Abstract
A T→G mutation at nucleotide 705 of human β-globin intron 2 creates an aberrant 5′ splice site and activates a cryptic 3′ splice site upstream. In consequence, the pre-mRNA is spliced via aberrant splice sites, despite the presence of the still functional correct sites. Surprisingly, when IVS2-705 HeLa or K562 cells were cultured at temperatures below 30°C, aberrant splicing was inhibited and correct splicing was restored. Similar temperature effects were seen for another β-globin pre-mRNA, IVS2-745, and in a construct in which a β-globin intron was inserted into a coding sequence of EGFP. Temperature-induced alternative splicing was affected by the nature of the internal aberrant splice sites flanking the correct sites and by exonic sequences. The results indicate that in the context of thalassemic splicing mutations and possibly in other alternatively spliced pre-mRNAs, temperature is one of the parameters that affect splice site selection.
INTRODUCTION
The mechanisms involved in alternative splicing of pre-mRNAs are not completely understood, but a large body of work suggests that selection of splice sites is determined by the balance of many splicing factors interacting with splice sites and/or other related sequence elements involved in splicing (1–3). This balance is affected by, for example, the presence or absence of gene-specific splicing factors, especially well defined in Drosophila (4), or by differences in relative concentration of antagonistic constitutive splicing factors (5,6). Both gene-specific and constitutive splicing factors that play a role in alternative splicing usually belong to the SR family of polypeptides (6). Evidence shows that SR proteins as well as other splicing factors are extensively phosphorylated (7). Thus phosphorylation and dephosphorylation of splicing factors may add an additional regulatory layer to alternative splicing (8–11). There is also evidence that cyclin E–cdk2 interacts with the splicing machinery providing a link between splicing, and possibly alternative splicing, and the cell cycle (12).
Elements of pre-mRNA that affect splice site selection include exon splicing enhancers (13), secondary structures (14) and the relative match of the 3′ and 5′ splice sites and branch points to their respective consensus sequences (1–3). The latter is well illustrated by the fact that mutations within natural splice sites lead to activation of cryptic sites or to skipping of exons bordered by the affected splice sites, a phenomenon frequently seen in splicing mutations causing genetic diseases (15). Experimental mutations that increase the match of a splice site or a branch point to their respective consensus sequence shift the splicing pattern and lead to inclusion of an otherwise ignored exon (16,17). It seems likely that these mutations either decrease or increase the affinity of the affected sequences for the appropriate splicing factors. The multiple mechanisms involved in splice site selection suggest that a dynamic network of interacting components determines the structure and function of spliceosomes and that spliceosomes assembled on different splice sites may not be identical. One can then hypothesize that changes in the physiology of the cell might variably affect splicing of different messages, especially those that are alternatively spliced.
We have tested this hypothesis using thalassemic mutations IVS2-654, IVS2-705 and IVS2-745 in intron 2 of the human β-globin gene (18) as a clinically relevant model for studies of the mechanisms involved in pre-mRNA splicing and splice site selection. The three mutations create aberrant 5′ splice sites at nucleotides 652, 706 and 745 of intron 2, respectively, and activate a common cryptic 3′ splice site at nucleotide 579. Splicing of mutated β-globin pre-mRNAs is shifted either completely or partially to aberrant pathways, even though the correct splice sites remain potentially functional. Portions of the intron sequences between the newly activated splice sites are recognized by the splicing machinery as exons and are retained in the spliced mRNAs. The retained exon-like sequences of either 73, 126 or 165 nt generate a stop codon that prevents proper translation of the mRNA (Fig. 1) and causes a deficiency in β-globin leading to β-thalassemia (18–22). The aberrant splicing patterns of mutated β-globin pre-mRNAs can be seen not only in patients’ erythroid cells (18) but also in 3T3, HeLa and K562 cell lines that stably express the thalassemic β-globin genes (19,23,24).
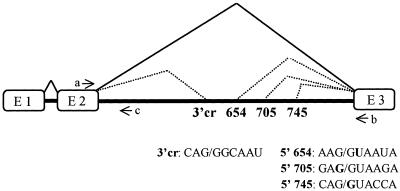
Figure 1.
Splicing of human β-globin IVS-2 mutant pre-mRNAs. Boxes, exons; solid heavy lines, introns; thin solid and dotted lines above intron 2, correct and aberrant splicing pathways, respectively; arrows, primers used in RT–PCR detecting spliced (a and b) and unspliced (a and c) β-globin RNAs. The aberrant 5′ splice sites (654, 705 and 745) and the 3′ cryptic splice sites (3′cr) are indicated; their sequences are shown below the scheme (mutated nucleotides in bold).
