Abstract
The sequence of the unique short (US) region of monkey B virus (BV) was determined. The 13 genes identified are arranged in the same order and orientation as in herpes simplex virus (HSV). These results demonstrate that the BV US region is entirely colinear with that of HSV type 1 (HSV-1), HSV-2, and simian agent 8 virus.
Cercopithecine herpesvirus 1 (monkey B virus [BV]) is a member of the alphaherpesvirus subfamily indigenous in Asiatic macaque monkeys. The natural history of BV in macaques is similar to that of the human herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) in humans, of simian agent 8 virus (SA8) in green monkeys, and of Herpesvirus papio 2 (HVP-2) in baboons (11, 12, 38). When transmitted to nonmacaque primates, BV produces severe infections which usually involve the central nervous system and are frequently fatal (31, 37, 38). The molecular basis for the extreme neuropathology of BV in nonmacaque species is an intriguing question that remains unanswered.
While sequences for a number of genes of BV have been reported (3, 9, 23, 34, 35, 37), genomic characterization of BV has largely been limited to restriction analysis and gene mapping by hybridization with HSV gene probes (10, 16, 17). Such studies have suggested that, for the most part, the BV genome is colinear with that of HSV. However, it has been reported that the unique short (US) region of the BV genome may not be colinear with that of HSV (16). Here we present the sequence of the US region of the BV genome and its genetic layout relative to that of HSV and SA8.
The DNA sequence of the BV rhesus genotype (BVrh) strain E2490 (35) US region was determined by a combination of cloning restriction fragments of the genome and PCR amplification of small gaps in the sequence with genomic BV DNA as a template. The BVrh US region (GenBank accession no. AB 077432) is 14,447 nucleotides long, slightly longer than the 12,980 bp reported for HSV-1 strain 17 (26) but close to the 14,329-bp HSV-2 US region (6). The BVrh US sequence presented here does not include sequences aligning with the N-terminal 2 codons of the US12 open reading frame (ORF). The overall G+C content of the BVrh US region is 73.2%, close to the 75% G+C estimated for the entire BV genome (19). The coding sequences in the US region have a combined G+C content of 74.4%, a value somewhat higher than that for the noncoding intergenic regions (68.9%). BV exhibits a strong bias toward use of codons with G or C in the third position (89.6%) and Arg and Leu codons with C in the first position (93.4%). While this GC bias is strong, it is not as extreme as in the SA8 US region (76.8% G+C overall, 92.9% GC in the third position, 97.2% C in the first position of the Arg and Leu codons [13]).
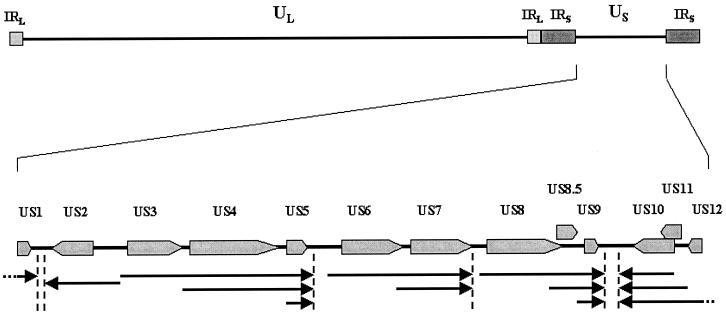
Analysis of the BVrh US sequence for ORFs, homology of predicted amino acid sequences with HSV polypeptides, promoter and transcriptional initiation sites, and mRNA termination sites [consensus poly(A) signals associated with mRNA termination motifs] indicates that the genetic layout of the BVrh US region is very similar to those of HSV-1 and HSV-2 and is identical to that of SA8 (Fig. 1). As summarized in Table 1, 13 ORFs corresponding to HSV US1 to US12 and US8.5 are all present in BV. All ORFs occur in the same order and orientation as in HSV and SA8. This is in contrast to results reported by others (16), which indicated that while ORFs analogous to those of HSV were present in the BV cynomolgus genotype (BVcy) US region, they were arranged in a different order. Based on poly(A) and mRNA termination signals, the transcriptional grouping of BV US mRNAs is identical to that reported for SA8 (13) and is probably identical to those for all BV genotypes and HVP-2 as well (9, 35). Thus, mRNAs for US3/US4/US5 and US6/US7 form two 3′-coterminal transcriptional sets in all of the simian viruses, while in HSV the transcriptional groupings of these genes are US3/US4 and US5/US6/US7. Transcriptional groupings of all other BV US genes are identical to that found in HSV. Analysis of the BV US sequence for additional ORFs associated with transcriptional elements and/or homologous unidentified genes in SA8 did not indicate that any such unidentified genes exist.
