Abstract
MutS, a member of the ABC ATPases superfamily, is a mismatch DNA-binding protein constituent of the DNA post-replicative mismatch repair system (MMRS). In this work, it is shown that the ATPase activity of Pseudomonas aeruginosa and Escherichia coli MutS is inhibited by ortho- and decavanadate. Structural comparison of the region involved in the ATP binding of E.coli MutS with the corresponding region of other ABC ATPases inhibited by vanadate, including the myosin– orthovanadate–Mg complex, showed that they are highly similar. From these results it is proposed that the orthovanadate inhibition of MutS ATPase can take place by a similar mechanism to that described for other ATPases. Docking of decavanadate on the ATP-binding region of MutS showed that the energetically more favorable interaction of this compound would take place with the complex MutS– ADP–Mg, suggesting that the inhibitory effect could be produced by a steric impediment of the protein ATP/ADP exchange. Besides the effect observed on the ATPase activity, vanadate also affects the DNA-binding capability of the protein, and partially inhibits the oligomerization of MutS and the temperature-induced inactivation of the protein. From the results obtained, and considering that vanadate is an intracellular trace component, this compound could be considered as a new modulator of the MMRS.
INTRODUCTION
The mismatch repair system (MMRS) is one of the cellular mechanisms present in prokaryotes and eukaryotes by which genome alterations are prevented. After DNA replication, this system corrects point mutations or small insertions/deletions that escape from the proofreading of the DNA polymerase activity. Some components of the repair system also inhibit DNA recombination between partially homologous sequences preventing chromosomal rearrangements (1,2). The function of the MMRS is critical for the stability of the cell phenotype since it has been shown that deficiency of its activity produces a hypermutator phenotype in bacteria and tumor development in humans (1–5).
At the molecular level, the assembly of the MMRS to repair DNA biosynthetic errors is initiated by MutS, which recognizes and binds to mispaired nucleotides. Besides the DNA-binding capability, MutS binds adenine nucleotides and displays an ATPase activity. These biochemical properties have been shown to be important for the correct functioning of the protein, since cells harboring mutated versions of the mutS gene that produces an inactive ATPase resulted in a hypermutator phenotype (6,7). Several roles have been attributed to ATP binding and hydrolysis in the DNA repair mechanisms (8–13). However, it is difficult to obtain a precise picture of the role of adenine nucleotides in the MMRS, as the addition of nucleotides can produce variable and drastic effects on the MutS state (14).
Recent results on the crystal structure of truncated Escherichia coli (15) and Thermus aquaticus (16) MutS brought new insights into the protein region involved in DNA and nucleotide interactions. Several modular domains constitute each monomer. Among these, the N-terminal region is involved in the DNA mismatch binding, whereas the C-terminal region contains the ATPase domain, which is followed by a helix–u-turn–helix domain responsible for dimerization (17).
MutS is a member of the ABC ATPases superfamily and is characterized by the presence of four conserved motifs involved in the ATPase domain (15,16), which are located at the dimer-interface region (17). In E.coli MutS, two motifs (GPNMGGKS, residues 614–621, and DE, residues 693–694) correspond to the Walker A and B motifs and constitute the ATP and magnesium-binding sites, respectively. The other two motifs (ST, residues 668–669, and TH, residues 727–728) are unique to the ABC ATPase superfamily, which includes MutS, UvrA, Rad50, and ABC transporters (18). Another common characteristic of MutS and other members of the ABC superfamily is that the dimer state of the protein is necessary for the ATPase activity (11,19).
The important role of MutS ATPase activity in the mismatch repair function makes it relevant to analyze potential inhibitors of this activity that can in turn be useful as biochemical tools to further understand the regulation of this repair mechanism. The fact that the ATPase activity of several members of the ABC superfamily is inhibited by vanadate (20–27) prompted us to analyze the effect of this compound on the ATPase activity of MutS. In this work, we show that, in fact, decavanadate and to a lesser extent orthovanadate inhibit the ATPase activity of Pseudomonas aeruginosa and E.coli MutS. It is also shown that this compound affects the MutS DNA-binding capability and partially preserves the activity of the protein by inhibiting the MutS aggregation. As in several ATPases, it has been described that the Walker A motif is involved in the binding of orthovanadate; we compared the structure of a region of 17 amino acids including the Walker A motif of MutS with several ABC ATPases inhibited by orthovanadate and found an excellent fit among these structures. We also undertook a theoretical study and proposed a model for the interaction of decavanadate with the MutS region containing the Walker A motif.
MATERIALS AND METHODS
Preparation of MutS
Recombinant mutS gene from E.coli (28) (kindly provided by P. Hsieh, NIH, USA) and P.aeruginosa (29) were overexpressed in E.coli and the protein obtained was purified by affinity chromatography (29).
ATPase activity assay
The ATPase activity was determined in 20 mM Tris–HCl buffer, pH 7.5, 3 mM CaCl2, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 100 µg/ml BSA, 200 µM ATP and 10 nCi/µl [γ-32P]ATP (300 Ci/mmol; NEM Life Sciences) in a total volume of 100 µl. The reaction was started by the addition of 700 ng of purified P.aeruginosa or E.coli MutS protein and incubated at 37°C for 15 min. The ATPase activity of MutS was assayed by measuring the free radioactive phosphate as previously described (30,31).
Preparation of vanadate solutions
Orthovanadate and decavanadate solutions were prepared by adjusting a 100 mM sodium orthovanadate aqueous solution to pH 11 or 7.4, respectively (32). We refer to these solutions as ‘orthovanadate’ or ‘decavanadate’. Pervanadate solution was prepared as previously described (33). Determination of the decavanadate content in decavanadate solutions was performed by differential decavanadate precipitation by guanidine as follows: guanidine at final concentration of 0.2 M was added to a vanadate solution and after 10 min on ice, the precipitated material was centrifuged and the pellet suspended in water. Total vanadate and decavanadate content in the supernatant and in the suspended pellet was determined by measuring the absorbance of both solutions at 265 and 400 nm (32,34). The data were corrected for the amount of the remaining decavanadate in the supernatant.
