Abstract
Introns and polyadenylation (pA) sites are known to improve transcript stability and nuclear-cytoplasmic transport and are normally present in efficient gene expression vectors. Standard retroviral vectors, however, do not allow the inclusion of such sequence elements, as mRNA processing at internal splice and pA sites interferes with the production of functional full-length vector genomes. In this report we examined the capability of hybrid vaccinia/retroviral vectors to transduce complex gene cassettes with nuclear RNA processing signals within the retroviral genome. A retroviral vector was constructed that contains a gene of interest (the human coagulation factor IX [FIX] cDNA), including an intron and an internal pA site. The modified proviral vector genome was cloned downstream of a vaccinia virus promoter and was inserted into the vaccinia virus genome. Infection of a packaging cell line with the recombinant vaccinia virus vector resulted in secretion of retroviral particles at average titers of 105 CFU per ml of cell culture supernatant. Due to the cytoplasmic transcription and the nonrecognition of nuclear transcription signals in the vaccinia virus system, full-length transcripts were obtained that still contained the intron. In the retrovirally transduced cell lines the FIX transcripts were terminated at the internal pA site. The transcripts were quantitatively spliced, and FIX was secreted. Recombinant cell lines with stable single-copy inserts containing sequence elements necessary for efficient gene function could be generated. Thus, a relatively simple cytoplasmic system for the generation of complex retroviral vectors is described. Retroviral vectors transducing intron-containing gene cassettes may play a further role in gene therapy applications.
Retroviral vectors are presently used as tools in biological research, mainly in virology and cell biology, and as gene therapy vectors (15). The vectors are usually produced by transfection of plasmids containing the vector genomes into packaging cell lines which provide the structural components. During infection the retroviral vector enters a host cell, reverse transcribes its RNA genome into proviral DNA, and integrates into the genomic DNA of the host cells, a process termed transduction (6). The transduced vector genome is transcribed in the nucleus into new RNA molecules and is packaged in the cytoplasm into infectious particles. Due to the nuclear location of the cellular transcription machinery, it is usually not possible to obtain intron-containing retroviral vectors by standard procedures. Gene cassettes used to overexpress proteins in cell lines and transgenic animals, however, usually contain introns that are necessary for efficient gene expression and nuclear-cytoplasmic transport of mRNA (4, 5, 9, 11, 19).
One approach to overcome this limitation is the use of retroviral vectors having the engineered gene cassette in reverse orientation to the 5′ long terminal repeat (LTR) (7, 13, 16). More recently, alphaviruses were shown to permit the production of retroviral vectors with introns and polyadenylation (pA) sites. For instance, Semliki forest virus vectors were used to package intron-containing chloramphenicol acetyltransferase genes into retroviruses (14), and human clotting factor IX (FIX) minigenes containing retroviral vectors were produced in an alphavirus/retrovirus system (22). For construction of transduction-competent particles by alphavirus-based vectors, the desired vectors are usually produced by delivery of in vitro-transcribed RNAs into the production cell by electroporation. This procedure may not be easy to scale up. A further development to obtain intron-containing retroviral vectors is the use of split-intron vectors which generate an intron in the provirus upon reverse transcription (RT) of an engineered splice donor site inserted between the U3 and R region of the 3′ LTR (12).
Vaccinia virus is a large double-stranded DNA virus with unspliced genes transcribed by its own transcriptional apparatus in the cytoplasm of the host cells (18), allowing the production of standard transduction-competent retroviral particles by poxviral/retroviral hybrid vectors (10). Therefore, the vaccinia virus system is an excellent alternative to produce RNAs in the cytoplasm, avoiding nuclear RNA processing signals. In this report we describe hybrid vaccinia virus vectors that can be used to produce such complex retroviral vectors containing introns and internal pA sites. The human coagulation FIX cDNA was used as the gene of interest in this study, because the FIX gene is mutated in hemophilia B patients, for whom efficient gene therapies are needed.
MATERIALS AND METHODS
Virus and cell lines.
The African green monkey kidney cell line CV-1 (ATCC CCL-70), the NIH 3T3 cell line (ATCC CRL-1658), the Chinese hamster ovary cell line CHO (ATCC CRL-9096), the thymidine kinase-negative cell line 143B (ATCC CRL-8303), and the Western Reserve (WR) strain of vaccinia virus (ATCC VR119) were obtained from the American Type Culture Collection. The PT67 cell line was obtained from Clontech Laboratories, Inc.
Construction of plasmids.(i) pUC-I9pA.
To substitute the two TTTTTNT signals (vaccinia virus early transcription stop signals [24]) in the FIX gene cassette present in pCMV-FIX (U. Schlokat, unpublished data), a PCR mutagenesis procedure was applied with the Quickchange mutagenesis kit (Stratagene, Inc.). In two subsequent steps the TTTTTNT signals, one in the FIX cDNA and one in the simian virus 40 (SV40) intron, were modified by using the primers pairs 5′-GTGCTGAATG TACAGTATTT CTTGATCATG-3′ and 5′-CATGATCAAG AAATACTGTA CATTCAGCAC-3′ and 5′-GTTGACTGGT AAGTTTAGTC TATTTGTC-3′ and 5′-GACAAATAGA CTAAACTTAC CAGTCAAC-3′, respectively. A 1.48-kb EcoRI fragment containing the FIX cDNA and a 430-bp HindIII fragment (containing the small SV40 intron and pA signal) were subcloned into the pUC18 vector (Stratagene, Inc.), resulting in the plasmids pUC-FIX and pUC-IpA. In the next step the pUC-FIX EagI fragment (containing the FIX cDNA) was inserted into the EagI site of pUC-IpA between the SV40 intron and the pA signal, resulting in pUC-I9pA.
(ii) pR-Xi9pASN.
To construct pR-Xi9pASN the multiple cloning site of pR-XSN (10) was modified, introducing additional single restriction sites by annealing the oligonucleotides 5′-AATTCCGCGG GCAGGCAATT G-3′ and 5′-GATCCAATTG CCTGCAGGCT CGAGTACGTA CGCGTCCGCG G-3′ and ligating them into the BamHI/EcoRI-cleaved pR-XSN, resulting in pR-XSNmod. The SnaBI/MunI fragment from pUC-I9pA, containing the SV40 intron, the FIX cDNA, and the pA signal, was then inserted in the SmaI/MunI-cleaved pR-XSNmod, resulting in the plasmid pR-Xi9pASN.