It has been shown that abnormal splicing of thalassemic β-globin pre-mRNA can be corrected by the use of antisense oligonucleotides and RNAs directed against the aberrant splice sites. Antisense oligonucleotides restored correct splicing in cell-free extracts from HeLa cells (25), in cultured mammalian cells expressing the mutated β-globin genes (19,23,24), in ex vivo treated bone marrow cells from thalassemic mice (26) and in erythroid cells from thalassemic patients (27). Modification of splicing by antisense oligonucleotides was also accomplished by targeting mutant human CFTR (28) and murine dystrophin (29) pre-mRNAs and the alternatively spliced human Bcl-x pre-mRNA (30,31). Here we report that the shift in splicing of mutated pre-mRNA from aberrant to correct is affected not only by antisense agents but also by a decrease in the temperature of cell culture from 37 to 21°C.
MATERIALS AND METHODS
Cells
Cell lines were obtained by stable transfection of HeLa or K562 erythroblastoid cells with the IVS2-654 or IVS2-705 thalassemic β-globin genes cloned under the cytomegalovirus promoter as previously described (19,23). The IVS2-705 K562 cell line was a gift from Dr L. Gorman. HeLa cells expressing the enhanced green fluorescent protein (EGFP) coding sequence interrupted by the thalassemic IVS2-654 or IVS2-705con β-globin intron 2 were generated as described in Sazani et al. (32).
HeLa cell lines were grown in S-MEM supplemented with 5% fetal calf and 5% horse sera, 50 µg/ml gentamicin, 200 µg/ml kanamycin and 15 mM HEPES buffer (pH 7.3). IVS2-705 K562 cells were grown in Dulbecco’s modified Eagle’s medium, 10% filtered Colorado calf serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 15 mM HEPES buffer (pH 7.3). Cells were maintained at 37°C and 5% CO2.
Temperature treatment
For all experiments, cells were plated in 25 ml flasks at 2.5 × 105 cells/2.5 ml medium per flask and cultured under standard conditions (i.e. with loose caps at 37°C and 5% CO2). After 24 h the flasks were tightly closed and placed at various temperatures (16, 21, 25, 30 and 37°C). Incubations at these temperatures ranged from 16 to 36 h. For recovery after the temperature treatment, the flasks were returned to the incubator at 37°C with 5% CO2 with loosened caps for periods ranging from 16 to 24 h.
RNA analysis
RNA was isolated from cells with TRI-Reagent (MRC, Cincinnati, OH). HeLa cells were lysed in flasks, whereas K562 cells were collected by centrifugation prior to TRI-Reagent treatment. Approximately 200 ng of RNA was analyzed by RT–PCR using rTth DNA polymerase as suggested by the manufacturer (Perkin-Elmer). The RT–PCR was carried out with [α-32P]dATP for 18 cycles and the PCR products were separated on 7.5% non-denaturing polyacrylamide gels. Under these conditions the amount of the PCR product was proportional to the amount of input RNA, as were the relative amounts of PCR products generated from aberrantly and correctly spliced RNA (23,27,33). For analysis of aberrantly and correctly spliced products, forward and reverse primers spanning, respectively, positions 21–43 of exon 2 (primer a) and positions 6–28 of exon 3 (primer b) of the human β-globin gene were used. For RT–PCR analysis of the expression of β-globin pre-mRNAs in HeLa and K562 cells, primer a was used in combination with primer c, spanning positions 119–143 of β-globin intron 2 (Fig. 1) (23).
Protein analysis
The preparation of total protein and immunoblot analysis with polyclonal affinity-purified chicken anti-human hemoglobin antibody was described previously (19,23). The blots were developed with the ECL detection system (Amersham). All autoradiograms were captured by a DAGE-MTI (Michigan City, IN) CCD72 video camera and computer prints of final figures were generated. For EGFP detection, fluorescent images of HeLa cells were taken with an Olympus inverted fluorescence microscope as described in Sazani et al. (32).