Oligonucleotide labeling and annealing
The following 39mer oligonucleotide (BioSynthesis, USA) 5′-CCTGGTACCTCGAGCCATCGAGCTTGGTGGAATTCGCCG-3′ and the mismatched complementary sequence were used to produce the mispaired G/T at the G position indicated in bold. Oligonucleotide labeling and annealing were carried out as previously described (35) with slight modifications (29). The annealed duplexes were used immediately or stored at –20°C.
Binding of MutS to double-stranded oligonucleotides and DNA mobility shift assay
The binding reaction was carried out in a total volume of 25 µl containing the following: 4 pmol of labeled oligonucleotide duplex, 3 µg of P.aeruginosa MutS, 20 mM Tris–HCl, pH 7.4, 1 mM DTT and 5 mM MgCl2. Following the incubation at 37°C for 15 min, 3 µl of a 50% sucrose solution was added and the mixture was electrophoresed in a non-denaturing 5% polyacrylamide gel (acrylamide–bisacrylamide, 30:1) in 40 mM Tris–AcH, pH 8.4, 1 mM EDTA (TAE buffer). Electrophoresis was performed at 4°C using TAE as a running buffer. Gels were run at 10 V/cm until the bromophenol blue dye migrated ∼6 cm from the top. The radioactive complex was visualized by autoradiography.
Sequence alignment, secondary structure prediction and three-dimensional fitting
The amino acid sequence alignment of P.aeruginosa MutS with the amino acid sequences of several ATPases was generated with the Multi Sequence Alignment 1.0 program (Infomax Bio Suite). Protein primary sequence and structural information were from GenBank and the Protein Data Bank (PDB) (Research Collaboratory for Structural Bioinformatics), respectively. The elements of protein secondary structure were obtained using the ‘J-predict’ program (http//jura.ebi.uk:8888) and designed as E (β sheet), H (helix) and C (β turn). The three-dimensional fits of the structures were performed with the GROMACS 3.0 (36) software package. The fitted structures were displayed with the graphic program RasMol.
Ligands docking
The molecular docking was performed with the AUTODOCK program (37). The structure of the E.coli MutS-bound ADP–Mg was taken from the PDB, while the structure for the decavanadate was from the Cambridge Structural Data Base. In all the docking runs, the protein and the ligands (vanadate and ADP–Mg) were treated as rigid bodies. For the protein, the lack of conformational flexibility is implied in the algorithm implemented in the current version (3.05) of the AUTODOCK program. The decavanadate is fairly rigid since it is a highly coordinated ion. The ADP–Mg could have conformational flexibility in principle, but a superposition of five randomly selected ADP–Mg taken from different ADP–protein complexes in the PDB clearly indicates that ADP–Mg adopts only one preferred conformation. The van der Waals and solvation parameters for vanadium were the same as for iron in the AUTODOCK database. This appears to be a good approximation, since the vanadium atoms are very deep in the decavanadate ion and the van der Waals and solvation interactions are short ranged. The formal charges were assigned for all the ligands. The docking was accomplished by a ‘genetic algorithm’ (37). The docking zone was a cube of 4 nm of size centered in the Lys620 (Walker A).
RESULTS
Inhibition of P.aeruginosa MutS ATPase activity by vanadate
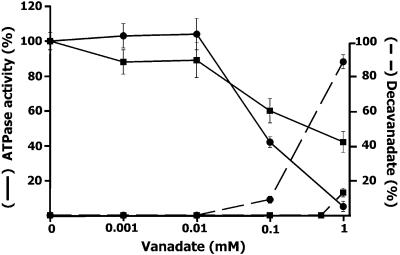
Vanadate is an oligoelement typically used as a tool to distinguish several types of ATPases because some are not inhibited by this compound (38). It is known that MutS is a member of the ABC ATPases superfamily and that several ATPases belonging to this group are inhibited by vanadate (20–27). For this reason, we tried to find out if the P.aeruginosa MutS ATPase activity was affected by vanadate. The MutS ATPase assayed at various concentrations of vanadate showed that the enzyme activity is progressively inhibited with the increase of vanadate concentration (Fig. 1). It is known that aqueous solutions of vanadium contain different molecular species of vanadate oxoanions, mainly orthovanadate and decavanadate, and that each species shows different inhibitory capability on different ATPases (20–27,39). Since the formation of these molecular species depends on the vanadium concentration, ionic strength, pH and temperature of the solution, we analyzed the correlation between the degree of inhibition of MutS ATPase and the presence of vanadate species in the solution where the activity assay was performed. Determination of the concentration of orthovanadate (VO43–), and the polymeric decavanadate (V10O286–) was carried out by differential guanidine decavanadate precipitation as described in Materials and Methods. An increase of the total vanadate concentration in the ATPase activity assay solution, either using an orthovanadate or decavanadate stock solution (see Materials and Methods) produces a shift of the species in equilibrium toward the decameric vanadate species (32). Using decavanadate stock solution, we detected the presence of the decameric polymer above 0.01 mM, reaching a maximum (92% of the total) at 1 mM vanadate (Fig. 1). As expected, when orthovanadate stock solution was used, the decameric vanadate species was detected only above 0.5 mM of total vanadate concentration and in a much lower proportion (15% of the total) than that present in decavanadate stock solution (Fig. 1).
Figure 1.
Effect of vanadate on MutS ATPase activity. ATPase activity (solid line) is expressed as percentage of radioactive 32P hydrolyzed from [γ-32P]ATP with respect to the value obtained in the absence of vanadate. The activity assay was carried out in the presence of orthovanadate (squares) or decavanadate (circles) solution as described in Materials and Methods. The concentration of decavanadate (dashed line) was determined in the ATPase assay mixture containing orthovanadate (squares) or decavanadate (circles) as described in Materials and Methods, and expressed as a percentage of total vanadate.