FIG. 1.
Genetic organization of the BV strain E2490 US region. ORFs predicted to encode proteins and their orientations are indicated by large arrows. The locations of transcriptional termination signals (AATAAA, followed by GT tracts) are indicated by vertical dashed lines. The proposed mRNA transcriptional map of the region is indicated by arrows located below the ORF map. IRL, inverted long repeat; IRS, inverted short repeat.
TABLE 1.
Properties of BVrh ORFs located in the US region
| BVrh E2490a
|
Function of the encoded protein | % Identity/% similarity of BV proteins with homologous proteinsb of:
|
||||
|---|---|---|---|---|---|---|
| ORF | Size (aa) | Predicted MW | HSV-1 | HSV-2 | SA8 | |
| US1 | 373 | 40,700 | Immediate-early protein ICP22 | 32/42 (420) | 34/45 (413) | NA |
| US2 | 302 | 32,900 | Unknown | 52/58 (291) | 48/57 (291) | NA |
| US3 | 379 | 41,600 | Serine/threonine protein kinase | 51/59 (481) | 52/59 (481) | 83/86 (377) |
| US4 | 673 | 67,900 | Glycoprotein gG | 9/12 (238) | 34/44 (699) | 53/61 (602) |
| US5 | 121 | 12,100 | Glycoprotein gJ | 26/36 (92) | 18/30 (92) | 51/58 (106) |
| US6 | 395 | 42,600 | Glycoprotein gD | 56/67 (394) | 55/68 (393) | 80/84 (395) |
| US7 | 401 | 42,000 | Glycoprotein gI | 43/51 (390) | 46/54 (372) | 66/69 (399) |
| US8 | 539 | 58,000 | Glycoprotein gE | 44/55 (550) | 44/55 (545) | 72/78 (540) |
| US8.5 | 122 | 12,600 | Unknown | 28/40 (159) | 31/43 (146) | 60/61 (102) |
| US9 | 90 | 9,900 | Envelope phosphoprotein | 57/69 (90) | 57/69 (89) | 75/84 (91) |
| US10 | 311 | 34,000 | Tegument protein | 36/46 (312) | 42/50 (302) | 84/91 (93)c |
| US11 | 145 | 16,300 | RNA-binding protein | 41/45 (161) | 38/40 (151) | NA |
| US12 | >78 | Immediate-early protein ICP47 | 21/33 (88) | 20/30 (86) | NA | |
aa, amino acids; MW, molecular weight.
Values are percentages of amino acid identity and percentages of amino acid similarity in pairwise alignment with the BV homologue. Numbers in parentheses represent the numbers of amino acid residues comprising the homologous viral protein. NA, sequence not available.
Only a partial sequence was available.
The US1 ORF encodes the immediate-early (alpha) regulatory protein ICP22 (6, 26). The BV US1 protein is somewhat smaller than the HSV-1 and HSV-2 proteins (Fig. 2). Although the overall amino acid sequence homology is low, there are several distinct regions that are strongly conserved. Similarly, the N-terminal 70% of the US2 protein is strongly conserved among BV, HSV-1, and HSV-2, having regions of identical sequence separated by stretches of more variant sequence. This is consistent with the noted conservation of the US2 gene in mammalian and avian alphaherpesviruses (21, 28).