RESULTS
Temperature-dependent shift in splicing of IVS2-705 pre-mRNA
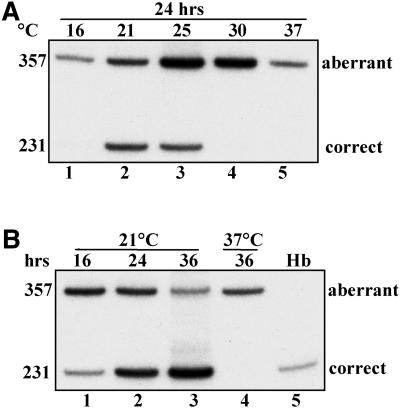
The IVS2-705 HeLa cell line stably expresses the mutated β-globin gene in which a T→G mutation at nucleotide 705 of intron 2 activated an aberrant 5′ splice site in the β-globin pre-mRNA (GAU/GUAAGA→GAG/GUAAGA). Due to the IVS2-705 mutation and activation of the 3′ splice site upstream, the cells produce an aberrantly spliced β-globin mRNA that retains a 126 nt portion of the intron (Fig. 1). As shown previously (19,23), under normal growth conditions (37°C) the only detectable β-globin mRNA was aberrantly spliced; RT–PCR with primers flanking intron 2 (primers a and b, Fig. 1) resulted in only one product (357 nt; Fig. 2A, lane 5). However, in cells grown at 21 and 25°C, an additional correctly spliced β-globin mRNA was generated (231 nt; Fig. 2A, lanes 2 and 3, respectively). The amounts of correct β-globin mRNA were negligible both at lower (16°C) and higher (30°C) temperatures (Fig. 2A, lanes 1 and 4, respectively). The shift in splicing from aberrant to correct was time dependent; at 21°C the amount of correctly spliced β-globin mRNA increased progressively between 16 and 36 h of incubation (Fig. 2B, lanes 1–3). Within this period the cell count remained constant (data not shown), indicating that the cells slowed, if not stopped, their growth and progression through the cell cycle.
Figure 2.
Temperature-dependent alteration of splicing of β-globin pre-mRNA in IVS2-705 HeLa cells. Analysis of total RNA by RT–PCR with primers a and b shown in Figure 1. (A) Temperature effects. Lanes 1–4, cells incubated for 24 h at increasing temperatures (indicated at the top); lane 5, control cells incubated at 37°C. The sizes (in nucleotides) of PCR bands representing aberrantly and correctly spliced mRNAs are indicated on the left. (B) Time course. Lanes 1–3, cells incubated at 21°C for the times indicated at the top; lane 4, control cells incubated at 37°C; lane 5, RNA from human blood (Hb). Unless otherwise indicated, similar designations were used in subsequent figures.
Figure 3 indicates that reduction of the cellular growth rate at 21°C did not prevent translation of newly generated correct β-globin mRNA. Immunoblot analysis with an anti-human hemoglobin antibody showed that the level of β-globin protein present in the cells increased over background (Fig. 3, lane 1) during prolonged incubation at low temperature, consistent with the shift in splicing from aberrant to correct (Fig. 3, lanes 2–4). This result shows that the low temperature is not lethal to the cells and that the translational machinery of the cell is functioning apparently unaffected by the low temperature treatment (see also below). Importantly, increased translation of β-globin peptide confirms that the appearance of correct β-globin mRNA is a consequence of its de novo synthesis and therefore must be due to a shift in splicing. Note also that the cell quiescence and the shift in the splicing pattern were reversed when the cells were returned from 21 to 37°C (data not shown). These results indicated that after incubation at 21°C the IVS2-705 HeLa cells remained viable and that the splicing machinery shifted to its pattern of recognition of aberrant slice sites.
Figure 3.
Restoration of β-globin protein translation in IVS2-705 HeLa cells. Immunoblot of total protein with anti-human hemoglobin antibody (see Materials and Methods). Lane 1, control cells incubated at 37°C; lanes 2–4, cells incubated at 21°C for 16, 24 and 36 h, respectively; lane 5, human β-globin polypeptide.
Expression of IVS2-705 pre-mRNA at low temperature
It was not clear why the increased production of correct β-globin mRNA at 30, 25 and 21°C was not maintained at 16°C. To address this question we re-assayed the RNAs obtained in the experiments described in the previous section using a pair of RT–PCR primers that detected only the β-globin pre-mRNA (primers a and c; Fig. 1). Figure 4A shows that significant levels of IVS2-705 pre-mRNA accumulated in the cells grown at 16, 21 and 25°C (lanes 1–3), but not at 30 and 37°C (lanes 4 and 5). Furthermore, incubation of the cells at 21°C for 16–36 h (Fig. 4B, lanes 1–3) generated increasing amounts of IVS2-705 pre-mRNA within this period, whereas in the control culture at 37°C, pre-mRNA was barely detectable (Fig. 4B, lane 4). Although it cannot be formally excluded that the decrease in temperature from 37 to 21°C activated the synthesis of pre-mRNA, thereby increasing its intracellular concentration, a more likely explanation is that at lower temperatures the rate of processing of the newly transcribed pre-mRNA was slowed down more than the rate of transcription. It appears then that at 25 and 21°C splicing was slowed down and, in addition, shifted from aberrant to correct, while at 16°C it was completely arrested, resulting in lack of correct β-globin mRNA.
