Abstract
The availability of a draft human genome sequence and ability to monitor the transcription of thousands of genes with DNA microarrays has necessitated the need for new computational tools that can analyze cis-regulatory elements controlling genes that display similar expression patterns. We have developed a tool designated EZ-Retrieve that can: (i) retrieve any particular region of human genome sequence from the NCBI database and (ii) analyze retrieved sequences for putative transcription factor-binding sites (TFBSs) as they appear on the TRANSFAC database. The tool is web-based, user-friendly and offers both batch sequence retrieval and batch TFBS prediction. A major application of EZ-Retrieve is the analysis of co-expressed genes that are highlighted as expression clusters in DNA microarray experiments.
INTRODUCTION
Genomic sequencing projects and the vast amount of expression data generated by DNA microarray technology have provided a framework that may seed our understanding and modeling of cellular physiology (1–4). In a typical data series derived from DNA microarray experiments, distinct groups of genes with similar expression profiles are identified when a suitable clustering algorithm is applied. Common cis-regulatory modules (CRM) shared among genes within a cluster are thought to be instrumental in dictating the observed similarity in the gene expression pattern (5,6). The expression of any particular gene is dependent on interactions of multiple transcription factors binding cooperatively to distinct promoter and enhancer elements. In the case of higher eukaryotic organisms, CRMs are typically located upstream and/or downstream of the protein coding sequences (7). Therefore, it is necessary to obtain these sequences from the genes of interest and identify common regulatory regions that may define an expression cluster.
In model eukaryotes such as Drosophila melanogaster and Saccharomyces cerevisiae, upstream and/or downstream gene sequences can be retrieved from GadFly at the Berkeley Drosophila Genome Project (BDGP: http://www.fruitfly.org/) and Gene/Sequence Resources at the Saccharomyces Genome Database (SGD: http://genome-www.stanford.edu/Saccharomyces/), respectively. Users can retrieve the sequence of interest either by specifying the flanking sequence length of the gene or by giving certain coordinates on the corresponding chromosome. However, such tools are not available for the retrieval of specified regions of the human genome sequence with respect to a specific gene. NCBI Entrez offers a batch retrieval tool for GenBank sequences, but the user cannot specify coordinates for sequences to be retrieved. Here we present EZ-Retrieve (http://www.cag.icph.org/bioinformatics.html), a web-based tool that can retrieve any particular region of a gene from the draft human genome sequence by specifying coordinates relative to the gene’s transcriptional start site. Furthermore, EZ-Retrieve can batch submit a cluster of sequences to the TFSEARCH server (http://www.cbrc.jp/research/db/TFSEARCH.html), which in turn can search for the putative transcription factor-binding sites (TFBSs) in the query sequences based on the database known as TRANSFAC (8). Search results are summarized in a tabular form and presented directly on the client’s web browser. This tool is not only useful for the retrieval and analysis of co-expressed genes from a set of DNA microarray experiments but also for studying the structural features of the 5′ regulatory regions of all the genes in a genome. As an example of retrieving and analyzing human sequences using the EZ-Retrieve tool, we present the retrieval of regulatory sequences and TFSEARCH of a family of genes that are believed to be under the control of the E2F transcription factor. We also demonstrate the utility of using EZ-Retrieve to submit multiple sequences from other organisms such as Drosophila to TFSEARCH in order to underscore putative TFBSs.
