Abstract
The rat cytomegalovirus (RCMV) R33 gene is conserved among all betaherpesviruses and encodes a protein (pR33) that shows sequence similarity with chemokine-binding G protein-coupled receptors (GPCRs). Previously, the physiological significance of the R33 gene was demonstrated by the finding that an RCMV strain with R33 deleted is severely attenuated in vivo and is unable to either enter or replicate in the salivary glands of infected rats. Here, we report that RCMV pR33 is expressed as a functional GPCR that signals in an agonist-independent manner in both COS-7 and Rat2 cells. Transient expression of pR33 in COS-7 cells results in constitutive activation of phospholipase C (PLC) due to coupling to G proteins of the Gq class. Interestingly, PLC activation is partially inhibited by cotransfection with Gα-transducin subunits, which indicates the involvement of Gβγ as well as Gα subunits in pR33-mediated signaling. Surprisingly, PLC activation is also partially inhibited by addition of pertussis toxin (PTX), suggesting that pR33 activates not only Gq but also Gi/0 proteins. The constitutive activation of Gi/0 proteins by pR33 is further demonstrated by the PTX-sensitive decrease of CRE-mediated transcription and the PTX-sensitive increase of both NF-κB- and SRE-mediated transcription. In contrast to its homolog of human herpesvirus 6B (pU12), pR33 does not bind RANTES.
Cytomegaloviruses (CMVs) are species-specific betaherpesviruses that cause acute, persisting, and latent infections in both humans and animals. The ability of CMVs to induce lifelong, latent infections implies that these viruses are highly adapted to their hosts. Specifically, they will have to employ sophisticated strategies in order to remain hidden from their host’s immune system. Among the CMV genes that are likely to be involved in these strategies are those encoding homologs of crucial immune effector or regulatory proteins of the host, such as G protein-coupled receptors (GPCRs). These genes have probably been pirated by the virus during the long coevolution of pathogen and host.
GPCRs form a large and diverse family of receptors that function in signal transduction through cell membranes. They are composed of a central core domain consisting of seven transmembrane helices connected by three intracellular and three extracellular loops. The majority of these receptors activate G proteins and are capable of transducing a wide variety of messages. Within the genomes of all sequenced CMVs, genes have been identified that potentially encode homologs of host cellular GPCRs. Human CMV (HCMV) carries four of these genes: US27, US28, UL33, and UL78 (13, 14). Only two of these, UL33 and UL78, were found to have counterparts in rat CMV (RCMV) (R33 and R78, respectively [4, 5, 52]) as well as murine CMV (MCMV) (M33 and M78, respectively [17, 41]). Of the predicted amino acid sequences derived from the CMV GPCR-like genes, those encoded by HCMV US27 and US28 were found to have the highest similarity to chemokine-binding GPCRs (chemokine receptors). HCMV US28 was demonstrated to encode a functional chemokine receptor (pUS28) which binds the CC (or β-) chemokines RANTES, MCP-1, MCP-3, MIP-1α, and MIP-1β (7, 8, 22, 31, 37) as well as the only known member of the CX3C (or δ-) chemokine family, CX3CL1 (also known as fractalkine or neurotactin), which is a membrane-associated chemokine with a cytoplasmic domain (29). Upon CC-chemokine binding, pUS28 was shown to engage in intracellular signaling by the mobilization of intracellular calcium and activation of extracellular signal-related kinase 2 (7). Remarkably, pUS28 was also found to signal in the absence of exogenously added chemokines, by activating phospholipase C (PLC) as well as NF-κB in transiently transfected COS-7 cells (11). This constitutive, agonist-independent activity of pUS28 can be modulated by fractalkine, which acts as a partial inverse agonist, whereas CC-chemokines appear to function as neutral antagonists (11). Another intriguing property of pUS28 was identified by Streblow and colleagues (49). They demonstrated that HCMV infection of primary arterial smooth muscle cells (SMCs) resulted in cellular migration, whereas an HCMV recombinant that lacks a functional US28 gene was unable to promote migration. The expression of pUS28 in SMCs in the presence of CC-chemokines was sufficient to induce migration. It was hypothesized by Streblow et al. that HCMV may be using SMCs as a cellular vehicle to disseminate the virus in vivo (49).