Figure 4.
Temperature-dependent alteration of expression of β-globin pre-mRNA in IVS2-705 HeLa cells. (A) Temperature effects. Lanes 1–5, cells incubated for 24 h at temperatures indicated at the top. (B) Time course. Lanes 1–3, cells incubated at 21°C for the times indicated; lane 4, control cells maintained at 37°C; lane 5, size marker; DNA from a plasmid carrying the human β-globin gene subjected to PCR. RT–PCR and PCR were carried out with primers a and c (Fig. 1).
Splicing of IVS2-654, IVS2-745 and IVS2-705con pre-mRNAs at low temperature
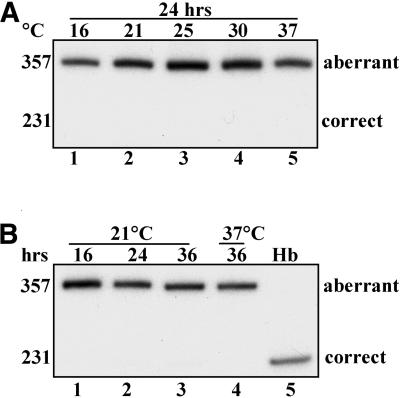
Temperature-dependant splicing was tested in HeLa cells that expressed the IVS2-654 thalassemic mutant of the β-globin gene. In IVS2-654 pre-mRNA the aberrant splice site is created by a C→U mutation at nucleotide 654 of β-globin intron 2 (AAG/GCAAUA→AAG/GUAAUA). Aberrantly spliced IVS2-654 mRNA retains a 73 nt fragment of the intron. This mutation was previously characterized as less susceptible to correction of splicing by an antisense oligonucleotide than IVS2-705 (23) and therefore it seemed possible that it might be less susceptible to changes in temperature. Indeed, treatment of IVS2-654 HeLa cells for 24 h at temperatures ranging from 16 to 37°C did not result in a shift from aberrant to correct splicing at any temperature (Fig. 5A).
Figure 5.
Aberrant splicing of IVS2-654 pre-mRNA is resistant to temperature change. (A) IVS2-654 HeLa cells were incubated for 24 h at the temperatures indicated at the top (lanes 1–5). Lane 6, RNA from human blood. (B) IVS2-745 HeLa cells were incubated for the indicated times at 21°C (lanes 1–4). Lane 5, RNA from human blood.
The IVS2-745 mutation leads to aberrantly spliced β-globin mRNA that retains a 165 nt long intron fragment. The IVS2-745 5′ splice site (CAG/GUACCA) is more distant from the consensus than its 654 and 705 counterparts, resulting in partially correct splicing in the HeLa cell line (see Fig. 5B, lane 1) that was very susceptible to antisense oligonucleotide treatment (23). Accordingly, treatment of IVS2-745 HeLa cells at 21°C led to disappearance of the aberrant mRNA after only 1 h treatment (Fig. 5B, lane 2). The transcribed pre-mRNA continued to be spliced correctly in a time-dependent manner, as evidenced by the increase in the intensity of the correctly spliced RT–PCR product after 5 and 24 h at room temperature (Fig. 5B, lanes 3 and 4).
Since the IVS2-654, IVS2-705 and IVS2-745 RNAs differ not only in the sequences of the aberrant 5′ splice sites (AAG/GUAAUA, GAG/GUAAGA and CAG/GUACCA, respectively) but also in the length (73, 126 and 165 nt) and hence the sequence of the retained intron fragment, we have tested another cell line expressing IVS-705con β-globin pre-mRNA. In the IVS2-705con construct, a GAG/GUAAGA→GAG/GUAAGU mutation increased the match of the aberrant 5′ splice site to the consensus splice site sequence. Note that this mutation does not alter the position of the aberrant 5′ splice site and therefore the product of aberrant splicing of IVS2-705con pre-mRNA also retains 126 nt of the intron. Thus, splicing of IVS2-705 and IVS2-705con results in identical aberrant and correct β-globin mRNAs.
In contrast to IVS2-705 cells, correct splicing of β-globin mRNA was not induced in IVS2-705con cells either by 24 h incubation at low temperatures (Fig. 6A, lanes 1–4) or by prolonged incubation at 21°C (Fig. 6B, lanes 1–3). Note also that the four ∼1600 nt long pre-mRNAs are identical except for single point mutations at the aberrant 5′ splice sites, making it unlikely that the pre-mRNAs, the actual substrates for the splicing reaction, differed significantly in their secondary and/or tertiary structures. Thus it appears that temperature-dependent changes in the ratio of aberrant to correct β-globin mRNAs are due to alteration of the splicing process itself and not to temperature effects unrelated to splicing, such as variations in stability of different mRNAs.