MATERIALS AND METHODS
Sequence retrieval from NCBI
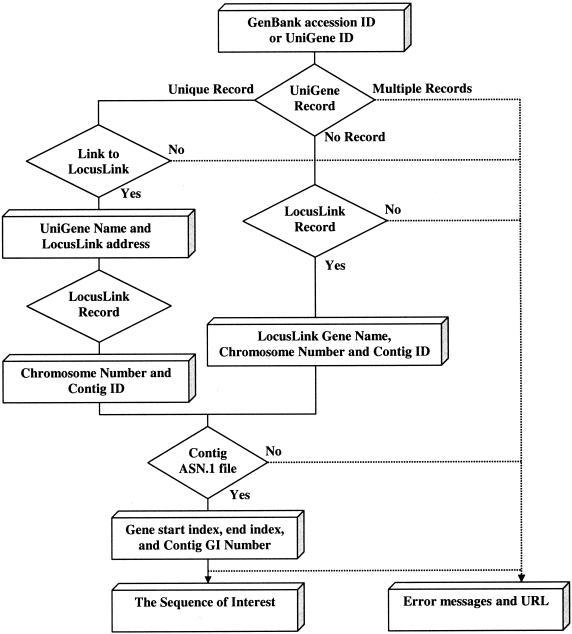
EZ-Retrieve can perform both single and multiple sequence retrievals. The retrieving strategy is based on the NCBI data model and NCBI UniGene (http://www.ncbi.nlm.nih.gov/UniGene) and LocusLink annotations (9). It mainly utilizes the annotation of the corresponding gene as it appears in the Abstract-Syntax-Notation-One (ASN.1: http://www.ncbi.nlm.nih.gov/Sitemap/Summary/asn1.html) file of the contig it locates. This provides information about the gene’s coordinates within the contig that defines the corresponding region of the human genome sequence. For genes that are not annotated in LocusLink and have not been successfully mapped back to the chromosomes, the corresponding requested sequences will not be retrieved and EZ-Retrieve will automatically report this to the end user. The start position (index ‘+1’) is determined by the ‘from’ feature for a gene in the ASN.1 file, if the gene is located on the ‘plus’ strand, or by the ‘to’ feature if the gene is located on the ‘minus’ strand. The sequence retrieved is in draft genomic context. When performing multiple sequence retrievals, a time interval of 5 s is given between queries to reduce network traffic at NCBI. The retrieval process is illustrated in Figure 1. Retrieval results are displayed via an Hyper Text Markup Language (HTML) web page where the retrieved sequences can be downloaded.
Figure 1.
Scheme of sequence retrieval from NCBI. The retrieval process (solid lines) for one gene is shown. Different NCBI resources (diamonds) are queried and useful information (boxes) is obtained. Error messages (dashed lines) generated during the retrieval process are returned to the client. The name of the target gene is derived from either UniGene or LocusLink annotations. If the target gene has not been annotated by either UniGene or LocusLink, error messages are returned. Chromosome Number and Contig ID are based on LocusLink annotation. Gene start index, end index and Contig GI numbers are assigned by the Contig ASN.1 file.
In the current version of EZ-Retrieve, the retrieved sequences are not RepeatMasked (A. F. A. Smit and P. Green, RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html). Thus, sequences retrieved for downstream analysis should be used with caution because interspersed DNA repeats and low complexity DNA might be present. This functionality will be provided in the next version of EZ-Retrieve.
Searching and underscoring TFBSs
EZ-Retrieve also provides a multiple or batch TFSEARCH tool where the user can input a cluster of distinct sequences in FASTA format and submit them to TFSEARCH using the TRANSFAC database (release 3.3). Search results are summarized and presented as a table that lists all TFBSs present in the entire group of submitted sequences. The total number of occurrences of each TFBS in each sequence is also presented in the table. This allows an overview of the possible common regulatory elements within a cluster of sequences. TFSEARCH can also be submitted directly with the sequence(s) retrieved using the single or multiple retrieval tools.