Using an orthovanadate stock solution, 50% inhibition was produced at total vanadate concentration of 300 µM. Because at this concentration the amount of decavanadate is negligible, we assumed that the orthovanadate species is inhibitory. On the other hand, using a decavanadate stock solution, a 50% inhibition of the ATPase activity was obtained at a total vanadate concentration of 90 µM (9 µM decavanadate) (Fig. 1). At this point the decavanadate and orthovanadate concentration is ∼10 and ∼80 µM, respectively. Moreover, at 1 mM vanadate >90% inhibition is produced at variance with ∼60% inhibition obtained with an orthovanadate solution. These results indicated that in these experimental conditions MutS ATPase is mainly inhibited by decavanadate, although orthovanadate also contributes to the inhibitory properties of vanadate.
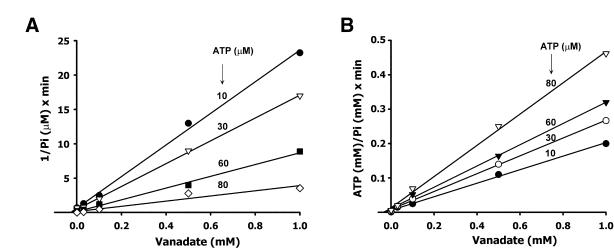
To analyze the type of inhibition produced by vanadate (using a decavanadate stock solution), we determined the vanadate inhibition on the MutS ATPase activity at different concentrations of ATP. A Lineweaver–Burk and Eadie– Hofstee analysis of vanadate inhibition was carried out and the inhibition constant calculated (Ki = 38 ± 5 µM). The inhibition of ATP hydrolysis by vanadate was consistent with a non-competitive mechanism described by the following equation:
V = (Kcat [MutS] [ATP] / Km) / (1 + [ATP] / Km + [vanadate] / Ki + [ATP] × [vanadate] / Km × Ki)
In order to verify this result, the inhibition values were plotted as 1 / v against [vanadate] (40) and [ATP] / v against [vanadate] (41) (Fig. 2A and B). In both cases, the pattern indicated a non-competitive inhibition with a Ki = 22 ± 8 µM and Ki′ = 28 ± 6 µM, respectively. Although a non-competitive inhibition appears to be operating, it is difficult to describe a precise mechanism of inhibition since more than one species of vanadate coexists in solution with an apparently different inhibitory capability (Fig. 1).
Figure 2.
Determination of the vanadate inhibition type of MutS ATPase activity. (A) The plot was constructed as 1 / v versus [vanadate] to obtain the Ki value from the intercept on the abscissa. (B) The plot was constructed as [ATP] / v versus [vanadate] to obtain the Ki′ value from the intercept on the abscissa.
It is important to point out that when the effect of vanadate was tested on MutS from E.coli, the inhibition of the ATPase activity observed (data not shown) was almost identical to that obtained for P.aeruginosa MutS.
We also determined the effect of ADP on the MutS ATPase activity and found a competitive inhibition with a KiADP = 61 ± 8 µM in agreement with previous results (42). On the other hand, orthophosphate (Pi) at concentrations as high as 5 mM did not show any effect on the ATPase activity, whereas pyrophosphate (PPi) showed a weak inhibitory effect (I50 = 2.3 mM). No effect was observed on the MutS ATPase activity with other known ATPases inhibitors (38) such as azide (0.2 mM), N-ethilmaleimide (1 mM) and oligomycin (5 µg/ml).
Sequence and structure homology of the ATP-binding domain of MutS with ABC ATPases inhibited by vanadate
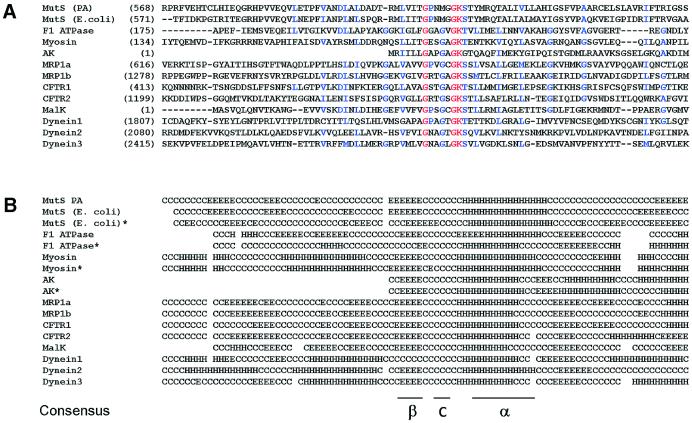
It has been demonstrated that in several ABC ATPases inhibited by vanadate the Walker A motif, which is part of the ATP-binding domain, is involved in the orthovanadate binding. For this reason we investigated the degree of similarity of the primary structure and the predicted secondary structure over 100 amino acids surrounding the Walker A motif present in each ATPase of this group (Fig. 3A and B). For this study, we have selected membrane-associated and cytoplasmic ATPases: F1-ATPase (20), myosin (21), adenylate kinase (22), MRP1 (23), CFRT (24,25), MalK (26) and flagellar dynein (27). Comparison of the predicted elements of the secondary structure of the ATPase domains of E.coli MutS, F1-ATPase, myosin and adenylate kinase with the secondary structure of these proteins obtained by X-ray diffraction indicated that the structural prediction is fairly good (Fig. 3B). Despite the very low extent of primary sequence similarity among the analyzed proteins, they are all folded into β-turn–helix motifs over approximately 25 residues including the Walker A motif. Thus, the conserved domain in the proteins analyzed might adopt a similar three-dimensional structure that probably allows vanadate to interact with MutS in a similar way to that described for other studied ATPases. To test this possibility we performed a three-dimensional fitting of the region involved in the orthovanadate binding present in the crystal structure of three proteins inhibited by vanadate with the ATP-binding domain of E.coli MutS. This analysis was performed with the crystallographic structure of the E.coli MutS since the structure of P.aeruginosa MutS has not been resolved experimentally. For every protein analyzed, we selected a region of 17 amino acids including the Walker A motif centered at the Lys residue present in this motif (GxxxxGKS/T). It was observed (Fig. 4) that the Walker A is a highly adapted anion-binding domain, since there is a confluence of three different sources of strong positive potential, namely the helix dipole, the charge of the conserved Lys residue, and the dipoles of the first three amino acids of the loop. The best fit of the structures was accomplished by minimizing the root mean square deviation (RMSD) of backbone atoms in the central loop (Fig. 4). The low RMSD value (0.05 nm on average) indicates an excellent fit between the structures and therefore a highly conserved local structure. The tridimensional structure observed in E.coli MutS could be extended to P.aeruginosa MutS because these proteins showed a 100% homology in the 17 amino acid sequence around the Walker A motif (E.coli, residues 613–629; P.aeruginosa, residues 612–628) as well as a considerable extent of secondary structure homology in a region of approximately 100 amino acids around the Walker A motif (Fig. 3).