Figure 6.
Temperature-dependent alteration of splicing of β-globin pre-mRNA in IVS2-705con HeLa cells. All designations in (A) and (B) are as in the legend to Figure 2.
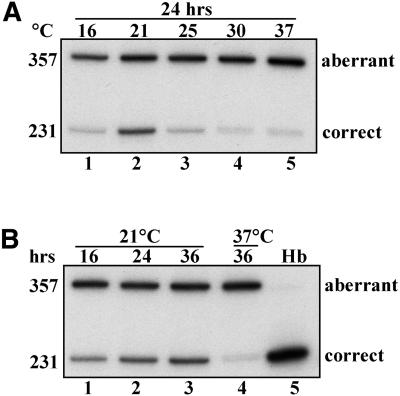
Temperature-dependent shift in splicing of IVS2-705 pre-mRNA in K562 cells
To ascertain that the observed change in splicing of β-globin RNA is not a peculiarity of HeLa cells, we have subjected a different cell line, K562 IVS2-705, to analogous temperature treatments. K562 cells are of particular interest since they are of human erythroid origin and can be induced to express the α- and γ-globin genes (34). It is conceivable that these cells may have developed a specialized globin-related splicing machinery that differs from that of HeLa cells. In fact, comparison of splicing patterns in IVS2-705 HeLa and K562 cells (Fig. 2A, lane 5 versus Fig. 7A, lane 5) shows a detectable level of correct β-globin mRNA in the latter but not the former cell line. However, in spite of this difference, IVS2-705 K562 cells exhibited a temperature- and time-dependent shift in β-globin splicing (Fig. 7A and B) similar to that seen in HeLa cells. Thus, the temperature sensitivity of the IVS2-705 splicing mutation exists in at least two different cellular backgrounds.
Figure 7.
Temperature-dependent alteration of splicing of β-globin pre-mRNA in IVS2-705 K562 cells. All designations in (A) and (B) are as indicated in the legend to Figure 2.
Temperature-dependent shift in splicing of the IVS2-705con EGFP analog
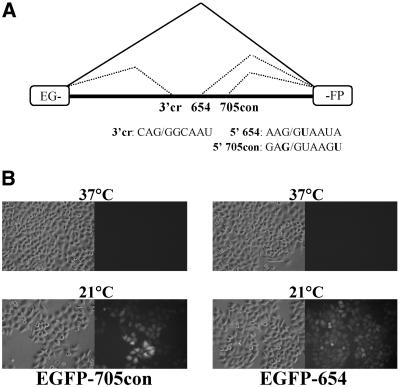
A recently developed cellular assay for shifting splicing is based on constructs in which the thalassemic β-globin introns (IVS2-654 and IVS2-705) were inserted into the coding sequence for EGFP (Fig. 8A) (32). Similarly to mutated β-globin pre-mRNA, these mutations prevent proper splicing and translation of EGFP, and dose-dependent increases in EGFP production in response to treatment with antisense oligonucleotides that block the aberrant splicing pathway were seen (32). Insertion of IVS2-654 into the EGFP coding sequence yielded a construct that exhibited no correct splicing, as was expected from studies with IVS2-654 in β-globin. However, splicing of IVS2-705 yielded a significant amount of background correct splicing (S.H.Kang and R.Kole, unpublished results). The latter result is most likely due to that fact that the correct 5′ splice site at the end of the inserted intron is closer to the consensus in the EGFP construct than in β-globin (GAG/GTGAGT and AGG/GTACAG, respectively) (35). When the spliced in IVS2-705 insert was strengthened to the consensus (IVS2-705con, GAG/GTAAGT) the resulting construct showed no background EGFP expression (Fig. 8B) and was sensitive to correction of splicing by antisense oligonucleotides (P.Sazani and R.Kole, unpublished results).
Figure 8.
Temperature-dependent alteration of splicing of EGFP-654 and 705con in HeLa cells. (A) Splicing of EGFP IVS-2 mutant pre-mRNAs. Designations are as in Figure 1. (B) Bright field (left) and fluorescence images (right) of cells treated at the indicated temperature.