Construction of the hybrid vaccinia virus (vR-Xi9pASN).
Twenty-five micrograms of pR-Xi9pASN plasmid DNA was transfected into CV-1 cells by using Lipofectamine 2000 (Life Technologies, Inc.) prior to infection with the wild-type vaccinia virus WR and was further processed as described previously (8). Plaque purifications were done twice under guanine phosphoribosyl transferase (gpt) selection in the CV-1 cell line followed by two rounds under bromodeoxyuridine selection in the 143B cell line. One isolate was grown to large scale on CV-1 cells, and cell-bound virus was prepared by trypsinization and centrifugation through a 36% sucrose cushion.
Genomic characterization of the virus.
Two micrograms of DNA of the recombinant virus and of the wild-type WR strain were digested with XhoI and HindIII, separated on 1% agarose gel, and transferred to a nylon membrane (Hybond N; Amersham Biotech) by capillary transfer. For hybridization the FIX EcoRI fragment was α-32P-radiolabeled by random priming using the High Prime system (Roche Molecular Biochemicals).
Colony-forming assay (CFA).
PT67 packaging cells were infected with the virus vR-Xi9pASN at a multiplicity of infection of 1 for 12 h. Supernatants were filtrated through 0.2-μm- or 0.1-μm-pore-size sterile filters (Schleicher & Schuell) to remove the vaccinia virus vector. Polybrene was added to a final concentration of 4 μg/ml. NIH 3T3 and CHO cells were infected with retroviral particles essentially as described in the user manual of the RetroXpress System (Clontech, Inc.). Selection was performed with G418 (500 μg/ml; PAA Laboratories, Linz, Austria). For titer determination, colonies were stained with crystal violet after 14 days. For further analysis cell clones were picked and grown to large scale under G418 selection.
Provirus analysis by PCR.
Genomic DNA from the transduced cell clones was isolated, and PCR products were generated from 1 μg of genomic DNA as a template by using Taq DNA polymerase (Roche Molecular Biochemicals). PCR amplification of the neomycin resistance gene (neo) was carried out at 95°C for 45 s, 62°C for 45 s, and 72°C for 1 min and was repeated for 35 cycles. The forward (5′-GCTATTCGGC TATGACTGGG CA-3′) and reverse (5′-TCGGCAAGCA GGCATCGCCA TG-3′) primers were designed to bind within the neo open reading frame. The PCR for the intron-containing part of the gene cassette was carried out at 95°C for 45 s, 59°C for 45 s, and 72°C for 90 s and was repeated for 35 cycles. The forward primer (5′-CCTGCCCAGG GACCACCGAC-3′) hybridizes in the packaging signal upstream of the retroviral splice donor, and the reverse primer (5′-CATGATCAAG AAATACTGTA CATTCAGCAC-3′) binds 100 bp downstream of the FIX start codon. The PCR products were analyzed on a 1% agarose gel.
Southern blotting of DNA of transduced cell clones.
Total DNAs from the transduced cell clones and from the wild-type NIH 3T3 cells as a negative control were isolated by standard methods. Thirty micrograms of DNA was digested with EcoRI and SacI, separated on a 1% agarose gel by field inversed gel electrophoresis, and transferred to a nylon membrane (Hybond N; Amersham Biotech Inc.) by capillary transfer. The positive control was wild-type NIH 3T3 DNA spiked with 10 pg of pR-Xi9pASN plasmid DNA cleaved with EcoRI and wild-type NIH 3T3 DNA spiked with 500 pg of vR-Xi9pASN viral DNA cleaved with SacI, respectively. For hybridization the FIX EcoRI fragment from pCMV-FIX was used as a probe.
Analysis of FIX gene transcription.
Transcripts of the FIX gene were analyzed by (RT)-PCR. Total RNA from the transduced NIH 3T3 cell clones, from wild-type NIH 3T3 cells (as a negative control), and from wild-type NIH 3T3 cells infected with vR-Xi9pASN for 2 h at a multiplicity of infection of 10 as a positive control were isolated with the RNAzol Reagent (Tel-Test, Inc.).
Analysis of splicing. From 500 ng of RNA the first-strand DNA was synthesized by Superscript II reverse transcriptase (Life Technologies) at 42°C for 45 min. Next the RNA/DNA hybrids were denatured and the reverse transcriptase was inactivated by incubation at 95°C for 5 min. PCR amplification was then performed by using Taq DNA polymerase (Roche Molecular Biochemicals). PCR amplification was carried out at 95°C for 45 s, 59°C for 45 s, and 72°C for 90 s and was repeated for 35 cycles. The forward primer P1 (5′-GGCCC TATCG AGGAA CTGAA AAACC-3′) hybridizes 110 bp upstream of the SV40 intron, and the reverse primer P2 (5′-CATGATCAAG AAATACTGTA CATTCAGCAC-3′) hybridizes 100 bp downstream of the FIX start codon. The PCR products were analyzed on a 1% agarose gel.
Analysis of transcriptional termination at the internal pA site. From 500 ng of RNA the first-strand DNA was synthesized by Superscript II reverse transcriptase (Life Technologies) at 42°C for 45 min by using the oligonucleotide 5′-GAGCAAATTC CTGTACTGAC TTTTTTTTTTT TTTTTTTTTT TTTTTTTTT NA-3′ as the oligo(dT) primer. Next the RNA/DNA hybrids were denatured and the reverse transcriptase was inactivated by incubation at 95°C for 5 min. PCR amplification was then performed with Taq DNA polymerase (Roche Molecular Biochemicals). PCR amplification was carried out at 95°C for 20 s, 54°C for 30 s, and 72°C for 70 s and was repeated for 35 cycles. The forward primer P3 (5′-CAGAAAACCAGAAGTCCTGTG-3′) hybridizes in the FIX open reading frame upstream of the internal pA site, and the reverse primer P4 (5′-GAGCAAATTC CTGTACTGAC-3′) binds to the 3′ end of the reverse-transcribed DNA strand. The PCR products were analyzed on a 1% agarose gel, purified by using the Qiaquick PCR Purification Kit (Qiagen), and subsequently sequenced on an Applied Biosystems Model 373A Sequencer using the cycle sequencing method with dye terminators (DNA Sequencing Kit, ABI PRISM, and BIG Dye Terminator Cycle Sequencing Ready Reaction Kit 4303153; Applied Biosystems) using the sequencing primer (5′-ACCAAGGTAT CCCGGTATG-3′).