Figure 3.
(A) Sequence alignment of the Walker A region of P.aeruginosa and E.coli MutS and several ABC ATPases inhibited by vanadate. A region of approximately 100 amino acids around the ‘Walker A’ cassette of each protein was selected and aligned using the multiple alignment algorithm (see Materials and Methods). Proteins analyzed and the corresponding GenBank accession numbers are: MutS PA, P.aeruginosa Hex1T MutS (7108493); MutS E.coli (1592569); F1 ATPase, human ATP synthase β chain (BAB22236); Myosin, human myosin heavy chain (1827758); AK, P.aeruginosa adenylate kinase (PDB 1ANK); MRP1a and MRP1b, two Walker A cassettes of the human multidrug resistance protein 1 (CAC69553); CFRT1 and CFRT2, two Walker A cassettes of the human cystic fibrosis transmembrane conductance regulator 1 (P13569); MalK, P.aeruginosa maltose transporter (146701); Dynein1, Dynein2 and Dynein3, three Walker A cassettes of the Chlamydomonas reinhardtii beta dynein heavy chain (118965). Identical residues present in all sequences are marked with red and identical or chemically similar amino acids present only in some sequences are marked with blue. Number in parentheses on the left refers to the amino acid position in each protein sequence. Since the adenylate kinase (AK) Walker A motif is very close to the N-terminus of the protein, the alignment of this protein was performed manually. (B) Secondary structure prediction from primary aligned sequences. The prediction was carried out as described in Materials and Methods. Elements of the secondary structure are indicated as follows: E, beta; H, helix; and C, coil. Secondary structures of crystallized proteins were obtained from the PDB (E.coli MutS, 1e3m; F1 ATPase, 1mab; Myosin, 1vomt; AK, 1ank) and are indicated by an asterisk.
Figure 4.
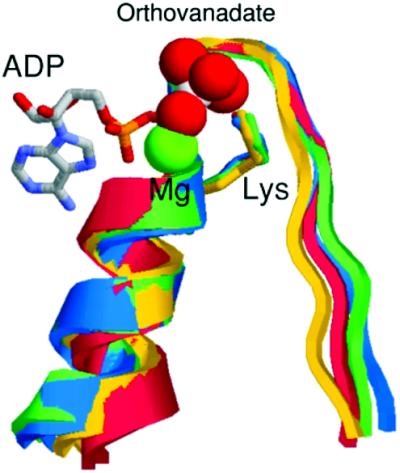
Three-dimensional fitting of 17 amino acids containing the Walker A region of crystallized ATPases inhibited by vanadate. Escherichia coli MutS, residues 613–628 (green); myosin, residues 178–194 (blue); F1-ATPase, residues 210–226 (yellow); and adenylate kinase, residues 5–21 (red). The ADP–Mg–orthovanadate corresponds to the crystallized complex structure of myosin. Structures were obtained from the PDB, accession numbers are given in Figure 3.
The high similarity between the structure of MutS and the myosin–orthovanadate complex suggests that the mechanism of orthovanadate inhibition of MutS ATPase is similar to that observed for other ABC ATPases inhibited by this compound. As in our experimental system, decavanadate is the main inhibitory species of vanadate; we docked it on the ATPase domain of MutS to gain insight into the interactions that result in the inhibitory action of this compound.
Decavanadate docking into MutS
The docking of decavanadate was performed on the crystallographic structure of the E.coli MutS because, as mentioned before, the structure of P.aeruginosa MutS has not been solved experimentally.
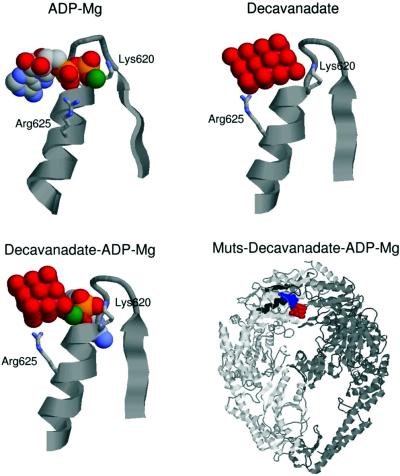
We performed three docking runs. First, and as a test of the quality of the docking that can be achieved in this system, we docked the ADP–Mg, which is already present in the crystallographic structure of E.coli MutS (15). The best-docked structure (Fig. 5) was in excellent agreement with the crystallographic structure (RMSD of only 0.04 nm with respect to the crystal complex) and a free energy of binding of –13 kcal/mol was estimated. Next, we performed the docking of the decavanadate ion. The best-predicted complex was located in the ADP-binding site as expected for the great positive electrostatic potential of this site. Due to the bigger size of the decavanadate ion, as compared to the ADP, it cannot penetrate into the center of the loop (Fig. 5), where the stronger attractions lie. This geometrical restriction makes the protein–decavanadate complex less stable with a predicted free energy of binding of –0.61 kcal/mol. It is important to point out that because the structure of E.coli MutS without ligands is not available, the docking was performed on the structure of MutS crystallized with ADP and DNA bound (15). Therefore, the free energy value obtained from the docking of decavanadate on MutS should be considered as approximate since it is known that both adenine nucleotides and DNA may produce changes in the protein structure (43,44).
Figure 5.
Resulting structures of docking ADP–Mg, decavanadate and decavanadate–ADP–Mg on the nucleotide-binding domain of E.coli MutS. Docking was carried out as described in Materials and Methods. The three-dimensional structure of the different ligands with the Walker A region complex is shown except the right-hand lower panel where a panoramic view of the MutS dimer–ADP–Mg–decavanadate complex is shown.