Figure 8B shows the results of incubation of HeLa EGFP-705con cells at 21°C for 24 h followed by subsequent analysis by fluorescence microscopy. While untreated cells did not exhibit any detectable fluorescence, the majority of cells subjected to low temperature were strongly fluorescent, indicating significant production of EGFP. Interestingly, HeLa EGFP-654 cells exposed to decreased temperature also displayed correction of splicing, reflected in production of EGFP protein. Clearly, temperature-dependent shifts in splicing take place not only in β-globin pre-mRNA but also when the β-globin intron is flanked by foreign, non-globin sequences. Moreover, the susceptibility of both EGFP-705con and EGFP-654 to lower temperature indicates that the outcome is influenced by the flanking splice sites and possibly sequences of the flanking exons. As in the β-globin experiments, de novo synthesis of EGFP protein indicates that the temperature effects were due to newly spliced EGFP mRNA and not because of a change in stability of pre-existing transcripts.
DISCUSSION
It is clear that there is no single determining sequence element responsible for splice site selection. Rather, splicing of a pre-mRNA is the end result of the effects of many parameters on the formation of the spliceosome. The current data indicate that for splicing of the IVS2-654, IVS2-705 and IVS2-745 mutations, temperature can affect the interplay between the aberrant splice sites and other splicing elements and their interactions with splicing factors. For example, both the IVS2-654 and IVS2-705con introns in their native β-globin gene were unresponsive to decreased temperature, but when inserted into the EGFP coding sequence, which provided stronger flanking splice sites, the aberrant exon within the introns was skipped in a temperature-dependent fashion. Thus, the stronger correct 5′ splice site in the EGFP construct diminished but did not eliminate recognition of the aberrant splice sites by the spliceosome. Likewise, lower temperature may have decreased the utility of an enhancer element in the aberrant exon, the existence of which has been strongly suggested by mutational analysis of that region (36). The enhancer element, in turn, was likely influenced by the sequence and the length of the internal aberrant exons in the different IVS2 mutants.
The above conclusions are also supported by our previous findings of varying susceptibility of the thalassemic β-globin pre-mRNAs to treatment with antisense oligonucleotides (23). Treatment of HeLa cells expressing IVS2-654, IVS2-705 and IVS2-745 thalassemic β-globin genes with antisense oligonucleotides targeted to the common aberrant 3′ splice site resulted in correction of splicing in all three cases, but the effective concentrations of the oligonucleotides required for 50% correction (EC50) were 1560, 31 and 2 nM, respectively (i.e. up to a 700-fold difference). These data agree with the results of the present work, since aberrant splicing of IVS2-705 and IVS2-745 pre-mRNAs was prevented by both the temperature shift and the oligonucleotides, while both treatments were relatively ineffective for IVS2-654 pre-mRNA. Similarly, the resistance of IVS2-705con to changes induced by low temperature is in agreement with its increased resistance to correction by the antisense oligonucleotides (23). Thus, it appears that both treatments interfere with proper assembly of the spliceosomes at the aberrant splice sites of IVS2-705 and IVS2-745, but not IVS2-654 and IVS2-705con, pre-mRNAs.
Interestingly, low temperature cell culture may also bring about inclusion rather than skipping of an exon. For example, several different mutations in the 5′ splice sites of the human collagen genes, which are responsible for the genetic disorders termed Ehlers–Danlos and osteogenesis imperfecta syndromes, led to exon skipping in patient’s fibroblasts cultured at 37°C. However, the rates of exon skipping markedly decreased when the cells were incubated at 31°C (37–40). Similarly, in the murine retrovirus MuSVts110, the splicing of viral RNA was reduced at 37°C and increased at 33°C (41), while exon skipping was prevented in a mutant floral homeotic gene AP3 of Arabidopsis thaliana when cell culture temperature was shifted from 28 to 16°C (42). It was concluded in these reports that exon inclusion at low temperature was due to improved hybridization of U1 snRNA to the mutated 5′ splice sites (37–40) while improved splicing of MuSVts110 retrovirus appeared to be due to enhanced hybridization of U2 to a weak branch point (41). The fact that in the β-globin pre-mRNAs the lower temperature decreased rather than increased inclusion of the aberrant exon indicated that different mechanisms must have operated during splicing of those RNAs. The findings of the present study also suggest the intriguing possibility of lowering the temperature of bone marrow, perhaps in the extremities, as a treatment for β-thalassemia arising from the IVS2-705 or IVS2-745 mutations (18). Hypothermic treatments have been successfully used to reduce deleterious consequences of cardiac arrest (43).