There are two parameters that need to be set for TFSEARCH: (i) taxonomy matrix and (ii) threshold. When doing human sequence retrieval and analysis, the default taxonomy matrix is set to ‘vertebrate’. However, when using the batch TFSEARCH tool for analysis of sequences from other species, an appropriate taxonomy matrix needs to be chosen. The threshold parameter is dependent on the score calculated by the following formula:
Score = 100.0 ∗ (‘weighted sum’ – min)/(max – min)
where ‘max’ and ‘min’ are the sum of possible maximum or minimum values of each position of the weighted matrix, respectively. The ‘weighted sum’ is the value calculated by comparing the sequence being evaluated to the weighted matrix. The scoring scheme is a gauge of how well a string matches with the pattern specified by the weighted matrix. The default value for the threshold is 85.0. Setting the threshold as high as 100.0 will force TFSEARCH to find only sequences that perfectly match letters that will give maximum scores at each position within the weighted matrix. Since there is no probability involved, the score does not reflect a statistical significance. A detailed explanation and an example of a TFSEARCH score calculation can be found on the EZ-Retrieve web sever.
Table 1 gives an overview of available tools at EZ-Retrieve. When doing multiple queries on TFSEARCH, a time interval of 2 s is set between each search to reduce network traffic on the TFSEARCH server. Results are displayed in a tabular form via an HTML web page and can also be downloaded as Microsoft Excel files.
Table 1. Tools at EZ-Retrieve.
| Tools | Description |
|---|---|
| Single retrieval | Input one GenBank accession ID, retrieve a single sequence, result can be submitted to TFSEARCH directly |
| Multiple retrievals | Input a batch of GenBank accession IDs, retrieve a batch of sequences, results can be submitted to TFSEARCH directly |
| Multiple TFSEARCH | Submit a batch of sequences to TFSEARCH |
Programming
Programming was performed using JAVA™ (build 1.3.1_01, http://java.sun.com/). Website construction was performed using JSP™ (JavaServer Pages™), HTML, and javascript. The servlet container is powered by Jakarta™ Tomcat 4.0 (http://jakarta.apache.org/tomcat/tomcat-4.0-doc/index.html).
Availability
EZ-Retrieve is available at http://www.cag.icph.org/ bioinformatics.html. The web service is interactive and relies on NCBI and TFSEARCH for sequence retrievals and TFBS searches, respectively.
RESULTS
Sequence retrievals and TFSEARCH
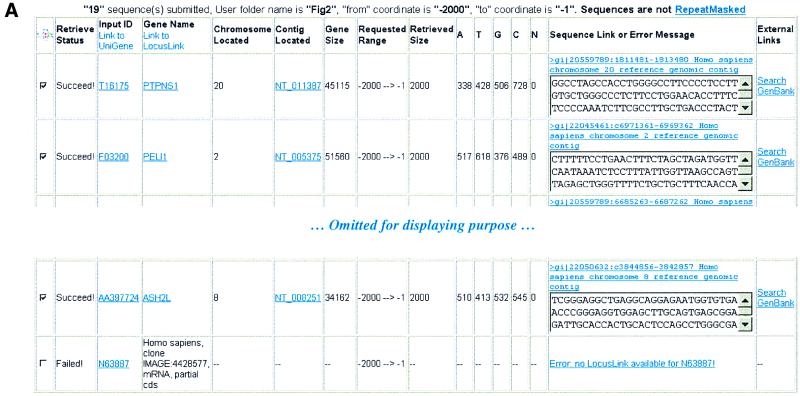