Finally, we docked the decavanadate ion into the protein– ADP–Mg complex. In the best-docked structure the decavanadate ion was located next to the ADP-binding site. There were two salt bridges that stabilized the ion in this position, one with the Mg2+ ion of the ADP–Mg, and the other with the Arg625 located in the helix. The estimated free energy of binding of decavanadate to the protein–ADP–Mg complex was –2.66 kcal/mol with respect to the docking of ADP–Mg. The relative free energy values estimated for each complex could indicate that once the complex decavanadate– ADP–Mg–protein is established, the ATP/ADP exchange is blocked. It is important to point out that the crystal structure of E.coli MutS (15) showed the presence of a disordered loop (amino acids 659–668) close to the interdimer region that might be involved in catalysis. Since this region is not present in the crystallographic structure, it was not possible to take it into account for the docking assays.
DNA binding to P.aeruginosa MutS in the presence of vanadate
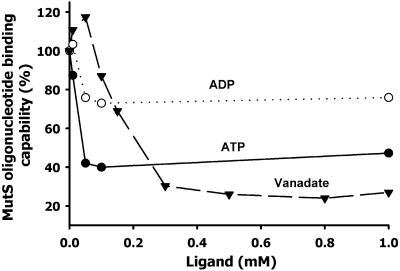
The results described in the previous section indicated that both orthovanadate and decavanadate probably interact at or close to the adenine nucleotide-binding site of MutS. It has been well established that the mismatch DNA affinity of bacterial and eukaryotic MutS homologs is reduced by the presence of adenine nucleotides, especially ATP (1). To test whether vanadate displays, similarly to nucleotides, some effect on the MutS capability to bind heteroduplex DNA, we performed 32P-39 bp T/G mismatch oligonucleotide binding experiments by mobility shift assay (see Materials and Methods) in the presence of different concentrations of vanadate, and compared it with the effects produced by ATP and ADP.
A slight activating effect of the MutS oligonucleotide binding was produced by vanadate at 10–50 µM (Fig. 6). At a concentration >80 µM, an inhibitory effect was observed with a maximum binding inhibition (71%) at 0.3 mM. In comparison, 50 µM ADP or ATP produced a 15 and 60% inhibition, respectively, with no further increase of the inhibition at higher concentrations similar to previous results (29).
Figure 6.
Binding of mismatch oligonucleotide to MutS in the presence of vanadate (decavanadate solution), ATP and ADP. 32P-39 bp mismatch (G/T) oligonucleotide binding reaction to MutS and determination of the oligonucleotide–MutS complex by gel-shift assay were performed as described in Materials and Methods. The extent of the binding is expressed as a percent of the complex formed in the presence of ligand with respect to the value obtained in the absence of ligand. Quantification of the complex was carried out by scanning the autoradiography of the gel band corresponding to the radioactive oligonucleotide–MutS complex.
Vanadate partially inhibits the oligomerization of P.aeruginosa MutS and preserves the protein from inactivation
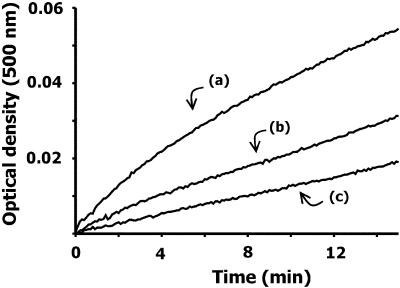
By using native gel electrophoresis, light scattering, gel filtration chromatography and electron microscopy, we previously demonstrated that bacterial MutS is able to oligomerize after a brief period of incubation at 37°C (29). The oligomerized protein showed the presence of regular structures probably conformed by four MutS dimers and other aggregates of higher molecular weight (29). Concomitant with the oligomerization process, we found that the ATPase activity and the oligonucleotide-binding capability of MutS are inactivated. However, the presence of ATP, ADP or heteroduplex DNA but not homoduplex DNA during the incubation period inhibited the protein oligomerization and preserved the protein activities (29). Assuming that the interaction of vanadate with MutS is related to the nucleotide- binding site of the protein, we analyzed the effect of vanadate on the oligomerization and inactivation of MutS. Analysis of the time course of the protein aggregation by light scattering showed that, in comparison with the protein without vanadate addition [Fig. 7, (a)], the oligomerization of MutS is partially inhibited by vanadate at 0.1 and 1 mM [Fig. 7, (b) and (c), respectively]. Next, it was analyzed whether vanadate was also capable of preserving the oligonucleotide-binding capability of MutS. For these experiments we used a vanadate concentration that does not produce or that only partially inhibits the oligonucleotide-binding capability of MutS (vanadate 0.01–0.1 mM, see Fig. 5). As was previously shown (29), pre-incubation of MutS for 15 min at 37°C produced a complete inhibition of the MutS–heteroduplex-binding capability (Fig. 8). However, the incubation of MutS in the presence of 0.01 and 0.1 mM vanadate partially preserved the binding capability of the protein (Fig. 8). In relation to the MutS ATPase activity, we previously showed that the presence of ATP or ADP also preserved the ATPase activity from temperature inactivation (29). However, the incubation of MutS at 37°C in the presence of vanadate at those concentrations that do not inhibit the ATPase activity showed no effect on the inactivation of the ATPase activity (data not shown). The failure of vanadate to preserve the ATPase activity is at variance with that observed for the oligonucleotide binding and it can be related to the fact that ATPase activity is more sensitive to temperature inactivation than the oligonucleotide-binding capability (29).
Figure 7.
Effect of vanadate on the oligomerization of MutS monitored by light scattering. The oligomerization reaction was initiated by adding a purified sample of MutS in 20 mM Tris–HCl buffer, pH 7.5, 5 mM MgCl2, 0.1 mM EDTA and 1 mM DTT to temperature pre-equilibrated cuvettes. The reaction was carried out for 15 min at 37°C and monitored by absorbance at 500 nm in a Shimadzu-UV-2401 PC (model 7CC-240A). The reaction was carried out with 1 µM MutS in the absence (a) or presence of vanadate (decavanadate solution) at 0.1 (b) and 1 mM (c).
Figure 8.
