RNAs isolated from male thalli (lanes MT), male sex organs (lanes MS), female thalli (lanes FT) and female sex organs (lanes FS) were reverse-transcribed, and the resulting cDNAs were used as templates (lanes +). The same amounts of poly(A)+ RNA but without the RT reactions were used for control (lanes –). Positive control PCRs were performed using 1 ng of pMM2D3 plasmid DNA as a template (P). Negative control PCRs were carried out without added DNA (N). Sizes of PCR products are given in bp on the right. Primer pairs for RT–PCR here were the same as those used for genomic PCRs.
RT–PCR products for M2D3.4 were found only in male tissues, although Southern blot analysis had suggested the presence of M2D3.4 homologous sequence(s) in the female genome, probably on an autosome (Fig. 6). In order to detect transcripts of the M2D3.4 homologous sequence in female, we further carried out northern blot analysis, since it is possible that the primer pairs for M2D3.4 do not detect the homologous sequence in RT–PCR. M2D3.4 seems to be expressed male specifically and, furthermore, preferentially in male mature sex organs (Fig. 8).
Figure 8.
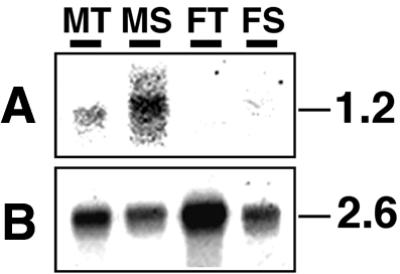
Northern blot analysis of M2D3.4. (A) Five micrograms of poly(A)+ RNA from male thalli (lane MT), male sex organs (lane MS), female thalli (lane FT) and female sex organ (lane FS) were blotted and probed with a 32P-labeled DNA covering M2D3.4. (B) The same membrane was reprobed with the CDPK gene, which is constitutively expressed in male and female. Sizes of signals are given in kb on the right.
DISCUSSION
In this study we provide comprehensive sequence information on ∼90 kb of the liverwort Y chromosome. In addition to a member of the Y chromosome-specific gene family, ORF162, we have identified five novel genes whose copies have been amplified uniquely on the Y chromosome, though four of them also have homologs present on the X chromosome and/or autosomes.
The integrity of the pMM2D3 sequence
Repeat sequences have always been one of the major problems in assembling sequence reads or genomic clones into the correct order. ‘Complete’ genome sequences of eukaryotes generally contain gaps of highly repetitive sequences (28). This is particularly true in the sex chromosomes in other species, as in M.polymorpha. For the region reported previously in pMM4G7, and now in pMM2D3, the Y chromosome-specific repeat sequences prevented us from reconstituting the sequences from these PAC clones. However, a copy of ORF162, embedded deeply within the repeat sequences, was successfully identified, demonstrating that it is feasible to uncover genes in highly repeated sequences without reconstituting the precise primary structure of the surrounding repetitive regions, provided that the non-repetitive region is fully represented by shotgun sequence reads. As far as the ‘net’ genetic content is concerned, we have indeed revealed the genetic content of the 90-kb Y chromosome fragment cloned in pMM2D3.
The Y chromosome-specific repeat sequences consist of a small number of repeat elements in various arrangements
The repeat sequences consist of unique units in structure and are distinct in their sequence from those in other species investigated so far (10). The previous genomic Southern blot analyses using one of the Y-specific repeat sequences, the 2.4-kb BamHI fragment, as a probe, had consistently detected signals of the 2.0-kb and 1.8-kb fragments in the male genomic DNA (10), confirming that the two novel 2.0-kb and 1.8-kb BamHI fragments found in pMM2D3 are also major repeat arrangements accumulated in the liverwort Y chromosome.
Multicopy genes on the Y chromosome
The five multicopy genes newly identified in this study seem to have one or more homologous sequences on autosomes, but the possibility that those homologous sequences in the female genome are present in the same restriction context on the X chromosome cannot be excluded. In either case, it is clear that these five genes have been amplified in the Y chromosome. Therefore, it is now evident that not only non-coding repeat sequences, but also embedded genes in the repeat sequences, contribute to the repetitive nature of the liverwort Y chromosome. Our preliminary estimation by dot blot analysis suggests that at least a few dozen copies of these genes have accumulated in the male genome (data not shown). Consistently, partial cDNA sequencing also revealed the presence of other copies of the genes with sequence variation (data not shown).
It is well established that the lack of recombination in most of the Y chromosome results in the accumulation of repeat sequences as well as of mutations (29). Y chromosomes have been reported to harbor unique repeat sequences in various organisms (9,10,30,31). A recent study reported that tandem duplication and inversion of large sequence blocks of 115– 678 kb, which contain active genes, resulted in an extensive palindromic complex and simultaneous amplification of the transcriptionally active genes in a portion of the human Y chromosome (32). Although no such long repeat units have thus far been identified in M.polymorpha, the same mechanism may be operating also in M.polymorpha and result in repetitive structure and the specific multicopy genes of the Y chromosome.