Northern blotting.
Twenty micrograms of total RNA was glyoxylated, resolved on a 1% agarose gel, and blotted onto a nylon membrane (Hybond N; Amersham Biotech Inc.). For hybridization, specific DNA fragments were α-32P-radiolabeled by random priming using the High Prime system (Roche Molecular Biochemicals). The blots were probed with DNA probes flanking the internal pA site and a probe specific for the packaging signal. PCRs were performed with plasmid pR-Xi9pASN as template using the primers 5′-GGTGAAGAGT GTGCAATGAA A-3′ and 5′-TTGTTGTTAA CTTGTTTATT GCAGC-3′ for probe 1 and 5′-TTGGATCCGG CTGTGGAA-3′ and 5′-TCCTCACTAC TTCTGGAATA GC-3′ for probe 2. PCRs were carried out at 95°C for 20 s, 56°C for 20 s, and 72°C for 30 s and were repeated for 35 cycles by using Taq DNA polymerase (Roche Molecular Biochemicals). The packaging probe was an 803-bp SpeI fragment from pR-Xi9pASN. PCR products and DNA fragments were purified from an agarose gel with the Sephaglas Bandprep Kit (Amersham Pharmacia Biotech Inc.) and were radiolabeled as described above.
Western blotting.
To analyze the cell clones expressing human FIX, Western blots of cell culture supernatants were performed. Ten microliters of supernatant was analyzed per lane; in the case of the CHO clones, supernatants were concentrated 20-fold. The blots were first incubated with rabbit anti-human FIX antibody (Accurate Chemical & Scientific Corp.) in a 1:1,000 dilution. The second antibody was alkaline phosphatase-coupled goat anti-rabbit immunoglobulin G (Bio-Rad, Inc.) at a 1:3,500 dilution.
FIX ELISA.
The FIX amount in the cell culture supernatant was determined by a FIX enzyme-linked immunosorbent assay (ELISA) using the Asserachrom IX:Ag Kit (Roche Molecular Biochemicals).
RESULTS
Structure of the proviral insert in the hybrid vaccinia virus vector.
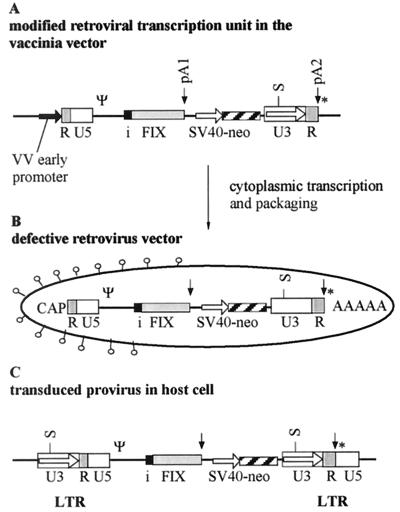
First, a provirus containing two gene cassettes was designed (Fig. 1). The first cassette consists of the retroviral promoter present in the 5′ LTR, a gene of interest with an intron and an internal pA site (pA1). The intron used, the small SV40 intron, is a known efficient genetic element that was introduced into the gene cassette upstream of the FIX cDNA. The natural splice donor site present in the parental vector pLXSN (15) was not removed because, in contrast to other concepts of intron-containing cassettes (12), particle yields in the vaccinia virus system should be tolerant to cryptic splice signals. A further important genetic element, the internal pA signal downstream of the FIX cDNA in the transduced gene cassette, should prevent the full-length transcription of the provirus, a new safety aspect in potential gene therapy vectors. The second transcription unit consists of the SV40 promoter and the open reading frame encoding the neomycin phosphotransferase (neo) which is followed by the retroviral 3′ LTR with its inherent pA site. The neo gene cassette enables selection of the transduced cell lines and titer determination of the retroviral vectors.
FIG. 1.
Schematic representation of the genetic elements in the viral genomes and transduced cells. (A) The modified retroviral transcription unit in the vaccinia virus (VV) vector. A vaccinia virus early promoter (bold arrow) is fused to the repeat region (R) of the LTR followed by the U5 region, the packaging signal, the small SV40 intron (i), and the FIX open reading frame, which is terminated by the internal pA site (pA1). The second transcription unit, consisting of the SV40 promoter and the neomycin open reading frame, is followed by the 3′ LTR that includes the terminal pA site (pA2). The SacI restriction site (S) present in the U3 region was used for genomic characterization. A vaccinia virus early transcription stop signal (*) is located immediately downstream of the second pA site. (B) The modified retroviral vector produced in the cytoplasmic vaccinia virus system retains the intron. The RNA genome has a CAP structure at the 5′end and a poly(A) tail (AAAAA) at the 3′ end. (C) In the transduced host cell, the LTR promoter (located in the U3 sequence) is reconstituted during RT and the integration process driving the FIX gene cassette.
Construction of the hybrid virus and production of the retroviral particles.
The plasmid pR-Xi9pASN, which harbors the proviral sequences, was constructed as described above; the plasmid further contains flanking regions derived from the vaccinia virus thymidine kinase (tk) gene that direct the insert into the viral tk locus and a gpt selection marker to allow for positive selection of vaccinia virus recombinants. The plasmid and the recombinant vaccinia virus, termed vR-Xi9pASN, were constructed by standard recombination techniques (see Materials and Methods). Characterization of the viral DNA by restriction endonuclease cleavage and Southern blotting revealed that the virus had the predicted structure and was free of wild-type virus (data not shown).