FIG. 2.
Alignment of immediate-early ICP22 (US1) polypeptides. Residues which are identical in all three sequences are shown with a black background; conservative residues are with a gray background. Degenerate repeats in the BV and HSV-1 sequences are identified by underlining. The Met in the HSV-1 sequence at which the proposed US1.5 protein initiates is indicated by “▴” below the aligned sequences. Note that neither BV nor HSV-2 have a Met residue near this HSV-1 Met codon.
In HSV-1, it has been reported that a second polypeptide, designated US1.5, is synthesized from the US1 ORF by initiation at an internal in-frame Met codon (4). There is no similar Met codon in BV or HSV-2 located near this HSV-1 Met codon (Fig. 2). Given the close phylogenetic relatedness of HSV-2 and BV to HSV-1, it is curious that HSV-1 is the only one of these three viruses which appears to synthesize a US1.5 protein.
Between the US2 and US3 ORFs is an intergenic region of 556 nucleotides in which should lie the promoters for both genes. Only one potential promoter is apparent, located ca. 440 bp 5′ of the US2 start codon. This same element likely serves as the US3 promoter on the complementary strand as well. Use of this promoter would result in a fairly long 5′ untranslated region (412 bp) on the US2 mRNA which is considerably longer than the 5′ untranslated sequence predicted for all other BV US genes (50 to 200 bp) or the HSV-1 and HSV-2 US2 mRNAs (∼290 bp).
The US3 proteins are serine/threonine protein kinases (25). Within the strongly conserved C-terminal 75% of the protein are the active sites for phosphorylation activity and ATP binding. Relative to the US3 proteins of HSV-1 and HSV-2, the simian virus US3 proteins are ca. 100 amino acids shorter. As for the US1 protein, ca. 80 of these “deleted” residues represent the N terminus of the polypeptide. Another 15 to 20 residues not present in the simian virus US3 polypeptide follow a 20- to 25-residue block of acidic residues located in the N-terminal region of the HSV US3 polypeptides. In this respect, the simian herpesvirus US3 polypeptides are more similar to those of varicella-zoster virus rather than to those of HSV (5, 15).
As in other primate alphaherpesviruses, US4 to US8 all encode glycoproteins. The US4 ORF encodes a glycoprotein designated gG (27), which varies considerably among these viruses. All of the simian virus gGs are more similar to the gG of HSV-2 than to that of HSV-1, having a hydrophobic N-terminal signal sequence, two distinct regions of the extracellular domain separated by a proteolytic cleavage site, a transmembrane domain, and a highly charged cytoplasmic tail (27, 36, 34). The more N-terminal of the extracellular domains is relatively conserved in sequence, with six Cys residues and one N-linked glycosylation site being positionally conserved. In contrast, the more membrane-proximal extracellular domain exhibits considerable variation in size and very little sequence identity between viruses. Nonetheless, all have a stretch of ca. 20 acidic residues located some 70 residues from the transmembrane domain, two to four additional N-glycosylation sites, and a region of high Pro, Ser, and Thr content typical of regions where O-linked carbohydrate residues are added that is located between the proteolytic cleavage site and the acidic domain.
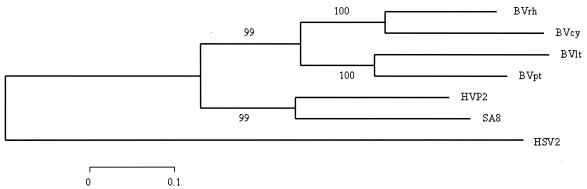
Sequencing of the US4 gene from cynomologous, pigtail, and lion-tailed macaque BV isolates (35, 37) indicates that, in addition to the high sequence variability between gGs of different viruses, the gG polypeptide also shows considerable sequence variation among different BV genotypes. The greatest genotypic variation occurs in the same regions where sequence variation occurs between BV and other primate virus gGs. Phylogenetic analysis of the US4 coding sequences (Fig. 3) predicts a relationship among BV genotypes very similar to that previously described based primarily on analyses of intergenic and US5 coding sequences (35, 37). The BVrh sequence also contains a six-times-repeated 6-residue motif (PAPTTT) located in the high-Pro/Ser/Thr-content region of the polypeptide, much like a similar repeat sequence observed in the US7 (gI) gene of HSV-1 (26). This repeat is not present in any of the other sequenced BV gGs. PCR amplification of BVrh genomic DNA indicates that this repeat is not a cloning artifact but rather is unique to the E2490 strain of BV.