As described above, the HCMV UL33 gene has homologs in all sequenced betaherpesviruses, which underlines the biological relevance of the UL33 gene family. At present, this family consists of six members: HCMV UL33 (13, 14), MCMV M33 (41), and RCMV R33 (5) and the U12 genes of human herpesvirus 6A (HHV-6A) (24), HHV-6B (19, 25), and HHV-7 (38). The predicted amino acid sequences of the proteins encoded by the UL33-like gene family were found to comprise several features characteristic of chemokine receptors (5, 17). In addition, we found limited but significant sequence similarity between the UL33 family members and mammalian chemokine receptors CCR3 and, in particular, CCR10 (54).
The biological significance of the UL33 family members has previously been demonstrated in studies using recombinant CMVs that carry either a disrupted UL33 (33), M33 (17), or R33 gene (5) in their genomes. In cell culture, each of these mutant viruses replicated with similar efficiency as the corresponding wild-type (WT) viruses (5, 17, 33). However, during in vivo infection, significant differences were observed between animals infected with the recombinants and those infected with the WT viruses. In contrast to their WT counterparts, viruses with M33 and R33 deleted could not be detected within the salivary glands of infected mice and rats, respectively (5, 17). This indicated that M33 and R33 play a role in virus dissemination to or replication in the salivary glands (5, 17). Furthermore, it was shown in the RCMV-rat model that R33 also has a more general function: a significantly lower mortality was seen among rats infected with RCMV from which R33 was deleted (RCMVΔR33) than among those infected with WT RCMV (5). These results stressed the importance of the UL33-like genes in the pathogenesis of CMV infection.
Although the HHV-6B member of the UL33 family, pU12, was reported to be a calcium-mobilizing receptor for several CC-chemokines (25), not much is known about the biochemical, immunological, and pharmacological processes in which the CMV members of this protein family may function. We therefore set out to study the putative signaling properties of the RCMV R33-encoded protein (pR33). We report here that pR33 is a functional GPCR that signals in an agonist-independent, constitutive fashion in COS-7 as well as Rat2 cells.
MATERIALS AND METHODS
Expression constructs.
The sequence containing RCMV open reading frames (ORFs) R32 (partially), R33, and R34 (partially) (position 24384 to 29545 of the RCMV genome sequence [53]) was cloned as a PstI fragment into vector pUC119, generating plasmid RPst5. This plasmid was digested with PvuI, and the resulting 2.9-kb fragment, which contains the complete R33 ORF, was treated with T4 DNA polymerase in the presence of deoxynucleoside triphosphates (dNTPs) in order to generate blunt ends. Subsequently, the blunt-ended fragment was cloned into the HincII restriction site of pUC119, generating plasmid p375. Then, a PCR-based procedure was used to change the sequence around the translation initiation codon of R33, 5′-CGGTATGGAC-3′, into the sequence 5′-CGCCATGGAC-3′, which contains an NcoI restriction site (indicated in italics). This was carried out as follows. First, the 5′ end of R33 was amplified by means of PCR using primers R33.N-F and R33.N-R (Table 1) and p375 as a template. Then, the 228-bp PCR product was treated with T4 DNA polymerase and cloned into the HincII site of pUC119, creating plasmid pre-379. The sequence upstream of R33 was amplified by PCR using primers R32.N-F and R32.N-R (Table 1) and p375 as a template. The 745-bp PCR product was treated with T4 DNA polymerase and subsequently digested with NcoI. The resulting NcoI-blunt fragment was cloned into pre-379 by using the SmaI site of the polylinker and the introduced NcoI site at the translation initiation codon of R33, resulting in plasmid p379. Finally, the 946-bp BglII-BsrGI fragment from p375 was exchanged for the BglII-BsrGI fragment from p379, which contains the introduced NcoI site at the start codon of R33, generating plasmid p383. The 2.1-kb BamHI-SmaI fragment from p383, containing ORF R33 with the introduced NcoI site, was cloned into BamHI- and HincII-digested pUC119. Subsequently, the 331-bp BamHI-NcoI fragment immediately upstream of R33 was deleted from the plasmid by digestion with BamHI and NcoI and treatment with the Klenow fragment of DNA polymerase I (Klenow) with dNTPs, followed by closing of the linear vector with T4 DNA ligase, resulting in plasmid pUC119/R33dN. The R33 expression vector pcDNA3/R33 was then generated by cloning the 1,680-bp KpnI fragment containing ORF R33 from pUC119/R33dN into KpnI-digested pcDNA3 (Invitrogen, Groningen, The Netherlands). The KpnI sites of pUC119/R33dN are derived from the pUC119 polylinker, located 5′ of the translation initiation codon of ORF R33, and the RCMV sequence downstream of the ORF R33 stop codon (corresponding to position 27950 of the RCMV sequence). The orientation of ORF R33 in pcDNA3/R33 was checked by restriction enzyme analysis.