The vaccinia virus vR-Xi9pASN was now used to infect the packaging cell line PT67. In the cytoplasm of the host cell the vaccinia virus RNA polymerase should start transcription at the upstream R region of the proviral DNA insert, transcribe the retroviral genome (including the intron), and ignore the internal nuclear pA site. Transcription stops at an engineered vaccinia virus stop signal (24) located immediately downstream of the terminal pA site (see Fig. 1). To obtain retroviral particles the packaging cell line PT67 was infected with 1 PFU of vaccinia virus per cell followed by incubation for a period of 8 to 12 h. In order to count the transducing particles, CFAs were performed. The supernatants of the vaccinia virus-infected cells were filtered through 0.2-μm-pore-size filters to remove cell debris and vaccinia virus particles and were titrated on NIH 3T3 cells. To analyze residual vaccinia virus, supernatants were titrated before and after filtration. Filtration through the 0.2-μm-pore-size filter removed 99.9% of the vaccinia virus vector. Filtration of the supernatants with a 0.1-μm-pore-size filter quantitatively removed the vaccinia virus, and the same amounts of colonies were found in the CFAs.
Neomycin selection was applied to obtain neomycin-resistant colonies (see Materials and Methods). Positive controls were performed in parallel with vdR-XSN, a hybrid vaccinia virus carrying a standard provirus genome consisting of the retroviral LTRs and an SV40-neomycin gene cassette (10). Negative controls, consisting of infections of the packaging cell line with wild-type vaccinia virus followed by the same transduction procedure on the indicator cell line, were also performed.
Surprisingly, titers in the range of 105 CFU per ml were obtained with the supernatants of the vR-Xi9pASN-infected packaging cells (Table 1), suggesting that transduction-competent complex retroviral vectors had formed. Since colony-forming titers were in the same range as that seen with the standard hybrid vector (Table 1), introns or internal pA sites do not impair the production of transduction-competent particles in the cytoplasmic vaccinia virus system. Neomycin-resistant colonies were not observed in the negative control, supernatants of packaging cells infected with vaccinia virus WR wild-type virus (Table 1).
TABLE 1.
CFAs for different cell lines
| Virus stocka | Cell line
|
|
|---|---|---|
| NIH 3T3 | CHO | |
| vR-Xi9pASN #4, cs | 1 × 105 | NDb |
| vR-Xi9pASN #4, cs | 8 × 104 | 8 × 102 |
| vR-Xi9pASN #4, pur | 1 × 105 | 7 × 102 |
| vR-Xi9pASN #4, pur | 2 × 105 | ND |
| vdR-XSN #4, pur | 7 × 105 | ND |
| vdR-XSN #4, pur | 1 × 105 | 7 × 102 |
| vdR-XSN #4, pur | 2 × 104 | 7 × 102 |
| vdR-XSN #4, pur | 2 × 105 | ND |
| Vaccinia virus WR | 0 | 0 |
cs, crude stock; pur, purified virus.
ND, not determined.
In addition to using NIH 3T3 cells for the transductions, the CFAs were also performed on CHO cells, an important cell line for the production of recombinant proteins. Cells lines could readily be selected by using 500 μg of G418 antibiotic/ml; however, titers were usually two orders of magnitude lower than titers in the standard NIH 3T3 indicator cell line (Table 1).
FIX expression in the transduced cell clones.
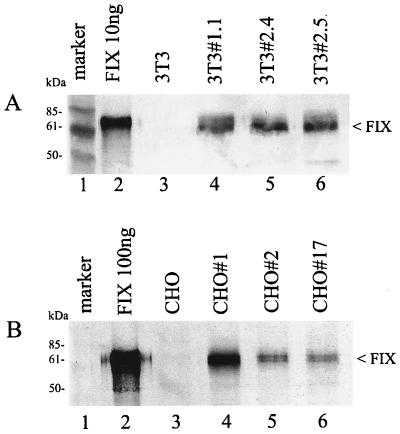
The transduced cell clones were screened for expression of human FIX. Several clones were expanded, and FIX levels in cell culture supernatants were determined by ELISA (see Materials and Methods). In the case of the NIH 3T3 cell clones, expression levels of 15 examined clones were in the range of 50 to 250 ng/24 h/106 cells; levels of 6 clones expressing the highest amounts are shown in Table 2. FIX was readily detectable in the cell culture supernatants by Western blot analysis. Supernatants of three NIH 3T3 clones were analyzed; a band comigrating with a recombinant FIX standard was visible in the correct size range of approximately 65 kDa (Fig. 2A, lanes 4 to 6). The supernatant of the parental NIH 3T3 cell line served as the negative control (lane 3).
TABLE 2.
FIX expression (ELISA data) of transduced NIH 3T3 and CHO cell clones
| NIH 3T3 clone | Amt of FIX (ng/106 cells/24 h) | CHO clone | Amt of FIX (ng/106 cells/24 h) |
|---|---|---|---|
| FIX#1.1 | 188 | FIX#1 | 25 |
| FIX#2.2 | 148 | FIX#2 | 7.2 |
| FIX#2.4 | 211 | FIX#17 | 4.6 |
| FIX#2.5 | 242 | FIX#19 | 4.1 |
| FIX#2.6 | 167 | ||
| FIX#3.6 | 169 | ||
| WT | <20 | WTa | <1 |
WT, wild type.
FIG. 2.
Western blot analysis of the FIX expression in the transduced 3T3 cell clones (A) and in the transduced CHO cell clones (B). (A and B) Lane 1, marker proteins; lane 2, FIX standard purified from plasma; lane 3, cell culture supernatants from nontransduced parental cell lines; lanes 4 to 6, cell culture supernatants from transduced cell clones. The arrowheads on the right point to the FIX band.
Several randomly picked transduced CHO clones were also analyzed for expression. FIX levels in this cell line were in the range of 0 to 25 ng/24 h/106 cells (Table 2) and thus were an order of magnitude lower than the levels observed in mouse NIH 3T3 cells. Since FIX protein was barely detectable in the initial Western blot analysis, the supernatants were concentrated 20-fold. The FIX band was detectable now as a band in the correct size range (Fig. 2B, lanes 4 to 6). Recombinant FIX-expressing cell lines were further subjected to genomic and RNA analyses.