FIG. 3.
Phylogenetic relationship of BV genotypes based on the US4 gG coding sequences. Predicted amino acid sequences were aligned and phylogenetic analyses performed as described previously (35). The tree shown was generated by using gamma distance estimates and neighbor-joining tree construction. Values represent bootstrap confidence intervals. BV genotypes isolated from rhesus (BVrh), cynomolgus (BVcy), lion-tailed (BVlt), and pigtailed (BVpt) macaques are indicated.
The US5 gene encodes a small glycoprotein designated gJ that has been reported to inhibit apoptosis (20, 40). Multiple alignment of 17 simian virus gJ sequences indicates that, unlike other herpesvirus glycoproteins, the two hydrophobic regions representing the signal sequence and transmembrane domain are the most conserved regions of the polypeptide. This is most evident when gJ sequences for different BV genotypes are aligned, since genotypic differences occur in the extracellular domain where two to three potential N-glycosylation sites occur. As for gG, one BVrh isolate was found to have reiterated sequences in the variable region of the extracellular domain (37).
The US6 to US8 genes encode glycoproteins gD, gI, and gE. The gD glycoprotein encoded by US6 is strongly conserved among the primate viruses and shows no significant genotypic variation among BV genotypes (3, 35, 37). The US7 gene encodes the gI glycoprotein, which complexes with the gE glycoprotein encoded by US8 to form an Fc receptor (22). Both gI and gE exhibit variable and conserved regions. In the extracellular domain of gI there is a region of highly variable sequence with a high content of Pro/Ser/Thr residues, suggesting modification by O-linked glycosylation. Although only the N-terminal 110 amino acids are available for the BVcy gI (3), only 1 amino acid difference exists between the BVrh and BVcy sequences, indicating that in this variable region there is not significant genotypic variation. A number of potential phosphorylation sites are present in the cytoplasmic tails of the BV and SA8 gI polypeptides, but only one is positionally conserved between the simian and human virus gI sequences (30). Interestingly, the domains of the gI and gE polypeptides that interact with immunoglobulin G and are involved in complexing of gI with gE (1, 2, 7) are not strongly conserved among the different primate virus gI polypeptides.
In contrast to gG, the N-terminal half of the gE extracellular domain is highly variable in sequence among the primate viruses, with the exception of a small, highly conserved stretch of 24 residues flanked by two Cys residues. Nearer the transmembrane region, the extracellular domain is more conserved, with seven of the nine conserved Cys residues being located within it. The transmembrane domain itself is fairly strongly conserved and, like the transmembrane domain of gI, has two conserved Cys residues near the cytoplasmic side. The long, highly charged cytoplasmic tail is less conserved, although both a series of basic residues adjacent to the transmembrane domain and an area rich in acidic residues are present in all of the primate virus gE polypeptides. In comparing the partial sequence for the BVcy US8 gene (23), it appears that genotypic variation does occur in the cytoplasmic tail of gE.
The function of the small protein encoded by the US8.5 gene is unknown, but it is phosphorylated and localized in the nucleoli of infected cells (14). As in SA8, the BV US8.5 ORF overlaps the 3′ end of the US8 ORF by 53 nucleotides. The N-terminal 70% of the US8.5 polypeptide is highly variable between the human and simian viruses but is fairly conserved between BV and SA8 (6). In contrast, sequence of the C-terminal region of the polypeptide is strongly conserved among all of the primate viruses. By deletion of one nucleotide that otherwise induces a frameshift, the published BVcy US8.5 sequence (23) is nearly identical to that of BVrh, suggesting that genotypic variation does not occur in US8.5.