TABLE 1.
PCR primers used in this study
| Primer | Sequencea | Restriction site(s) | Source | Positions | Orientationb |
|---|---|---|---|---|---|
| GFP.CN-F | 5′-ATAAGAATTCCAGATCCGCTAGCGCTACCG-3 | EcoRI | pEGFP-N1 | 574–603 | + |
| EGFP.N1-R | 5′-TATATCGCGACTCATGAACTTGTACAGCTCGTCC-3′ | NruI, BspHI | pEGFP-N1 | 1380–1403 | − |
| R32.N-F | 5′-TCGCAGATCTCCCGCCAGCCC-3′ | RCMV genome | 25535–25555 | + | |
| R32.N-R | 5′-TATACCATGGCGGTCCCGTCTCCGTC-3′ | NcoI | RCMV genome | 26253–26280 | − |
| R33.N-F | 5′-TAATCCATGGACGTGCTGCTGGGGAC-3′ | NcoI | RCMV genome | 26271–26290 | + |
| R33.N-R | 5′-GGTTCGTCATGTACAGGGTCGGCGTC-3′ | RCMV genome | 26474–26499 | − | |
| R33.C-F | 5′-TCCTGGTGGTGCAGACGCCGTTC-3′ | RCMV genome | 27025–27047 | + | |
| R33.C-R | 5′-CGTCTAGACAGTTCGGCGGCGGAC-3′ | XbaI | RCMV genome | 27415–27438 | − |
| R34.C-F | 5′-ACTGTCTAGACGGCAGATCGCCGCC-3′ | XbaI | RCMV genome | 27427–27451 | + |
| R34.C-R | 5′-GCTTCTCCGTCTCCCCGCGCC-3′ | RCMV genome | 28024–28044 | − |
Underlined sequences indicate restriction enzyme recognition sites; sequences in boldface type differ from that of the original sequence as described in the columns for source and positions.
+, orientation of the sequence is identical to the published sequence between the indicated positions; −, sequence is complementary to the published sequence between the indicated positions.
The expression vector pcDNA3/EGFP was generated by digestion of pEGFP-N1 (Clontech, Palo Alto, Calif.) with BamHI and NotI and cloning the resulting 739-bp fragment into BamHI- and NotI-digested pcDNA3. For the construction of expression vector pcDNA3/EGFP-R33, a BspHI site was introduced immediately downstream of the enhanced green fluorescent protein (EGFP) ORF. This was carried out by means of PCR with primers GFP.CN-F and EGFP.N1-R (Table 1), using pEGFP-N1 as a template. The amplified fragment of 829 bp was treated with T4 DNA polymerase and the blunt-ended fragment was subsequently cloned into HincII-digested pUC119, generating pUC119-EGFP. Then, the 720-bp NcoI-BspHI fragment from pUC119/EGFP was cloned into the introduced NcoI site at the translation initiation codon of R33 within plasmid p383, resulting in plasmid p383-EGFP. The orientation of the EGFP ORF was checked by restriction enzyme analysis. Then, p383-EGFP was digested with NcoI and treated with Klenow. Subsequently, the DNA was digested with ApaI to generate a 2.4-kb blunt-ApaI fragment which contains the EGFP ORF fused to R33. This fragment was cloned into EcoRV- and ApaI-digested pcDNA3, resulting in the expression vector pcDNA3/EGFP-R33.