The intron sequences are present in the transduced DNA of the host cell.
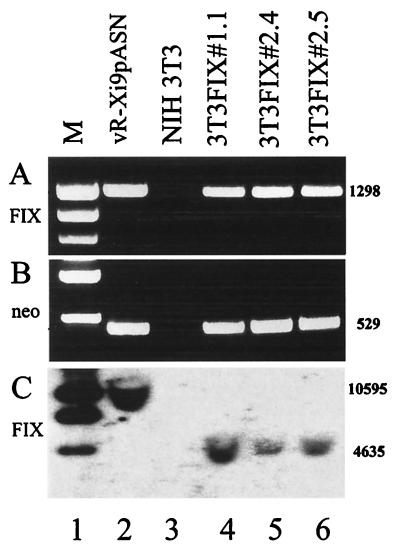
Total genomic DNAs of the retrovirally transduced cell clones previously analyzed by Western blotting were prepared and subjected to PCR analysis with primers that amplify parts of the FIX cDNA that include the intron. All cell clones examined gave rise to the expected amplicon of 1,298 bp, indicating the presence of the FIX and intron sequences (Fig. 3A, lanes 4 to 6). As a positive control, the DNA of the virus vR-Xi9pASN was amplified, resulting in the same-sized amplicon (lane 2). As a negative control, DNA prepared from the parental cell line NIH 3T3 was used (lane 3). A second set of primers was used to directly confirm the presence of the neomycin marker in the DNA of the cell clones. As expected, all transduced clones that grew under 500 μg of neomycin/ml were also positive in this PCR assay (Fig. 3B, lanes 4 to 6), while the negative control did not show this signal (lane 3) and the positive control, DNA of the virus vR-Xi9pASN, was also positive in the assay (lane 2).
FIG. 3.
Genomic analysis of the DNA of the cell clones by PCR for the presence of the FIX sequences (A) and for the neomycin selection marker (B) and by Southern blot for presence of the FIX and intron sequences (C). Lane 1, size markers (M); lane 2, viral DNA of the hybrid vaccinia virus vR-Xi9pASN; lane 3, negative control DNA of NIH 3T3 cells; lanes 4 to 6, DNA of the transduced clones 3T3FIX#1.1, 3T3FIX#2.4, and 3T3FIX#2.5. The numbers on the right are the sizes of the bands in base pairs.
To independently confirm the integrated provirus in the host cell DNA, a Southern blot analysis was performed. In the first Southern analysis, total cellular DNA of transduced cell lines and controls was digested with the enzyme SacI. This enzyme cuts once in the U3 region of the retroviral transcription unit and therefore, after RT and integration, releases a distinct fragment including most of the proviral sequences in the transduced clones (Fig. 3C, lanes 4 to 6). The positive control, consisting of total DNA of wild-type NIH 3T3 cells spiked with 500 pg of vR-Xi9pASN vaccinia virus DNA, shows a larger fragment resulting from a second SacI site in the vaccinia virus genome that is positioned about 6 kb upstream of the retroviral transcription unit (Fig. 3C, lane 2). In the negative control, total DNA from NIH 3T3 cells, this signal is not detectable (lane 2). This analysis demonstrates integration of the provirus and confirms the reconstitution of the U3 region by the presence of a new upstream SacI site.
A second Southern analysis using the same DNAs digested with EcoRI, an enzyme cutting once in the transduced provirus, revealed one hybridizing fragment of various size in the clones examined (data not shown). This analysis indicated that random single-copy insertions of the intron-containing provirus had occurred in the transduced cell clones. In summary, the genomic analysis of the transduced cell clones confirmed transduction of the full-length, intron-containing retroviral vectors. Therefore, complex gene cassettes containing introns and internal pA sites can be delivered into host cells by retroviral vectors produced in the cytoplasmic vaccinia virus system.
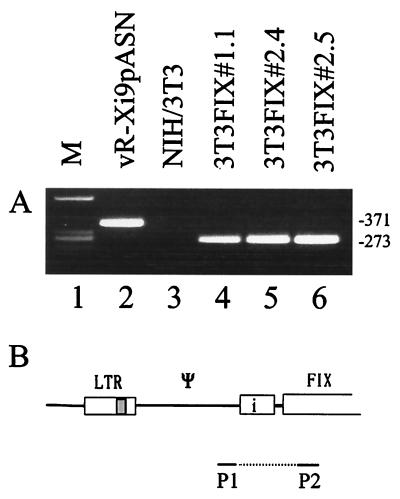
The transcripts are fully spliced in the small SV40 intron.
The transcripts of the cell clones and of the controls were analyzed by RT-PCR. Total RNA from the transduced NIH 3T3 cell lines and from vaccinia virus-infected cells were isolated and subjected to RT-PCR (Fig. 4). The primer pair was designed to frame the SV40 intron sequence (Fig. 4B). Using total vaccinia virus RNA as the template, an amplicon of 371 bp was formed, reflecting the unspliced RNA. By using the RNA of the transduced NIH 3T3 clones, a single amplicon of 273 bp was obtained, indicating correct splicing of the message (Fig. 4A, lanes 4 to 6). Splicing occurred quantitatively, as indicated by the absence of a 371-bp band.
FIG. 4.
Analysis of the transcripts of the transduced cell clones and the vaccinia virus-infected cells by RT-PCR (A) and outline of the primers (P1-P2) and the template used (B). (A) Lane 1, size markers (M); lane 2, RNA of cells infected with the hybrid virus vR-Xi9pASN; lane 3, negative control RNA of NIH 3T3 cells; lanes 4 to 6, RNA of the transduced clones 3T3FIX#1.1, 3T3FIX#2.4, and 3T3FIX#2.5. The numbers on the right are the sizes of the bands in base pairs.