The US9 gene encodes a small type II membrane protein found in the virion envelope and, as previously noted (23), is conserved among the primate viruses, especially in the region surrounding the tyrosine kinase phosphorylation sites (YY). Again, the BVcy and BVrh US9 sequences show little variation, being 93.3% identical and 97.8% similar to each other.
The US10 gene encodes a phosphoprotein of unknown function that is located in the virion tegument and infected cell nuclear matrix (39). The BVrh US10 protein is nearly identical to the HSV-1 US10 protein in size, but there is very little absolute sequence conservation in the N-terminal 70% of the polypeptide. In contrast, the C-terminal region of US10 is highly conserved among all of the primate viruses, including the putative zinc-finger motif (18). The amino acid sequences of the C-terminal halves of the BVrh and BVcy sequences are identical, reflecting the strong conservation of this protein.
The US11 gene encodes an RNA-binding protein which prevents phosphorylation of eukaryotic initiation factor 2 by binding to the cellular PKP kinase. The US11 ORF begins 145 nucleotides 5′ of the US10 ORF, resulting in the 3′ 65% of the US11 coding sequence overlapping the US10 ORF. The BV and HSV US11 polypeptides are strongly conserved in the C-terminal region, which contains a reiteration of 19 to 20 copies of the sequence RXP. RNA-binding activity, the nucleolar localization signal sequence, and the region responsible for association with 60S rRNA have all been mapped to this conserved region (32, 33). The 5′ portion of the gene encoding the nonoverlapping sequence shows very little conservation between BV and HSV.
The US12 gene encodes the small immediate-early (alpha) regulatory protein ICP47, which inhibits antigen presentation in infected cells by specifically binding to and blocking the transporter associated with processing (8, 24, 29). Although the start codon of the BVrh US12 ORF was not identified, what is probably the majority of the BV US12 ORF was determined. Surprisingly, alignment of the BV and HSV US12 sequences reveals very little sequence identity among these regulatory polypeptides. However, analysis of the polypeptides for various features of secondary structure indicates that the BV and HSV ICP47 proteins are likely quite similar in structure.
In conclusion, the sequence of the US region of BVrh clearly demonstrates that this region of the genome is entirely colinear with the US regions of HSV-1, HSV-2, and SA8. Except for the US1.5 ORF, homologues of all genes detected in HSV-1 and HSV-2 are present in the BV US region, and the BV proteins are predicted to be similar in size to the homologous HSV-2 polypeptides. While a number of genes do exhibit substantial sequence variation, these genes also exhibit regions of highly conserved sequence. Such highly conserved regions undoubtedly represent important structural and/or functional regions of these proteins, indicating that these proteins have structures and functions similar to those of their HSV homologs. This is also consistent with the extensive antigenic cross-reactivity observed between almost all HSV, SA8, HVP-2, and BV proteins (10, 12, 17). Finally, genotypic sequence variation among BV isolates was minimal in nonglycoproteins but readily evident in the glycoproteins except for the conserved gD glycoprotein.
Acknowledgments
This work was supported in part by PHS grants NCRR R01 RR07849 and P40 RR12317 (R.E.) and a Grant-in-Aid for Scientific Research B (grant 09400014) from the Ministry of Education, Culture, Sport, Science, and Technology of Japan (H.S.).