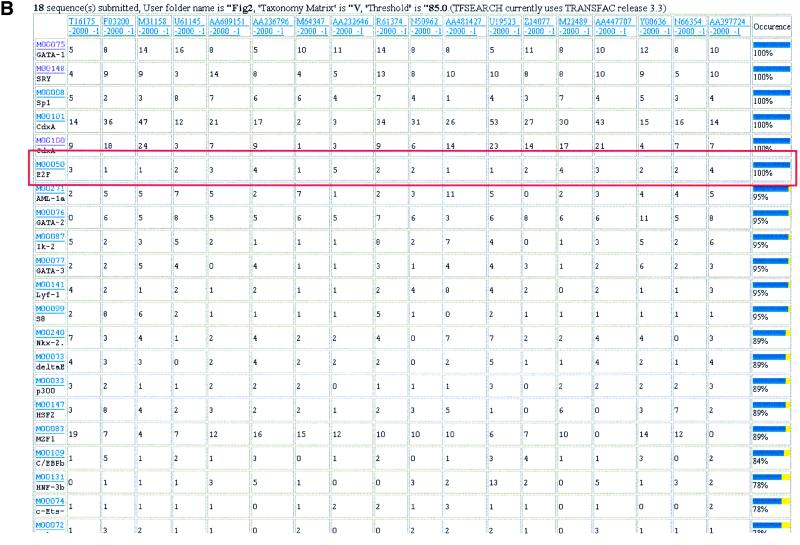
To exemplify the utility of EZ-Retrieve, we first focused attention to the E2F family of transcription factors since they are known to regulate the expression of several genes, especially those involved in cell cycle progression and DNA synthesis (10). These site-specific DNA-binding proteins recognize the consensus sequence 5′ TTT(G/C)(G/C)CGC-3′ present in the promoter region of the genes that they regulate. Among the many human transcripts that have been shown by microarray experiments to be induced by ectopic expression of E2F1, E2F2 and E2F3, 19 have been confirmed by northern blot analysis (11). In Figure 2A, we demonstrate the use of EZ-Retrieve to successfully retrieve 2 kb of DNA sequences located directly upstream of the transcriptional start site of all but one of these transcripts. The single failure was expected as this transcript represents a partial cDNA sequence, not an annotated gene. Furthermore, we searched for putative TFBSs on all these 18 retrieved sequences using the EZ-Retrieve multiple TFSEARCH tool. The results of this search are displayed in Figure 2B where all 18 of the retrieved 2 kb upstream regions display putative E2F-binding sites.
Figure 2.
Analysis of E2F family using EZ-Retrieve. (A) Output for multiple sequence retrievals. Relative information is shown in tabular form, including links to UniGene and LocusLink, the chromosome and the contig harboring the queried sequence, the gene size, range and size of retrieval, base composition, sequence of interest if the retrieval succeeds with discription from which human genome sequence the retrieved regions were derived, error message if it fails, and a link to GenBank records. In this example, 19 queries were submitted and 18 were successfully retrieved (only four rows are shown for display purposes). (B) TFSEARCH output following submission of the 18 successfully retrieved sequences. Predicted TFBSs are represented by rows, and queried sequences are represented in columns. TFBSs are searched in all sequences. Occurrences for each predicted TFBS are counted and shown in each cell of the table. The frequency of occurrence of each putative TFBS in all sequences are calculated and shown in the last column. Rows are ranked by the frequency of occurrence of the corresponding TFBS. Links to both transcription factor description pages and to original TFSEARCH results for each sequence are provided. Only putative TFBSs with a frequency of occurrence >78% are displayed. The result row of E2F is highlighted by a red box.