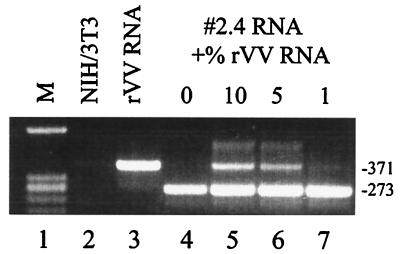
In order to estimate the detection limit of potentially unspliced RNA in the transduced clones, mixing experiments were performed. Increasing amounts of unspliced virus RNA derived from vR-Xi9pASN-infected cells was added to the RNA of cell clone 2.4 followed by RT-PCR as described above. With no virus RNA added, only the 273-bp band was seen (Fig. 5, lane 4), while addition of as low as 1% unspliced RNA resulted in an additional band in the 371-bp size range (lanes 5 to 7), indicating that at least 99% of the RNA in the cell clones was spliced.
FIG. 5.
RT-PCR analysis of the transcripts of a transduced cell clone spiked with unspliced vaccinia virus RNA. Lane 1, marker (M); lane 2, RNA of the NIH 3T3 negative control; lane 3, RNA of cells infected with the recombinant vaccinia virus (rVV) vR-Xi9pASN; lanes 4 to 7, RNA of the transduced cell clone 2.4 spiked with the indicated amounts of unspliced viral RNA. The numbers on the right are the sizes of the bands in base pairs.
The transcripts of the gene cassette of interest terminate at the engineered internal pA site.
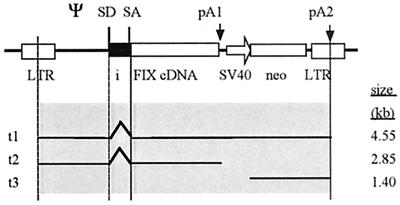
The transduced provirus is complex; its genome consists of the FIX transcription unit that includes the intron flanked by an internal pA site. The second transcription unit consists of the SV40 promoter and the neo selection marker followed by the pA signal of the 3′ LTR. In order to analyze termination of the transcripts in the transduced cell clones, Northern blots with total cellular RNAs were performed. Figure 6 shows the detailed structure of the proviral insert and depicts the transcripts expected if the internal pA site is inactive (t1) or active (t2); a 1.4-kb independent transcript (t3) of the second transcription unit, the SV40 promoter neomycin gene cassette, is also expected.
FIG. 6.
Schematic representation of the retroviral transcription unit and the expected transcripts (t1 to t3) in the transduced host cells. SD1, splice donor; SA, splice acceptor. The two pA sites (pA1 and pA2) are indicated by arrows. The sizes of the predicted transcripts are on the right.
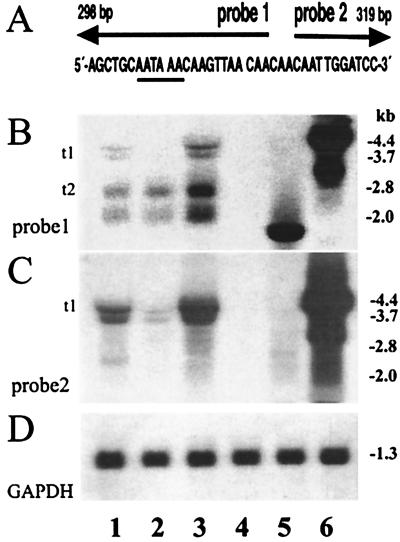
The blots were hybridized with four different probes: probes 1 and 2 (see also Fig. 7A), the neomycin gene, and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes. The latter probe was used to control the quality of the RNA and to confirm that equal amounts of RNA had been loaded onto the gels (Fig. 7D). Assuming correct splicing and termination at the internal pA site, a single transcript of approximately 2.8 kb was expected after hybridization with probe 1; this probe consists of a DNA sequence located immediately upstream of the internal pA site and should pick up all transcripts starting at the 5′ LTR promoter. However, four transcripts of approximately 4.5, 3.7, 2.8, and 2.0 kb were detected (Fig. 7B, lanes 1 to 3). The larger-sized transcripts of approximately 4.5 and 3.7 kb (t1) are, due to their size and their hybridization with both probes 1 and 2 (Fig. 7B and C, lanes 1 to 3), read-through messages ignoring the internal pA site (see Fig. 6). The smaller-sized major transcripts of 2.8 and 2.0 kb (Fig. 7B, lanes 1 to 3) are probably, due to their size and hybridization with probe 1 (and not with probe 2, located immediately downstream of the internal pA site), correctly terminated FIX transcripts, with the 2.8-kb transcripts being the expected FIX message (Fig. 7B, t2). The smaller transcript of about 2.0 kb is presumably an aberrant splice product, most probably involving the natural retroviral splice donor. This interpretation is supported by an additional Northern blot that used a packaging signal probe in which only the larger transcripts (t1 and t2) were detectable (data not shown). Hybridization with probe 2, derived from the region downstream of the internal pA site, essentially revealed only the two larger read-through transcripts of 4.5 and 3.7 kb (Fig. 7C, lanes 1 to 3). The neomycin gene probe revealed the expected 1.4-kb-sized independent transcripts originating from the internal SV40 promoter (data not shown).
FIG. 7.
Northern blot analysis of the transcripts derived from the transduced cell clones and the controls. (A) A schematic representation of the binding regions of probes 1 and 2. Probe 1 is complementary to the region upstream of the AATAAA signal (underlined) of the FIX transcripts. Probe 2 is complementary to the region downstream of the processing signal. (B and C) The Northern blots obtained with probe 1 (B) and probe 2 (C). The transcripts (t1-t2, left side) and their respective sizes in kilobases (right side) are shown. Lanes 1 to 3, analysis of RNAs from transduced cell clones 3T3FIX#1.1, 3T3FIX#2.4, and 3T3FIX#2.5; lane 4, negative control RNA from NIH 3T3 cells; lane 5, positive control RNA from a classically transfected and amplified CHO cell clone expressing human FIX; lane 6, control RNA from CV-1 cells infected with the vaccinia virus vR-Xi9pASN. (D) A control blot obtained with a GAPDH probe confirming equal RNA loading of the gel.
Furthermore, the positive control consisting of an unrelated, traditionally transfected and amplified CHO clone expressing FIX hybridizes only with probe 1 (which shares FIX sequences), showing an expected single transcript of 1.8 kb (Fig. 7B, lane 5) and confirming the specificity of the hybridizations. As expected, the vaccinia virus RNA control containing the total retroviral genome hybridizes with all probes (lanes 6). In summary, the Northern analysis suggests that the majority of the FIX transcripts are correctly internally terminated. Splicing of the SV40 intron is complete, and additional aberrant splice products with this prototype vector are found.