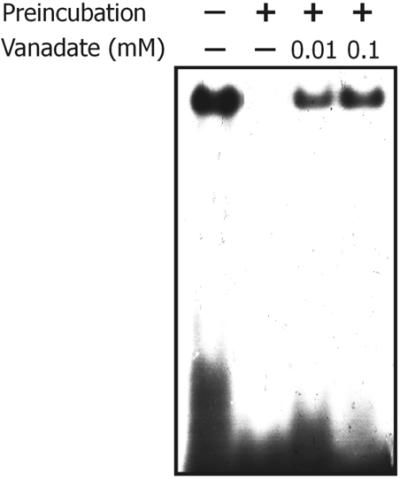
Effect of vanadate on the inactivation of the oligonucleotide- binding capability of MutS. An autoradiogram of the 32P-39 bp oligonucleotide G/T–MutS complex analyzed by native PAGE (see Materials and Methods) is shown. Pre-incubation of MutS prior to the binding reaction was performed for 15 min at 37°C in the binding buffer in the absence or presence of vanadate (decavanadate solution) at the concentrations indicated.
We concluded that, although somewhat less efficiently, vanadate behaves in a similar way to adenine nucleotides when it comes to hinder the oligomerization of MutS and to preserve the oligonucleotide binding of the protein. In addition to the foregoing results, these results also suggest that the nucleotide-binding site of MutS is probably involved in the interaction with vanadate.
Vanadate affects the conformation of P.aeruginosa MutS tested by limited proteolysis
In order to verify that vanadate interacts with MutS in the absence of nucleotides, we analyzed whether the presence of vanadate affects the partial digestion of the protein by trypsin. Figure 9 shows an SDS–PAGE analysis of the peptide pattern generated by limited trypsin digestion of MutS, which was previously incubated in the absence (lane 2) or the presence of vanadate (lanes 3–5). An increase of vanadate concentration produces a general protection in the degradation pattern of MutS with a clear difference in the relative intensity of some peptide bands. Although no striking differences were observed, a similar moderate change in the peptide pattern generated by limited proteolysis was observed in bacterial MutS in the presence of adenine nucleotides and DNA (43,44). From this experiment we concluded that vanadate interacts with MutS affecting the protein conformation.
Figure 9.
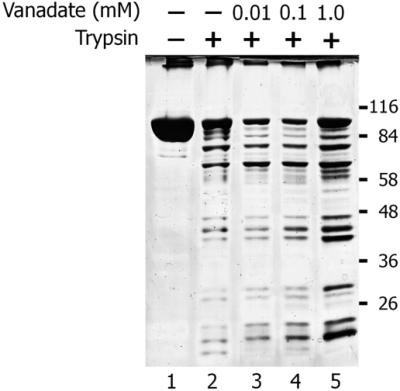
Limited proteolysis of P.aeruginosa MutS by trypsin. MutS (50 pmol) in 20 µl of 20 mM Tris–HCl buffer, pH 7.4, 1 mM dithiotreithol, 50 mM KCl and 5 mM MgCl2 was incubated in the presence or absence of different concentrations (mM) of ligands for 10 min at 37°C as indicated. After this incubation, 80 ng of trypsin (Sigma) was added and the mixture was incubated for an additional 60 s. The reaction was stopped by the addition of 6 µl of 30 mM PMSF and cooled on ice for 5 min. Samples were analyzed by 8% SDS–PAGE followed by silver staining.
DISCUSSION
MutS has been included as a constituent of the ABC ATPases superfamily, whose functions depend on the energy provided by ATP hydrolysis (11,45,46). The determination of the structure of bacterial MutS confirmed this classification (15,16). In this work, we show that P.aeruginosa and E.coli MutS ATPase, like other members of the ABC family, are inhibited by vanadate. It is known that vanadate solution contains different biologically active forms of oxoanions (32,39). Under our experimental conditions, decavanadate appears as the more effective inhibitory species, even though orthovanadate also produced an inhibitory effect. The kinetic analysis of inhibition showed that it is non-competitive, although it is difficult to determine the precise mechanism because both decavanadate and orthovanadate coexist in solution and the proportion of each compound depends on the total vanadate concentration. In several ABC ATPases the inhibition produced by orthovanadate involved a region that comprises the Walker A cassette (20–27). A non-competitive inhibition of dynein ATPase by vanadate has been described (47). In this case it was proposed that the non-competitive inhibition is compatible with a mechanism in which vanadate replaces Pi at the Pi site and involves the formation of a tertiary complex protein–ATP–vanadate with subsequent isomerization to protein–ADP–vanadate with loss of Pi. However, it has also been described that orthovanadate photo-cleavage also occurs in the absence of nucleotides (27). Considering that ABC ATPases inhibited by orthovanadate are phosphorylated, the low inhibitory effect of this compound on MutS ATPase could be attributed to the absence of a stable phosphorylation state of MutS. Very little is known about the mechanisms of inhibition of ATPases by decavanadate. It has been reported that the K channel interacts specifically with decavanadate and not with orthovanadate (48). Photo-oxidative cleavage experiments of orthovanadate and decavanadate bound to sarcoplasmic reticulum ATPase showed that whereas orthovanadate is bound to the phosphorylation site, decavanadate interacts with the nucleotide site (27,49).
To further understand the interaction of vanadate with MutS, we compared the MutS amino acid sequence region involved in the binding of adenine nucleotides with other ABC ATPases inhibited by vanadate. To extend the range of homology analysis, we also predicted the secondary structure of these proteins. The results obtained showed that, although the amino acid sequence homology was only limited to the Walker A cassette (GxxxxGKS/T), a region of approximately 30 amino acids adjacent to this cassette showed a fairly high secondary structure similarity. In addition, the comparison of the tridimensional structure of this region from crystallized MutS, adenylate kinase, F1-ATPase and myosin also showed an important structural similarity. Since the crystal structure of myosin–ADP–orthovanadate is highly similar to E.coli MutS, it leads us to postulate that the inhibition of the P.aeruginosa or E.coli MutS by orthovanadate is probably operating through a similar mechanism. In an attempt to obtain information about the action of decavanadate, we docked this compound on the ATPase domain of MutS. The results obtained indicated a higher affinity of the inhibitor for the enzyme–substrate complex when compared with the interaction with pure MutS, which suggests that the mechanism of inhibition could involve the stabilization of the complex containing the nucleotide. Unlike orthovanadate, decavanadate interacts outside the Walker A loop and is stabilized by the interaction with the Mg2+ ion, and the Arg625. However, this Arg residue is not conserved among different ABC ATPases except for MutS, myosin, MalK and dynein. Therefore, if the presence of this residue were essential for the binding stabilization, only some members of the ABC superfamily would be able to interact with decavanadate.