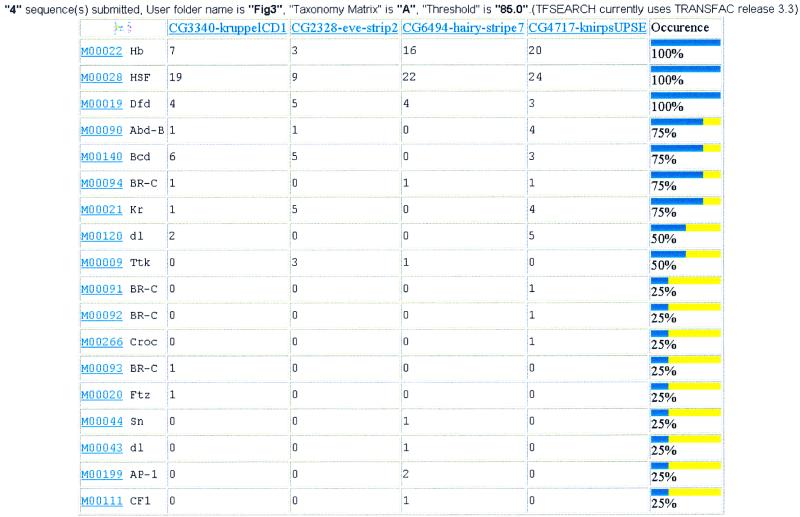
Although EZ-Retrieve is designed to retrieve only human sequences, the multiple TFSEARCH tool can be used to search for TFBSs in sequences derived from any species when those sequences are available in FASTA format. To the best of our knowledge, no such tool is available for multiple transcription factor searches. A sample output of a TFBS search and analysis using EZ-Retrieve multiple TFSEARCH tool is shown in Figure 3. In this example, four CRMs from Drosophila were retrieved individually from GadFly at BDGP and analyzed with the EZ-Retrieve multiple TFSEARCH tool for putative TFBSs. The result table displays seven TFBSs (Hb, HSF, Dfd, Abd-B, Bcd, BR-C, Kr) that appear in at least 75% of all CRMs. Three of these (Hb, Bcd, Kr) TFBSs have been experimentally shown to drive domain-specific expression within the Drosophila embryo as summarized in supporting Table 1 in Berman et al. (6) (http://www.pnas.org/cgi/content/full/99/2/757/DC1/1).
Figure 3.
Output of the EZ-Retrieve multiple TFSEARCH tool with Drosophila sequences. See legend of Figure 2B for a description of table results.
DISCUSSION
Gene predictions based on the draft human genome sequence suggest that only a very small fraction of the genome consists of protein coding sequences (12). A significant portion of the genome is presumably involved in controlling gene expression and the elementary building blocks of discrete regulatory regions are different TFBSs. It is the different combination and order of these building blocks that determine gene expression (13,14). By searching for similar DNA elements in the upstream region of all the genes in a sequenced genome, one can predict common regulatory motifs that may control a group of genes. However, the mere presence of a regulatory motif common to a group of genes does not mean co-regulation as most TFBSs occur in many promoters (15). This problem can be overcome by correlating the predicted TFBSs that are common to genes that show co-expression in a series of microarray experiments (16). In this context, EZ-Retrieve is a helpful tool both in terms of sequence retrieval and putative TFBS searches and alignments. Although many known TFBSs cluster in the vicinity of a transcription start site, higher eukaryotes such as humans often contain enhancer regions that extend to more distant locations (15). EZ-Retrieve thus offers flexibility in choosing the exact coordinates (length and position) of the desired sequences while searching for TFBSs.
EZ-retreive operates in terms of finding the requested regions even when gaps of unspecified length are encountered in the human genome sequence. Since gene loci are annotated within the context of genome contigs, it is the contig boundaries that define how far the requested region can be retrieved. If a gene is located at either end of a contig and a gap exists between this and the adjacent contig, the retrieved sequence will be shorter than requested and EZ-Retrieve will report this to the user.
A recent paper describes a program called PEG developed by Zhang and Zhang (17) that can extract promoter sequences for large sets of genes using information present in GenBank. PEG is a perl program that automatically extracts eukaryotic promoter sequences based on GenBank annotations. It functions to analyze promoter regions and is limited to retrieving 5 kb of upstream sequences. In addition to the PEG program itself in a UNIX environment, the user also requires a locally installed NCBI BLAST program and several other perl modules and expertise and experience to set up several thresholds. This requires users to run PEG several times for the same query.
EZ-Retrieve is based on NCBI UniGene, RefSeq and LocusLink projects. Because of the volume of work that has been done to correct and annotate data by these projects, sequences retrieved by EZ-Retrieve are much more accurate than those originally submitted to GenBank. This program can retrieve any region without a limit specified by coordinates relative to the transcription start site including both upstream and downstream sequences. It is web-based, user friendly and does not require installation. Thus, users can access EZ-Retrieve anytime, anywhere with a web browser.