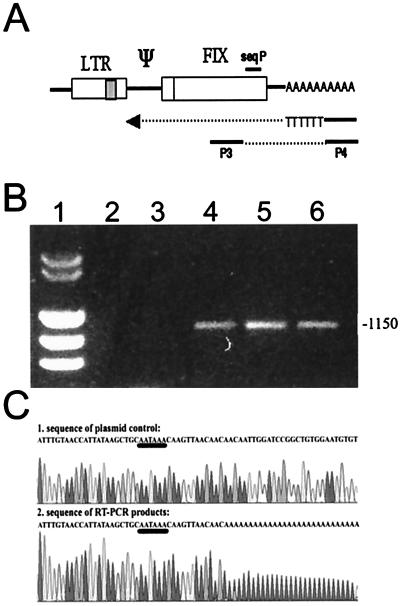
To further analyze termination, cDNAs of the putative internally terminated transcripts were amplified and partially sequenced. To isolate the cDNAs, an RT reaction using a tagged oligo(dT) primer was used. The amplification step was carried out with a primer complementary to the tag in the oligo(dT) primer (also present in the cDNA) and a FIX-specific primer (see Materials and Methods and Fig. 8A). Using the RNA of the NIH 3T3 clones, an amplicon of about 1,150 bp was obtained, indicating internally terminated polyadenylated transcripts (Fig. 8B, lanes 4 to 6). With total vaccinia virus RNA and RNA from wild-type NIH 3T3 cells, no specific amplicon was formed (lanes 2 and 3). Omitting the RT step in the RT-PCR did not result in amplicons, excluding false positives by DNA contamination (data not shown). Sequence analysis of the amplicons with a primer which binds about 250 bp upstream of the 3′ end (Fig. 8A, seqP) revealed the same sequence in all three analyzed cell clones. The poly(A) tail started 12 bp downstream of the internal pA site (Fig. 8C, AATAAA, underlined), confirming correct function of the pA site. The reference sequence (Fig. 8C, upper part) was obtained by using the corresponding plasmid DNA.
FIG. 8.
RT-PCR of transcripts terminating at the internal pA site and sequence analysis of cDNAs. (A) Schematic outline of the primers and the template used to map pA. (B) Agarose gel electrophoresis showing the results of the PCR analysis. Lane 1, size markers; lane 2, RNA of cells infected with the hybrid virus vR-Xi9pASN; lane 3, negative control RNA of NIH 3T3 cells; lanes 4 to 6, RNA of the transduced clones 3T3FIX#1.1, 3T3FIX#2.4, and 3T3FIX#2.5. The number on the right is the size of the bands in base pairs. (C) Sequence of plasmid control pR-Xi9pASN and the cDNAs derived from the internally terminated transcripts.
DISCUSSION
Retroviral vectors are still the most important gene therapy vectors, mainly because of their low immunogenicity and their stable integration into the host genome. The major goal of this study was to provide a system for the generation of complex retroviral vectors. This could be achieved by constructing a hybrid vaccinia virus vector harboring a proviral insert containing an intron and an internal pA site. Infection of a packaging cell line with the prototype vector resulted in high titers of complex transduction-competent particles. The cytoplasmic vaccinia virus system is therefore a suitable system to construct complex retroviral vectors. It is easy to handle, and scale-up for vector production, a problem not fully solved for retroviral vectors (2), seems to be feasible because at least vaccinia virus production is scalable. Vaccinia virus has a broad host range, infecting the majority of packaging cell lines, and can be grown by using high-cell-density fermentation on microcarriers in large fermenters (3). In addition, since vaccinia virus belongs to the largest animal viruses and retroviruses belong to the smallest, the production hybrid vector can be easily separated from the product, the retrovirus particle, by filtration.
Compared to the split-intron vectors (12), titers were approximately 10-fold lower but were in the same range as those obtained with comparable vectors produced in the alphavirus chimeric system, where an internal pA site also was included (14). However, optimizations to increase the yields of particles, such as screening of the most suitable packaging cell line and optimization of production conditions, are good possibilities to improve yields.
The transduced cell lines had incorporated the genetic elements that are not transducible by the classical retroviral vectors. As expected, the intron and the internal pA site, originally derived from the SV40 virus, did not affect the full-length transcription of the retroviral genome by the vaccinia virus. We also could demonstrate full integrity of the vector in all analyzed cell clones. However, we did not examine cell populations like Wahlfors and Morgan (22), who found partial vector rearrangements after transduction, did.
The internal pA site was active in the transduced cell clones. Mapping of the poly(A) tails in transcripts of three transduced clones showed correct terminal processing and pA. The specific hybridization patterns of the smaller-sized major transcripts and the quantitative distribution of all transcripts further suggests efficient termination at the internal pA site. Internal pA sites as part of a retroviral gene therapy vector may, therefore, increase safety because only complete retroviral vector transcripts containing both R regions of the LTR can contribute to the formation of transducing particles. While split-intron vectors efficiently create introns upon transduction (12), they are not suited to transduce internal pA sites. An excellent further way to prevent full-length transcripts and increase safety are self-inactivating vectors (17).
The prototype vector described in this report contains a FIX gene cassette with the efficient SV40 intron. Transcripts were fully spliced in this intron; however, in addition to the correctly spliced and terminated message aberrantly spliced transcripts were found in the Northern blots, probably splice variants induced by the original retroviral splice donor still present and a cryptic acceptor site in the packaging signal. This suggests that the prototype vector may be further improved as described for the split-intron vectors, for instance, by placing the packaging signal between efficient splicing sites.
Besides application as gene therapy vectors, complex retroviral vectors may be used to obtain production cell lines for recombinant proteins. Dihydrofolate reductase (dhfr)-negative CHO cells, a biotechnologically widely used production cell line (21), was easily transducible by the vectors. FIX expression levels in initial CHO clones were lower compared to the murine NIH 3T3 cell clones; they were, however, in a range similar to those in initial CHO clones obtained from transfections with traditional plasmid vectors. Since overexpression in CHO is usually achieved only after gene amplification using the dhfr marker, complex retroviral vectors using the dhfr marker rather than the neomycin gene may be a useful further development.