In humans, it has been shown that multicopy gene families on the Y chromosome are expressed specifically in testes and are involved in male reproductive functions such as spermatogenesis (33). Similarly, in M.polymorpha, the ORF162 family encoding RING-finger proteins is localized on the Y chromosome and expressed specifically in male sex organs, though its function is unknown (10). Similarly, transcripts of the multicopy genes represented by M2D3.4 are detectable by northern blot analysis in male sex organs at much higher levels than in male thalli (Fig. 8). On the other hand, there is no detectable mRNA in female thalli and sex organs (Fig. 8). Given its similarity to a protein localized in male gametic cells in lily (34), the M2D3.4 gene product may have a function in the male reproductive system in M.polymorpha.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Dr Axel Brennicke (University of Ulm, Germany) for his valuable suggestions and critical reading of the manuscript. We thank Choi Seung Hyouk, visiting student research fellow from Korea University, Korea, for his excellent technical support. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (no. 13460150 and no. 14011222) and a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists (K.I.).
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +81 75 753 6389; Fax: +81 75 753 6127; Email: AF542555–AF542560
REFERENCES
- 1.Ainsworth C., Parker,J. and Buchanan-Wollaston,V. (1998) Sex determination in plants. Curr. Top. Dev. Biol., 38, 167–223. [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto K., Shimomura,K., Komeda,Y., Kamada,H. and Satoh,S. (1995) A male-associated DNA sequence in a dioecious plant, Cannabis sativa L.Plant Cell Physiol., 36, 1549–1554. [PubMed] [Google Scholar]
- 3.Donnison I.S., Siroky,J., Vyskot,B., Saedler,H. and Grant,S.R. (1996) Isolation of Y chromosome-specific sequences from Silene latifolia and mapping of male sex-determining genes using representational difference analysis. Genetics, 144, 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buzek J., Koutnikova,H., Houben,A., Riha,K., Janousek,B., Siroky,J., Grant,S. and Vyskot,B. (1997) Isolation and characterization of X chromosome-derived DNA sequences from a dioecious plant Melandrium album.Chromosome Res., 5, 57–65. [DOI] [PubMed] [Google Scholar]
- 5.Scutt C.P., Kamisugi,Y., Sakai,F. and Gilmartin,P.M. (1997) Laser isolation of plant sex chromosomes: studies on the DNA composition of the X and Y sex chromosomes of Silene latifolia. Genome, 40, 705–715. [DOI] [PubMed] [Google Scholar]
- 6.Matsunaga S., Kawano,S., Michimoto,T., Higashiyama,T., Nakao,S., Sakai,A. and Kuroiwa,T. (1999) Semi-automatic laser beam microdissection of the Y chromosome and analysis of Y chromosome DNA in a dioecious plant, Silene latifolia. Plant Cell Physiol., 40, 60–68. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto K., Ohmido,N., Fukui,K., Kamada,H. and Satoh,S. (2000) Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol. Biol., 44, 723–732. [DOI] [PubMed] [Google Scholar]
- 8.Shibata F., Hizume,M. and Kuroki,Y. (2000) Differentiation and the polymorphic nature of the Y chromosomes revealed by repetitive sequences in the dioecious plant, Rumex acetosa. Chromosome Res., 8, 229–236. [DOI] [PubMed] [Google Scholar]
- 9.Shibata F., Hizume,M. and Kuroki,Y. (1999) Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma, 108, 266–270. [DOI] [PubMed] [Google Scholar]
- 10.Okada S., Sone,T., Fujisawa,M., Nakayama,S., Takenaka,M., Ishizaki,K., Kono,K., Shimizu-Ueda,Y., Hanajiri,T., Yamato,K.T., Fukuzawa,H., Brennicke,A. and Ohyama,K. (2001) The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl Acad. Sci. USA, 98, 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsunaga S., Kawano,S., Takano,H., Uchida,H., Sakai,A. and Kuroiwa,T. (1996) Isolation and developmental expression of male reproductive organ-specific genes in a dioecious campion, Melandrium album (Silene latifolia). Plant J., 10, 679–689. [DOI] [PubMed] [Google Scholar]
- 12.Guttman D.S. and Charlesworth,D. (1998) An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature, 393, 263–266. [DOI] [PubMed] [Google Scholar]