REFERENCES
- 1.Basu, S., G. Dubin, M. Basu, V. Nguyen, and H. M. Friedman. 1995. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J. Immunol. 154:260–267. [PubMed] [Google Scholar]
- 2.Basu, S., G. Dubin, T. Nagahunmugam, M. Basu, L. T. Goldstein, L. Wang, B. Weeks, and H. M. Freidman. 1997. Mapping regions of the herpes simplex virus type 1 glycoprotein I required for formation of the viral Fc receptor for monomeric IgG. J. Immunol. 158:209–215. [PubMed] [Google Scholar]
- 3.Bennett, A. M., L. Harrington, and D. C. Kelly. 1992. Nucleotide sequence analysis of genes encoding glycoproteins D and J in simian herpes B virus. J. Gen. Virol. 73:2963–2967. [DOI] [PubMed] [Google Scholar]
- 4.Carter, K. L., and B. Roizman. 1996. The promoter and transcriptional unit of a novel herpes simplex virus 1 alpha gene are contained in, and encode a protein in frame with, the open reading frame of the alpha 22 gene. J. Virol. 70:172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816. [DOI] [PubMed] [Google Scholar]
- 6.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubin, G., S. Basu, D. L. Mallory, M. Basu, R. Tal-Singer, and H. M. Friedman. 1994. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J. Virol. 68:2478–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easterfield, A. J., B. M. Austen, and O. M. Westwood. 2001. Inhibition of antigen transport by expression of infected cell peptide 47 (ICP47) prevents cell surface expression of HLA in choriocarcinoma cell lines. J. Reprod. Immunol. 50:19–40. [DOI] [PubMed] [Google Scholar]
- 9.Eberle, R., and D. Black. 1999. Molecular aspects of monkey B virus and implications for diagnostic test development. Recent Res. Dev. Virol. 1:85–94. [Google Scholar]
- 10.Eberle, R., D. Black, and J. K. Hilliard. 1989. Relatedness of glycoproteins expressed on the surface of simian herpesvirus virions and infected cells to specific herpes simplex virus glycoproteins. Arch. Virol. 109:233–252. [DOI] [PubMed] [Google Scholar]
- 11.Eberle, R., D. Black, T. Lehenbauer, and G. White. 1998. Shedding and transmission of baboon Herpesvirus papio 2 (HVP2) in a breeding colony. Lab. Anim. Sci. 48:23–28. [PubMed] [Google Scholar]
- 12.Eberle, R., and J. Hilliard. 1995. The simian herpesviruses. Infect. Agents Dis. 4:55–70. [PubMed] [Google Scholar]
- 13.Eberle, R., M. Zhang, and D. Black. 1993. Gene mapping and sequence analysis of the unique short region of the simian herpesvirus SA8 genome. Arch. Virol. 130:391–411. [DOI] [PubMed] [Google Scholar]
- 14.Georgopoulou, U., A. Kakkanas, V. Miriagou, A. Michaelidou, and P. Mavromara. 1995. Characterization of the US8.5 protein of herpes simplex virus. Arch. Virol. 140:2227–2241. [DOI] [PubMed] [Google Scholar]
- 15.Gray, W. L., B. Starnes, M. W. White, and R. Mahalingam. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123–130. [DOI] [PubMed] [Google Scholar]
- 16.Harrington, L., L. V. M. Wall, and D. C. Kelly. 1992. Molecular cloning and physical mapping of the genome of simian herpes B virus and comparison of genome organization with that of herpes simplex virus type 1. J. Gen. Virol. 73:1217–1226. [DOI] [PubMed] [Google Scholar]
- 17.Hilliard, J. K., D. H. Black, and R. Eberle. 1989. Simian alpha-herpesviruses and their relation to the human herpes simplex viruses. Arch. Virol. 109:83–102. [DOI] [PubMed] [Google Scholar]
- 18.Holden, V. R., R. R. Yalamanchili, R. N. Harty, and D. J. O’Callaghan. 1992. Identification and characterization of an equine herpesvirus 1 late gene encoding a potential zinc finger. Virology 188:704–713. [DOI] [PubMed] [Google Scholar]
- 19.Honess, R. W. 1984. Herpes simplex and ‘the herpes complex’: diverse observations and a unifying hypothesis. J. Gen. Virol. 65:2077–2107. [DOI] [PubMed] [Google Scholar]