The complex retroviral vectors with optimized expression cassettes are very promising in the context of gene therapy. In classical gene therapy, when retroviral vectors are infused directly into the patient smaller amounts of vector should be sufficient for a therapeutic effect. In somatic cell gene therapy protocols, including ex vivo transduction and selection of patients’ cells followed by reinfusion (see, for example, reference 20), the complex retroviral vectors may combine the desirable elements of nonviral vectors with stable transduction that is seen only with retroviral vectors. In addition, in the cytoplasmic production system aberrant splice sites in the transgene or elsewhere do not impair retroviral vector structure and hence do not lead to transduction of truncated genes as observed with a classically produced retroviral vector in a clinical study, in which most of the patients’ transduced cells had a truncated multidrug resistance type 1 gene due to an aberrant splice site (1, 23). In conclusion, a scalable production system for retroviral vectors with complex gene cassettes was presented which, optimized for expression and safety in humans, may add significant value to gene therapy.
Acknowledgments
We thank U. Schlokat for plasmid pCMV-FIX and CHO cell line 500-2, G. Antoine for oligonucleotide synthesis and sequencing, N. Makarov for the FIX ELISA assays, and B. Ober and I. Livey for critically reading the manuscript and for helpful discussions.
REFERENCES
- 1.Abonour, R., D. A. Williams, L. Einhorn, K. M. Hall, J. Chen, J. Coffman, C. M. Traycoff, A. Bank, I. Kato, M. Ward, S. D. Williams, R. Hromas, M. J. Robertson, F. O. Smith, D. Woo, B. Mills, E. F. Srour, and K. Cornetta. 2000. Efficient retrovirus-mediated transfer of the multidrug resistance 1 gene into autologous human long-term repopulating hematopoietic stem cells. Nat. Med. 6:652–658. [DOI] [PubMed] [Google Scholar]
- 2.Andreadis, S. T., C. M. Roth, J. M. Le Doux, J. R. Morgan, and M. L. Yarmush. 1999. Large-scale processing of recombinant retroviruses for gene therapy. Biotechnol. Prog. 15:1–11. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, P. N., A. Mitterer, W. Mundt, J. Eibl, M. Eibl, R. C. Gallo, B. Moss, and F. Dorner. 1989. Large-scale production and purification of a vaccinia recombinant-derived HIV-1 gp160 and analysis of its immunogenicity. AIDS Res. Hum. Retrovir. 5:159–171. [DOI] [PubMed] [Google Scholar]
- 4.Brinster, R. L., J. M. Allen, R. R. Behringer, R. E. Gelinas, and R. D. Palmiter. 1988. Introns increase transcriptional efficiency in transgenic mice. Proc. Natl. Acad. Sci. USA 85:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchman, A. R., and P. Berg. 1988. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 8:4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p.1767–1847. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3 ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 7.Dzierzak, E. A., T. Papayannopoulou, and R. C. Mulligan. 1988. Lineage-specific expression of a human beta-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature 331:35–41. [DOI] [PubMed] [Google Scholar]
- 8.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamer, D. H., and P. Leder. 1979. Splicing and the formation of stable RNA. Cell 18:1299–1302. [DOI] [PubMed] [Google Scholar]
- 10.Holzer, G. W., J. A. Mayrhofer, W. Gritschenberger, F. Dorner, and F. G. Falkner. 1999. Poxviral/retroviral chimeric vectors allow cytoplasmic production of transducing defective retroviral particles. Virology 253:107–114. [DOI] [PubMed] [Google Scholar]
- 11.Huang, M. T., and C. M. Gorman. 1990. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismail, S. I., S. M. Kingsman, A. J. Kingsman, and M. Uden. 2000. Split-intron retroviral vectors: enhanced expression with improved safety. J. Virol. 74:2365–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson, S., T. Papayannopoulou, S. G. Schweiger, G. Stamatoyannopoulos, and A. W. Nienhuis. 1987. Retroviral-mediated transfer of genomic globin genes leads to regulated production of RNA and protein. Proc. Natl. Acad. Sci. USA 84:2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, K. J., and H. Garoff. 1998. Packaging of intron-containing genes into retrovirus vectors by alphavirus vectors. Proc. Natl. Acad. Sci. USA 95:3650–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, A. D. 1997. Development and applications of retroviral vectors. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 16.Miller, A. D., M. A. Bender, E. A. Harris, M. Kaleko, and R. E. Gelinas. 1988. Design of retrovirus vectors for transfer and expression of the human beta-globin gene. J. Virol. 62:4337–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss, B. 1996. Poxviridae: the viruses and their replication. In B. N. Fields, D. M. Knipe, R. M. Chanock, J. Melnick, B. Roizman, and R. Shope (ed.), Fields virology. Raven Press, Philadelphia, Pa.
- 19.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730–732. [DOI] [PubMed] [Google Scholar]
- 20.Roth, D. A., N. E. Tawa, Jr., J. M. O’Brien, D. A. Treco, and R. F. Selden. 2001. Nonviral transfer of the gene encoding coagulation factor VIII in patients with severe hemophilia A. N. Engl. J. Med. 344:1735–1742. [DOI] [PubMed] [Google Scholar]
- 21.Urlaub, G., and L. A. Chasin. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. USA 77:4216–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahlfors, J. J., and R. A. Morgan. 1999. Production of minigene-containing retroviral vectors using an alphavirus/retrovirus hybrid vector system. Hum. Gene Ther. 10:1197–1206. [DOI] [PubMed] [Google Scholar]
- 23.Ward, M., P. Pioli, J. Ayello, R. Reiss, G. Urzi, C. Richardson, C. Hesdorffer, and A. Bank. 1996. Retroviral transfer and expression of the human multiple drug resistance (MDR) gene in peripheral blood progenitor cells. Clin. Cancer Res. 2:873–876. [PubMed] [Google Scholar]
- 24.Yuen, L., and B. Moss. 1987. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc. Natl. Acad. Sci. USA 84:6417–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]