- 13.Kejnovsky E., Vrána,J., Matsunaga,S., Soucek,P., Siroky,J., Dolezel,J. and Vyskot,B. (2001) Localization of male-specifically expressed MROS genes of Silene latifolia by PCR on flow-sorted sex chromosomes and autosomes. Genetics, 158, 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delichère C., Veuskens,J., Hernould,M., Barbacar,N., Mouras,A., Negrutiu,I. and Monéger,F. (1999) SlY1, the first active gene cloned from a plant Y chromosome, encodes a WD-repeat protein. EMBO J., 18, 4169–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atanassov I., Delichère,C., Filatov,D.A., Charlesworth,D., Negrutiu,I. and Monéger,F. (2001) Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol. Biol. Evol., 18, 2162–2168. [DOI] [PubMed] [Google Scholar]
- 16.Okada S., Fujisawa,M., Sone,T., Nakayama,S., Nishiyama,R., Takenaka,M., Yamaoka,S., Sakaida,M., Kono,K., Takahama,M., Yamato,K.T., Fukuzawa,H., Brennicke,A. and Ohyama,K. (2000) Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J., 24, 421–428. [DOI] [PubMed] [Google Scholar]
- 17.Takenaka M., Yamaoka,S., Hanajiri,T., Shimizu-Ueda,Y., Yamato,K.T., Fukuzawa,H. and Ohyama,K. (2000) Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L.Transgenic Res., 9, 179–185. [DOI] [PubMed] [Google Scholar]
- 18.Ono K., Ohyama,K. and Gamborg,O.L. (1979) Regeneration of the liverwort Marchantia polymorpha L. from protoplasts isolated from cell suspension culture. Plant Sci. Lett., 14, 225–229. [Google Scholar]
- 19.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 20.Nagai J., Yamato,K.T., Sakaida,M., Yoda,H., Fukuzawa,H. and Ohyama,K. (1999) Expressed sequence tags from immature female sexual organ of a liverwort, Marchantia polymorpha. DNA Res., 6, 1–11. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama R., Yamato,K.T., Miura,K., Sakaida,M., Okada,S., Kono,K., Takahama,M., Sone,T., Takenaka,M., Fukuzawa,H. and Ohyama,K. (2000) Comparison of expressed sequence tags from male and female sexual organs of Marchantia polymorpha. DNA Res., 30, 165–174. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., NY, USA.
- 23.Manabe T., Hasumi,A., Sugiyama,M., Yamazaki,M. and Saito,K. (1998) Allinase [S-alk(en)yl-l-cysteine sulfoxide lyase] from Allium tuberosum (Chinese chive). Purification, localization, cDNA cloning and heterologous functional expression. Eur. J. Biochem., 257, 21–30. [DOI] [PubMed] [Google Scholar]
- 24.Ulmasov T., Hagen,G. and Guilfoyle,T.J. (1997) ARF1, a transcription factor that binds to auxin response elements. Science, 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki M., Kao,C.Y. and McCarty,D.R. (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell, 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagaya Y., Ohmiya,K. and Hattori,T. (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res., 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brubaker K., Cowley,S.M., Huang,K., Loo,L., Yochum,G.S., Ayer,D.E., Eisenman,R.N. and Radhakrishnan,I. (2000) Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell, 103, 655–665. [DOI] [PubMed] [Google Scholar]
- 28.Mardis E., McPherson,J., Martienssen,R., Wilson,R.K. and McCombie,W.R. (2002) What is finished and why does it matter. Genome Res., 12, 669–671. [DOI] [PubMed] [Google Scholar]
- 29.Charlesworth B. (1991) The evolution of sex chromosomes. Science, 251, 1030–1033. [DOI] [PubMed] [Google Scholar]
- 30.Foote S., Vollrath,D., Hilton,A. and Page,D.C. (1992) The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science, 258, 60–66. [DOI] [PubMed] [Google Scholar]
- 31.Devlin R.H., Stone,G.W. and Smailus,D.E. (1998) Extensive direct-tandem organization of a long repeat DNA sequence on the Y chromosome of chinook salmon (Oncorhynchus tshawytscha). J. Mol. Evol., 46, 277–287. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda-Kawaguchi T., Skaletsky,H., Brown,L.G., Minx,P.J., Cordum,H.S., Waterston,R.H., Wilson,R.K., Silber,S., Oates,R., Rozen,S. and Page,D.C. (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet., 29, 279–286. [DOI] [PubMed] [Google Scholar]
- 33.Lahn B.T. and Page,D.C. (1997) Functional coherence of the human Y chromosome. Science, 278, 675–680. [DOI] [PubMed] [Google Scholar]
- 34.Xu H., Swoboda,I., Bhalla,P.L. and Singh,M.B. (1999) Male gametic cell-specific gene expression in flowering plants. Proc. Natl Acad. Sci. USA, 96, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]