- 20.Jerome, K. R., Z. Chen, R. Lang, M. R. Torres, J. Hofmeister, S. Smith, R. Fox, C. J. Froelich, and L. Corey. 2001. HSV and glycoprotein J inhibit caspase activation and apoptosis induced by granzyme B or Fas. J. Immunol. 167:3928–3935. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, Y. M., H. Yamada, F. Goshima, T. Daikoku, S. Oshima, K. Wada, and Y. Nishiyama. 1998. Characterization of the herpes simplex virus type 2 (HSV-2) US2 gene product and a US2-deficient HSV-2 mutant. J. Gen. Virol. 79:2777–2784. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. C., and V. Feenstra. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 61:2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killeen, A. M., L. Harrington, L. V. Wall, and D. C. Kelly. 1992. Nucleotide sequence analysis of a homologue of herpes simplex virus type 1 gene US9 found in the genome of simian herpes B virus. J. Gen. Virol. 73:195–199. [DOI] [PubMed] [Google Scholar]
- 24.Lacaille, V. G., and M. J. Androlewicz. 1998. Herpes simplex virus inhibitor ICP47 destabilizes the transporter associated with antigen processing (TAP) heterodimer. J. Biol. Chem. 273:17386–17390. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch, D. J., and A. J. Davison. 1986. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 25:1765–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1–13. [DOI] [PubMed] [Google Scholar]
- 27.McGeoch, D. J., H. W. M. Moss, D. McNab, and M. C. Frame. 1987. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J. Gen. Virol. 68:19–38. [DOI] [PubMed] [Google Scholar]
- 28.Meindl, A., and N. Osterrieder. 1999. The equine herpesvirus 1 Us2 homolog encodes a nonessential membrane-associated virion component. J. Virol. 73:3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann, L., W. Kraas, S. Uebel, G. Jung, and R. Tampe. 1997. The active domain of the herpes simplex virus protein ICP47: a potent inhibitor of the transporter associated with antigen processing. J. Mol. Biol. 272:484–492. [DOI] [PubMed] [Google Scholar]
- 30.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37–48. [DOI] [PubMed] [Google Scholar]
- 31.Palmer, A. E. 1987. B virus. Herpesvirus simiae: historical perspective. J. Med. Primatol. 16:99–130. [PubMed] [Google Scholar]
- 32.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215–11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 70:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slomka, M. J., L. Harrington, C. Arnold, J. P. N. Norcott, and D. W. Brown. 1995. Complete nucleotide sequence of Herpesvirus simiae glycoprotein G gene and its expression as an immunogenic fusion protein in bacteria. J. Gen. Virol. 76:2161–2168. [DOI] [PubMed] [Google Scholar]
- 35.Smith, A. L., D. H. Black, and R. Eberle. 1998. Molecular evidence for distinct genotypes of monkey B virus (Herpesvirus simiae) which are related to the macaque host species. J. Virol. 72:9224–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su, H. K., J. D. Fetherston, M. E. Smith, and R. J. Courtney. 1993. Orientation of the cleavage site of the herpes simplex virus glycoprotein G-2. J. Virol. 67:2954–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, S. A., J. K. Hilliard, D. Kittel, S. Lipper, W. E. Giddens, Jr., D. H. Black, and R. Eberle. 2000. Retrospective analysis of an outbreak of B virus infection in a colony of DeBrazza’s monkeys (Cercopithecus neglectus). Comp. Med. 50:649–657. [PubMed] [Google Scholar]
- 38.Weigler, B. J. 1992. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 15:555–567. [DOI] [PubMed] [Google Scholar]
- 39.Yamada, H., T. Daikoku, Y. Yamashita, Y. M. Jiang, T. Tsurumi, and Y. Nishiyama. 1997. The product of the US10 gene of herpes simplex virus type 1 is a capsid/tegument-associated phosphoprotein which copurifies with the nuclear matrix. J. Gen. Virol. 78:2923–2931. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by herpes simplex virus 1 mutants lacking intact genes expressing both glycoproteins. J. Virol. 74:11782–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]