Splicing of various mRNAs is affected in a different manner not only by a decrease but also by an increase in the temperature of cell culture. In our hands, a 3 h 42°C heat shock of IVS2-654 and IVS2-705 HeLa cells had no effect on aberrant splicing of the two thalassemic β-globin mRNAs (data not shown). However, activation of a 5′ splice site and a novel alternatively spliced murine HSP47 mRNA was detected in 3T3 cells subjected to heat shock at 42°C (44). A mutation in the 5′ splice site of intron 37 of type III procollagen, in contrast to the collagen mutations discussed above, led to a minor, but statistically significant, decrease in exon skipping when the temperature of the cell culture was raised from 31 to 37°C (45). These results reinforce our conclusion that variations in temperature affect the spliceosome and/or its interactions with specific splice sites and other sequence elements involved in splicing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elizabeth Smith for excellent technical assistance. This work was supported in part by an NHLBI, NIH grant (HL-51940) to R.K.
REFERENCES
- 1.Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 2.Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T. and Tasic,B. (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature, 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 4.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 5.Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 6.Hanamura A., Caceres,J.F., Mayeda,A., Franza,B.R.,Jr and Krainer,A.R. (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA, 4, 430–444. [PMC free article] [PubMed] [Google Scholar]
- 7.Stojdl D.F. and Bell,J.C. (1999) SR protein kinases: the splice of life. Biochem. Cell Biol., 77, 293–298. [PubMed] [Google Scholar]
- 8.Murray M.V., Kobayashi,R. and Krainer,A.R. (1999) The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev., 13, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misteli T. (1999) RNA splicing: what has phosphorylation got to do with it? Curr. Biol., 9, R198–R200. [DOI] [PubMed] [Google Scholar]
- 10.Prasad J., Colwill,K., Pawson,T. and Manley,J.L. (1999) The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol., 19, 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao S.H. and Manley,J.L. (1998) Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J., 17, 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seghezzi W., Chua,K., Shanahan,F., Gozani,O., Reed,R. and Lees,E. (1998) Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol. Cell. Biol., 18, 4526–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blencowe B.J. (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Selvakumar,M. and Helfman,D. (1997) Alternative pre-mRNA splicing. In Krainer,A. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 242–278.
- 15.Krawczak M., Reiss,J. and Cooper,D.N. (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet., 90, 41–54. [DOI] [PubMed] [Google Scholar]
- 16.Dominski Z. and Kole,R. (1992) Cooperation of pre-mRNA sequence elements in splice site selection. Mol. Cell. Biol., 12, 2108–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman B.E. and Grabowski,P.J. (1992) U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev., 6, 2554–2568. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz E. and Benz,E. (1995) Thalassemia syndromes. In Hoffman,R., Benz,E., Shattil,S., Furie,B., Cohen,H., Silberstein,L. and McGlave,P. (eds), Hematology: Basic Principles and Practice. Churchill Livingston, New York, NY.
- 19.Sierakowska H., Sambade,M.J., Agrawal,S. and Kole,R. (1996) Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 93, 12840–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treisman R., Orkin,S.H. and Maniatis,T. (1983) Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature, 302, 591–596. [DOI] [PubMed] [Google Scholar]
- 21.Dobkin C. and Bank,A. (1985) Reversibility of IVS 2 missplicing in a mutant human beta-globin gene. J. Biol. Chem., 260, 16332–16337. [PubMed] [Google Scholar]
- 22.Cheng T.C., Orkin,S.H., Antonarakis,S.E., Potter,M.J., Sexton,J.P., Markham,A.F., Giardina,P.J., Li,A. and Kazazian,H.H.,Jr (1984) beta-Thalassemia in Chinese: use of in vivo RNA analysis and oligonucleotide hybridization in systematic characterization of molecular defects. Proc. Natl Acad. Sci. USA, 81, 2821–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sierakowska H., Sambade,M.J., Schumperli,D. and Kole,R. (1999) Sensitivity of splice sites to antisense oligonucleotides in vivo. RNA, 5, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmajuk G., Sierakowska,H. and Kole,R. (1999) Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J. Biol. Chem., 274, 21783–21789. [DOI] [PubMed] [Google Scholar]