We found that vanadate not only alters the ATPase activity but also the DNA-binding capability of the protein. At concentrations >0.1 mM, vanadate produces a clear inhibition of the oligonucleotide binding, higher than that produced by ATP or ADP. The fact that these experiments were carried out in the absence of nucleotides leads us to think that vanadate is also able to interact with MutS in a nucleotide-independent way. The peptide pattern generated by trypsin digestion of P.aeruginosa MutS in the presence of vanadate confirmed that this compound interacts with the protein in the absence of nucleotide.
We recently found (29) that incubation of MutS at 37°C produces a rapid inactivation of the ATPase and DNA-binding capability with the concomitant oligomerization of the protein. Both the inactivation and the aggregation processes can be prevented by the presence of low concentrations of adenine nucleotides or mismatched oligonucleotide (29). In the present work, we show that similar effects are obtained by addition of low vanadate concentrations. Altogether, these results can be explained assuming that the interaction of vanadate in the MutS nucleotide-binding domain (orthovanadate) or close to this site (decavanadate) partially mimics the effect of nucleotides. Further experiments using MutS mutants that can bind DNA but are defective in ATP binding or hydrolysis would confirm this assumption.
The ABC ATPase superfamily is composed of members involved in different cellular functions (45). Recently, a common mechanism for ATP hydrolysis in the ABC transporter and helicase superfamily has been proposed (46). The effect of vanadate described in the present work could also be extended to other proteins related to DNA recombination and repair such as RecN, RecF, UvrA, UvrB and Rad50. All these proteins contain the Walker A domain and the predicted secondary structures are also arranged in β–c–α conformation (data not shown). It is then possible to speculate that their physiological activities could also be modified by interaction with vanadate. The vanadate interactions with proteins containing the Walker A cassette can also become a new element to functionally classify members of this superfamily.
Finally, considering that vanadium is distributed extensively in nature and that it is present in almost all living organisms including human beings (50), this trace element could be considered as a potential factor capable of regulating the MMRS through the interaction with MutS.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Andrea M. Smania and José L. Barra for helpful discussions and critical reading of the manuscript. This work was supported in part by grants from the SECyT (UNC), Agencia Córdoba Ciencia and ‘R. Carrilo-A. Oñativia’ fellowship, Ministerio de Salud de la Nación.
REFERENCES
- 1.Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev., 10, 1433–1442. [DOI] [PubMed] [Google Scholar]
- 3.LeClerc J.E., Baouguang,L., Payne,W.L. and Cebula,T.A. (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science, 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- 4.Oliver A., Canton,R., Campo,P., Baquero,F. and Blazquez,J. (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288, 1251–1254. [DOI] [PubMed] [Google Scholar]
- 5.Umar A. and Kunkel,T.A. (1996) DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur. J. Biochem., 238, 297–307. [DOI] [PubMed] [Google Scholar]
- 6.Wu T.H. and Marinus,M.G. (1994) Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol., 176, 5393–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alani E., Sokolsky,T., Studamire,B., Miret,J.J. and Lahue,R.S. (1997) Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol., 17, 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell L.J., Martik,D., Bjornson,K.P., Bjornson,E.S. and Modrich,P. (1998) Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem., 273, 32055–32062. [DOI] [PubMed] [Google Scholar]
- 9.Gradia S., Acharya,S. and Fishel,R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell, 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- 10.Gradia S., Subramanian,D., Wilson,T., Acharya,S., Makhov,A. and Griffith,J. (1999) hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell, 3, 255–261. [DOI] [PubMed] [Google Scholar]
- 11.Junop M.S., Obmolova,G., Rausch,K., Hsieh,P. and Yang,W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- 12.Sixma T.K. (2001) DNA mismatch repair: MutS structures bound to mismatches. Curr. Opin. Struct. Biol., 11, 47–52. [DOI] [PubMed] [Google Scholar]
- 13.Joshi A. and Rao,B.J. (2002) ATP hydrolysis induces expansion of MutS contacts on heteroduplex: a case for MutS treadmilling? Biochemistry, 41, 3654–3666. [DOI] [PubMed] [Google Scholar]
- 14.Blackwell L.J., Bjornson,K.P., Allen,D.J. and Modrich,P. (2001) Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem., 276, 34339–34347. [DOI] [PubMed] [Google Scholar]
- 15.Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H.K., Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- 16.Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 403, 703–710. [DOI] [PubMed] [Google Scholar]
- 17.Biswas I., Obmolova,G., Takahashi,M., Herr,A., Newman,M.A., Yang,W. and Hsieh,P. (2001) Disruption of the helix–u-turn–helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol., 26, 805–816. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya A.E. and Koonin,E.V. (1990) Superfamily of UvrA-related NTP-binding proteins implication for rational classification of recombination/repair systems. J. Mol. Biol., 213, 583–591. [DOI] [PubMed] [Google Scholar]
- 19.Holland I.B. and Blight,M.A. (1999) ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol., 293, 381–399. [DOI] [PubMed] [Google Scholar]
- 20.Ko Y.H., Bianchet,M., Amzel,L.M. and Pedersen,P.L. (1997) Novel insights into the chemical mechanism of ATP synthase. Evidence that in the transition state the gamma-phosphate of ATP is near the conserved alanine within the P-loop of the beta-subunit. J. Biol. Chem., 272, 18875–18881. [DOI] [PubMed] [Google Scholar]