In order to generate the pcDNA3/R33-EGFP expression vector, a pUC119 vector was created in which the SphI and XbaI sites of the polylinker were destroyed (pre-376). This was carried out as follows. pUC119 was digested with SphI and treated with T4 polymerase in the presence of dNTPs, and the linear vector was closed with T4 DNA ligase. Subsequently, the vector was digested with XbaI, treated with Klenow, and closed with T4 DNA ligase, generating plasmid pre-376. The previously described 2.9-kb PvuI fragment containing the complete R33 ORF was cloned into the HincII site of pre-376, generating plasmid p376. Then, a PCR-based procedure was used to modify the sequence around the stop codon of R33, 5′-TGCTGAGTCG-3′, into the sequence 5′-TGTCTAGACG-3′, which contains an XbaI restriction site (in italics) and changes the R33 stop codon to a leucine codon. This was performed as follows. First, the 3′ end of ORF R33 was amplified by PCR with primers R33.C-F and R33.C-R (Table 1), using p375 as a template. The 413-bp PCR product was digested with SphI and XbaI and cloned into SphI- and XbaI-digested pUC119, generating plasmid pre-380 (the SphI restriction site in R33 corresponds to position 27134 of the RCMV sequence). Then, the sequence downstream of the ORF R33 stop codon was amplified by PCR with primers R34.C-F and R34.C-R (Table 1), using p375 as a template. The amplified fragment of 617 bp was digested with Asp718 and XbaI and cloned into the corresponding sites of pre-380, resulting in plasmid p380 (the Asp718 site in R34 corresponds to position 27947 of the RCMV sequence). Finally, the 737-bp SphI-NruI fragment of p376 (the NruI restriction site corresponds to position 27872 of the RCMV sequence) was exchanged for the SphI-NruI fragment of p380, which contains the introduced XbaI site in the R33 ORF, resulting in plasmid p384. Before the EGFP ORF could be cloned immediately downstream of R33, a part of the sequence upstream of the EGFP ORF in pEGFP-N1 was deleted. This was carried out by digestion of plasmid pEGFP-N1 with BamHI and BglII and subsequent closing of the linearized vector, resulting in plasmid p368. Then, the 768-bp NheI-XbaI fragment from plasmid p368, which contains the EGFP ORF, was cloned into the introduced XbaI site of R33 in p384, resulting in plasmid p384-EGFP. The orientation of the EGFP ORF was checked by restriction enzyme analysis. Plasmid p384-EGFP was digested with BamHI and NotI, and the resulting 2.3-kb fragment containing the R33 ORF fused to EGFP was cloned into BamHI- and NotI-digested pcDNA3, resulting in the expression vector pcDNA3/R33-EGFP.
The cDNA encoding HCMV US28 (AD169 strain) in pcDNA3 (pcDNA3/US28) (11) was obtained from R. Doms (University of Pennsylvania, Philadelphia). The cDNA encoding human CCR10 (26) in pcDNA3 (pcDNA3/CCR10) was obtained from C. Gerard (Ina Sue Perlmutter Laboratory, Boston, Mass.). The cDNA of human CCR3 in pcDNA1 was provided by C. Tensen (Free University, Amsterdam, The Netherlands). The CCR3 cDNA was recloned in pcDNA3 by cutting the CCR3 fragment out of pcDNA1 using the flanking XbaI and HindIII sites in the pcDNA1 polylinker and cloning the fragment in the corresponding polylinker sites of pcDNA3, creating pcDNA3/CCR3. The pcDNA3-derived vectors expressing G protein receptor kinase 2 (GRK2) (pcDNA3/GRK2) and GRK2K220R (pcDNA3/GRK2K220R) (18) were provided by S. Cotecchia (University of Lausanne, Lausanne, Switzerland). The Gαt, Gαq, Gα11, and Gα16 expression vectors (pcDNA3/Gαt, pcDNA3/Gαq, pCMV5/Gα11, and pC1H1/Gα16, respectively) were kind gifts from B. Defize (Hubrecht Laboratory, Utrecht, The Netherlands), B. Conklin (University of California, San Francisco), H. Umemori (University of Tokyo, Tokyo, Japan), and S. Rees (University of Glasgow, Glasgow, United Kingdom), respectively. The reporter plasmids pTLNC-21CRE and pTLNC-3SRE (21) were obtained from W. Born (National Jewish Medical and Research Center, Denver, Colo.). The pNF-κB-Luc vector was purchased from Stratagene (La Jolla, Calif.). The integrity of all DNA constructs was verified by sequence analysis.
Cell culture and transfection.