- 25.Dominski Z. and Kole,R. (1993) Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 90, 8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwanmanee T., Sierakowska,H., Lacerra,G., Svasti,S., Kirby,S., Walsh,C., Fucharoen,S. and Kole,R. (2002) Restoration of human beta-globin gene expression in murine and human IVS2-654 thalassemic erythroid cells by free uptake of antisense oligonucleotides. Mol. Pharmacol., 62, 545–553. [DOI] [PubMed] [Google Scholar]
- 27.Lacerra G., Sierakowska,H., Carestia,C., Fucharoen,S., Summerton,J., Weller,D. and Kole,R. (2000) Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc. Natl Acad. Sci. USA, 97, 9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman K.J., Kole,J., Cohn,J.A., Knowles,M.R., Silverman,L.M. and Kole,R. (1999) Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem., 274, 36193–36199. [DOI] [PubMed] [Google Scholar]
- 29.Wilton S.D., Lloyd,F., Carville,K., Fletcher,S., Honeyman,K., Agrawal,S. and Kole,R. (1999) Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul. Disord., 9, 330–338. [DOI] [PubMed] [Google Scholar]
- 30.Taylor J.K., Zhang,Q.Q., Wyatt,J.R. and Dean,N.M. (1999) Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol., 17, 1097–1100. [DOI] [PubMed] [Google Scholar]
- 31.Mercatante D.R., Bortner,C.D., Cidlowski,J.A. and Kole,R. (2001) Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. Analysis of apoptosis and cell death. J. Biol. Chem., 276, 16411–16417. [DOI] [PubMed] [Google Scholar]
- 32.Sazani P., Kang,S.H., Maier,M.A., Wei,C., Dillman,J., Summerton,J., Manoharan,M. and Kole,R. (2001) Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res., 29, 3965–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen I.T. and Chasin,L.A. (1993) Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol. Cell. Biol., 13, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benz E.J. Jr, Murnane,M.J., Tonkonow,B.L., Berman,B.W., Mazur,E.M., Cavallesco,C., Jenko,T., Snyder,E.L., Forget,B.G. and Hoffman,R. (1980) Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc. Natl Acad. Sci. USA, 77, 3509–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senapathy P., Shapiro,M.B. and Harris,N.L. (1990) Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol., 183, 252–278. [DOI] [PubMed] [Google Scholar]
- 36.Gemignani F., Landi,S., DeMarini,D.M. and Kole,R. (2001) Spontaneous and MNNG-induced reversion of an EGFP construct in HeLa cells: an assay for observing mutations in living cells by fluorescent microscopy. Hum. Mutat., 18, 526–534. [DOI] [PubMed] [Google Scholar]
- 37.Bonadio J., Ramirez,F. and Barr,M. (1990) An intron mutation in the human alpha 1(I) collagen gene alters the efficiency of pre-mRNA splicing and is associated with osteogenesis imperfecta type II. J. Biol. Chem., 265, 2262–2268. [PubMed] [Google Scholar]
- 38.D’Alessio M., Ramirez,F., Blumberg,B.D., Wirtz,M.K., Rao,V.H., Godfrey,M.D. and Hollister,D.W. (1991) Characterization of a COL1A1 splicing defect in a case of Ehlers-Danlos syndrome type VII: further evidence of molecular homogeneity. Am. J. Hum. Genet., 49, 400–406. [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B., Vitale,E., Superti-Furga,A., Steinmann,B. and Ramirez,F. (1991) G to T transversion at position +5 of a splice donor site causes skipping of the preceding exon in the type III procollagen transcripts of a patient with Ehlers-Danlos syndrome type IV. J. Biol. Chem., 266, 5256–5259. [PubMed] [Google Scholar]
- 40.Weil D., D’Alessio,M., Ramirez,F., Steinmann,B., Wirtz,M.K., Glanville,R.W. and Hollister,D.W. (1989) Temperature-dependent expression of a collagen splicing defect in the fibroblasts of a patient with Ehlers-Danlos syndrome type VII. J. Biol. Chem., 264, 16804–16809. [PubMed] [Google Scholar]
- 41.Touchman J.W., D’Souza,I., Heckman,C.A., Zhou,R., Biggart,N.W. and Murphy,E.C.,Jr (1995) Branchpoint and polypyrimidine tract mutations mediating the loss and partial recovery of the Moloney murine sarcoma virus MuSVts110 thermosensitive splicing phenotype. J. Virol., 69, 7724–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sablowski R.W. and Meyerowitz,E.M. (1998) Temperature-sensitive splicing in the floral homeotic mutant apetala3-1. Plant Cell, 10, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safar P.J. and Kochanek,P.M. (2002) Therapeutic hypothermia after cardiac arrest. N. Engl. J. Med., 346, 612–613. [DOI] [PubMed] [Google Scholar]
- 44.Takechi H., Hosokawa,N., Hirayoshi,K. and Nagata,K. (1994) Alternative 5′ splice site selection induced by heat shock. Mol. Cell. Biol., 14, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., Kuivaniemi,H., Tromp,G., Strobel,D., Romanic,A.M. and Prockop,D.J. (1993) Temperature sensitivity of aberrant RNA splicing with a mutation in the G+5 position of intron 37 of the gene for type III procollagen from a patient with Ehlers-Danlos syndrome type IV. Hum. Mutat., 2, 28–36. [DOI] [PubMed] [Google Scholar]