- 21.Smith C.A. and Rayment,I. (1996) X-ray structure of the magnesium ADP.Vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry, 35, 5404–5417. [DOI] [PubMed] [Google Scholar]
- 22.Pai E.F., Sachsenheimer,W., Schirmer,R.H. and Schulz,G.E. (1997) Substrate position and induced-fit in crystalline adenylate kinase. J. Mol. Biol., 114, 37–45. [DOI] [PubMed] [Google Scholar]
- 23.Nagata K., Nishitani,M., Matsuo,M., Kioka,N., Amachi,T. and Ueda,K. (2000) Nonequivalent nucleotide trapping in the two nucleotide binding folds of the human multidrug resistance protein MRP1. J. Biol. Chem., 275, 17626–17630. [DOI] [PubMed] [Google Scholar]
- 24.Urbatsch I.L., Sankaran,B., Bhagat,S. and Senior,A.E. (1995) Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem., 270, 26956–26961. [DOI] [PubMed] [Google Scholar]
- 25.Szabo K., Szakacs,G., Hegeds,T. and Sarkadi,B. (1999) Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator. Different patterns in the two nucleotide binding domain. J. Biol. Chem., 274, 12209–12212. [DOI] [PubMed] [Google Scholar]
- 26.Sharma S. and Davidson,A.L. (2000) Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J. Bacteriol., 182, 6570–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W.Y. and Gibbons,I.R. (1987) Photosensitized cleavage of dynein heavy chain. Cleavage at the V2 site by irradiation at 365 nm in the presence of oligovanadate. J. Biol. Chem., 262, 17728–17734. [PubMed] [Google Scholar]
- 28.Feng G. and Winkler,M.E. (1995) Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of Escherichia coli K-12. Biotechniques, 19, 956–965. [PubMed] [Google Scholar]
- 29.Pezza R.J., Smania,A.M., Barra,J.L. and Argaraña,C.E. (2002) Nucleotides and heteroduplex DNA preserve the active conformation of Pseudomonas aeruginosa MutS by preventing protein oligomerization. Biochem. J., 361, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glynn I.M. and Clappell,J.B. (1964) A simple method for the preparation of 32P-labelled adenosine triphosphate of high specific activity. Biochem. J., 90, 147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Meis L. (1972) Phosphorylation of the membranous protein of the sarcoplasmic reticulum. Inhibition by Na+ and K+. Biochemistry, 11, 2460–2465. [DOI] [PubMed] [Google Scholar]
- 32.Csermely P., Martonosi,A., Levy,G.C. and Ejchart,A.J. (1985) 51V-n.m.r. analysis of the binding of vanadium (V) oligoanions to sarcoplasmic reticulum. Biochem. J., 230, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imbert V., Peyron,J.F., Farahi Far,D., Mari,B., Auberger,P. and Rossi,B. (1994) Induction of tyrosine phosphorylation and T-cell activation by vanadate peroxide, an inhibitor of protein tyrosine phosphatases. Biochem. J., 297, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varga S., Csermely,P. and Martinosi,A. (1985) The binding of vanadium (V) oligoanions to sarcoplasmic reticulum. Eur. J. Biochem., 148, 119–126. [DOI] [PubMed] [Google Scholar]
- 35.Jiricny J., Su,S.S., Wood,S.G. and Modrich,P. (1988) Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res., 16, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erik L., Berk,H. and van der Spoel,D. (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model., 7, 306–317. [Google Scholar]
- 37.Morris G.M., Goodsell,D.S., Halliday,R.S., Huey,R., Hart,W.E., Belew,R.K. and Olson,A.J. (1998) Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem., 19, 1639–1662. [Google Scholar]
- 38.Pedersen P.L. and Carafoli,E. (1987) Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem. Sci., 12, 146–150. [Google Scholar]
- 39.Baran E.J. (2000) Oxovanadium (IV) oxovanadium (V) complexes relevant to biological systems. J. Inorg. Biochem., 80, 1–10. [DOI] [PubMed] [Google Scholar]
- 40.Dixon M. (1953) The determination of enzyme inhibitor constants. Biochem. J., 55, 170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornish-Bowden A. (1974) A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J., 137, 143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjornson K.P., Allen,D.J. and Modrich,P. (2000) Modulation of MutS ATP hydrolysis by DNA cofactors. Biochemistry, 39, 3176–3183. [DOI] [PubMed] [Google Scholar]
- 43.Joshi A., Sen,S. and Rao,B.J. (2000) ATP-hydrolysis-dependent conformational switch modulates the stability of MutS-mismatch complexes. Nucleic Acids Res., 28, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biswas I. and Vijayvargia,R. (2000) Heteroduplex DNA and ATP induced conformational changes of a MutS mismatch repair protein from Thermus aquaticus. Biochem. J., 347, 881–886. [PMC free article] [PubMed] [Google Scholar]
- 45.Koonin E.V. (1993) A superfamily of ATPases with diverse function containing either classical or deviant ATP-binding motif. J. Mol. Biol., 229, 1165–1174. [DOI] [PubMed] [Google Scholar]
- 46.Geourjon C., Orelle,C., Steinfels,E., Blanchet,C., Deleage,G., Di Pietro,A. and Jault,J.M. (2001) A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem. Sci., 26, 539–544. [DOI] [PubMed] [Google Scholar]
- 47.Anderson S.A. and Purich,D.L. (1982) A reinvestigation of dynein ATPase kinetics and the inhibitory action of vanadate. J. Biol. Chem., 257, 6656–6658. [PubMed] [Google Scholar]
- 48.Proks P., Ashfield,R. and Ashcroft,F.M. (1999) Interaction of vanadate with the cloned beta cell K(ATP) channel. J. Biol. Chem., 274, 25393–25397. [DOI] [PubMed] [Google Scholar]
- 49.Hua S., Inesi,G. and Toyoshima,C. (2000) Distinct topologies of mono- and decavanadate binding and photo-oxidative cleavage in the sarcoplasmic reticulum ATPase. J. Biol. Chem., 275, 30546–30550. [DOI] [PubMed] [Google Scholar]
- 50.Jandhyala B.S. and Hom,G.J. (1983) Minireview: physiological and pharmacological properties of vanadium. Life Sci., 33, 1325–1340. [DOI] [PubMed] [Google Scholar]