COS-7 cells (ATCC CRL-1651) and Rat2 cells (TK−; ATCC CRL-1764) were cultured as described previously (4, 11). Transfection of Rat2 cells was performed on 80% confluently grown monolayers by using DEAE-dextran (final concentration, 0.4 mg/ml) in serum-free Eagle’s minimal essential medium with Earle’s balanced salt solution (EMEM) supplemented with 2 mM l-glutamine, and 1× nonessential amino acids (NEAA). After 2 h of incubation at 5% CO2 and 37°C, the transfection medium was aspirated and the cells were treated with 10% dimethyl sulfoxide in phosphate-buffered saline (PBS) for 1 min. Subsequently, the cells were washed three times with PBS and further grown at 5% CO2 and 37°C in serum-free EMEM containing 2 mM l-glutamine and 1× NEAA. Transfection of COS-7 cells was carried out as described previously (11).
Confocal imaging.
Transiently transfected cells (COS-7 or Rat2) were grown on glass coverslips and were fixed for 20 min with 3.7% Formol in PBS 48 h after transfection. To stain the trans-Golgi complex, cells were washed three times with PBS and incubated with 1 μM Bodipy TR ceramide (Molecular Probes, Leiden, The Netherlands) and 1% bovine serum albumin (BSA) (Sigma) in PBS for 30 min at 4°C. For staining of the endoplasmic reticulum (ER), cells were permeabilized with 0.05% NP-40 (Boehringer Mannheim, Mannheim, Germany) in PBS for 20 min and subsequently incubated at 37°C for 1.5 h with mouse monoclonal antibody IID8 (directed against SERCA 2) (ABR, Golden, Colo.) in PBS containing 1% BSA, washed three times with PBS, and incubated for 1.5 h at 37°C with rabbit anti-mouse–rhodamine (1:50 in PBS; DAKO, Glostrup, Denmark). Thereafter, cells were washed three times with PBS and the coverslips were mounted in 90% glycerol containing 0.02 M Tris-HCl (pH 8.0), 0.002% NaN3, and 2% DABCO [1,4-diazabicyclo-(2,2,2)-octane; Merck, Darmstadt, Germany]. Confocal images were collected at wavelengths of 488 and 568 nm using an MRC 600 confocal microscope equipped with an oil immersion objective (40× magnification, numerical aperture = 1.3; Bio-Rad, Hemel Hempstead, United Kingdom) as described previously (9). The digital images were projected and merged using Confocal Assistance software from Bio-Rad (Veenendaal, The Netherlands).
EGFP-based monolayer enzyme-linked immunosorbent assay (ELISA).
Transfected cells were seeded in 48-well plates (Costar). After 48 h of incubation, the monolayers were washed once with Tris-buffered saline (TBS) (50 mM Tris, 150 mM NaCl [pH 7.5]) and fixed for 30 min with 4% paraformaldehyde in PBS. Subsequently, cells were washed three times with TBS and permeabilized with 0.5% NP-40 in TBS. After 30 min, the permeabilization solution was replaced with blocking buffer (1% fat-free milk, 0.1 M NaHCO3 [pH 8.6]) and the cells were incubated for 4 h at room temperature. Then, the blocking buffer was replaced with the primary antibody solution containing a 1:100 dilution of anti-EGFP polyclonal antibody (Clontech) in TBS with 0.1% BSA. The cells were incubated overnight at 4°C with shaking. The monolayers were washed three times with TBS, after which the secondary antibody containing a 1:2,500 dilution of goat anti-rabbit–horseradish peroxidase conjugate (Bio-Rad) was added. After 2 h of incubation at room temperature, the cells were washed three times with TBS and the OPD substrate solution (5 mM O-phenylenediamine [Sigma], 0.03% H2O2 in 0.1 M citrate–phosphate buffer [pH 5.0]) was applied for approximately 10 min. The coloring reaction was stopped by the addition of 1 M H2SO4, samples were taken from the supernatants, and the optical density was measured in a Victor2 Wallac multilabel counter at 490 nm.
[3H]inositol phosphate production.
Inositol phosphate production in transfected COS-7 cells was determined as described previously (11).
Reporter assays.