PEG is slower and ‘noisier’ than EZ-Retrieve because it spends more computational power locating the promoter and possible alternative or orthologous promoter sequences. EZ-Retrieve only spends time on querying and parsing UniGene, LocusLink, and the ASN.1 file of the genomic contig that the gene is located on. Therefore, EZ-Retrieve is a faster and a more accurate tool to retrieve genomic sequences. Conversely, PEG is slower but a powerful tool for eukaryotic promoter discovery and comparative analysis. Thus, when doing promoter analysis, EZ-Retrieve is the recommended tool to obtain desired sequences if the coordinates for the region of interest are known. PEG is recommended if the genes of interest are not yet annotated by RefSeq and LocusLink or if the user is interested in possible alternative or orthologous promoters.
Future directions of EZ-Retrieve include: (i) utilizing locations of matrix matches to do position-specific cluster analysis of putative TFBSs; and (ii) providing batch job submissions to other downstream programs or servers so that EZ-Retrieve becomes a sequence-retrieval-centered pipeline for data analysis.
Acknowledgments
ACKNOWLEDGEMENTS
This work is supported by NIH grant CA83213 from the National Cancer Institute awarded to P.P.T. The Center for Applied Genomics is supported in part with R&D Excellence grant 00-2042-007-21 from the New Jersey Commission on Science and Technology awarded to P.P.T.
REFERENCES
- 1.Bussemaker H.J., Li,H. and Siggia,E.D. (2001) Regulatory element detection using correlation with expression. Nature Genet., 27, 167–171. [DOI] [PubMed] [Google Scholar]
- 2.Jakt L.M., Cao,L., Cheah,K.S. and Smith,D.K. (2001) Assessing clusters and motifs from gene expression data. Genome Res., 11, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujibuchi W., Anderson,J.S. and Landsman,D. (2001) PROSPECT improves cis-acting regulatory element prediction by integrating expression profile data with consensus pattern searches. Nucleic Acids Res., 29, 3988–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramanathan Y., Zhang,H., Aris,V., Soteropoulos,P., Aaronson,S.A. and Tolias,P.P. (2002) Functional cloning, sorting and expression profiling of nucleic-acid binding proteins. Genome Res., 12, 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M.Q. (1999) Large-scale gene expression data analysis: a new challenge to computational biologists. Genome Res., 9, 681–688. [PubMed] [Google Scholar]
- 6.Berman B.P., Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen A.G., Baldi,P., Chauvin,Y. and Brunak,S. (1999) The biology of eukaryotic promoter prediction—a review. Comput. Chem., 23, 191–207. [DOI] [PubMed] [Google Scholar]
- 8.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhauser,R., Pruss,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller H. and Helin,K. (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta, 1470, M1–M12. [DOI] [PubMed] [Google Scholar]
- 11.Muller H., Bracken,A.P., Vernell,R., Moroni,M.C., Christians,F., Grassilli,E., Prosperini,E., Vigo,E., Oliner,J.D. and Helin,K. (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation and apoptosis. Genes Dev., 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 13.Smale S.T. (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev., 15, 2503–2508. [DOI] [PubMed] [Google Scholar]
- 14.Werner T. (1999) Models for prediction and recognition of eukaryotic promoters. Mamm. Genome, 10, 168–175. [DOI] [PubMed] [Google Scholar]
- 15.Werner T. (2001) The promoter connection. Nature Genet., 29, 105–106. [DOI] [PubMed] [Google Scholar]
- 16.Pilpel Y., Sudarsanam,P. and Church,G.M. (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nature Genet., 29, 153–159. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T. and Zhang,M. (2001) Promoter extraction from GenBank (PEG): automatic extraction of eukaryotic promoter sequences in large sets of genes. Bioinformatics, 17, 1232–1233. [DOI] [PubMed] [Google Scholar]