COS-7 cells were transiently transfected with one of the following reporter plasmids: pNF-κB-Luc (NF-κB assay), pTLNC-21CRE (cyclic AMP [cAMP]-responsive element [CRE] assay), pTLNC-3SRE serum-responsive promoter element [SRE] assay) (5 μg/106 cells), and one of the GPCR-expressing plasmids. Transfected cells were seeded in 96-well white plates (Costar) in serum-free medium either in the presence or absence of pertussis toxin (PTX) (100 ng/ml) (all assays) or human or rat RANTES (100 nM, NF-κB and SRE assay). To cells cotransfected with the CRE reporter plasmid, 10 μM forskolin was added 24 h after transfection, either in the presence or absence of human or rat RANTES (100 nM). After a 6-h incubation, luciferase activity was assayed by aspiration of the culture medium and addition of 25 μl of luciferase assay reagent (0.83 mM ATP, 0.83 mM d-luciferin [Duchefa Biochemie B.V., Haarlem, The Netherlands], 18.7 mM MgCl2, 0.78 μM Na2H2P2O7, 38.9 mM Tris [pH 7.8], 0.39% [vol/vol] glycerol, 0.03% [vol/vol] Triton X-100, and 2.6 μM dithiothreitol). After a 5-min incubation, the relative luminescence was measured for 3 s using a Victor2 Wallac multilabel counter. For cells transfected with either the NF-κB or the SRE reporter constructs, luciferase activity was measured 48 h after transfection using the same protocol.
Chemokine binding assay.
Human and rat RANTES were labeled with 125I using the Iodo-gen method (Pierce, Rockford, Ill.) according to the manufacturer’s protocol. 125I-labeled RANTES was separated from free iodine (>99%) using a 25-cm, 10-ml Sephadex G-25 gel filtration column (ICN Pharmaceuticals Inc., Costa Mesa, Calif.). Iodine incorporation and specific activity were determined via precipitation of the protein by the trichloroacetic acid (TCA) method (6). Transiently transfected cells were seeded in 24-well plates. In saturation experiments, binding was performed 48 h after transfection on whole cells for 3 h at 4°C using different concentrations of 125I-labeled human or rat RANTES (from 0.1 to 6 nM) in binding buffer (50 mM HEPES [pH 7.4], 1 mM CaCl2, 5 mM MgCl2, and 0.5% BSA). Nonspecific binding was determined in the presence of 0.1 μM unlabeled competitor (human or rat RANTES). In competition experiments, cells were incubated with 0.6 nM 125I-labeled human RANTES and various amounts of unlabeled human or rat RANTES. After incubation, cells were washed four times with binding buffer supplemented with 0.5 M NaCl.
Statistical analysis.
All data shown are expressed as mean ± standard error. Statistical analysis was carried out by Student’s t tests. P values of <0.05 were considered to indicate a significant difference.
RESULTS
RCMV pR33 and pR33-EGFP signal constitutively in COS-7 cells.
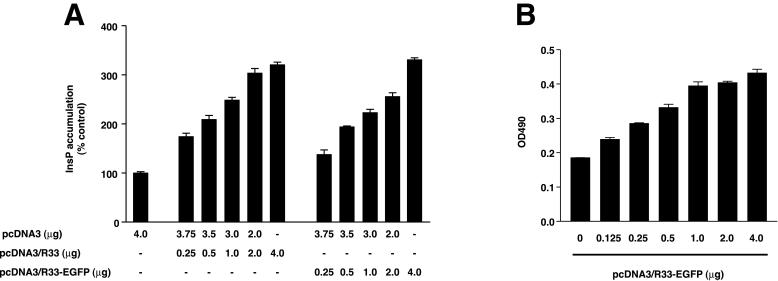
To test whether the RCMV R33-encoded protein (pR33) functions as a GPCR in vitro, we set out to examine the effect of pR33 expression on the accumulation of inositol phosphates (InsP) in COS-7 cells. To this end, the cells were transiently transfected with either of four different expression constructs, which encode native RCMV pR33 (pcDNA3/R33), pR33-EGFP (pcDNA3/R33-EGFP), pEGFP-R33 (pcDNA3/EGFP-R33) or no protein (pcDNA3). At 24 h after transfection, the cells were incubated with 3H-labeled inositol, and at 48 h after transfection, the accumulation of radiolabeled InsP was measured. As shown in Fig. 1A, transfection of increasing amounts of both pcDNA3/R33 and pcDNA3/R33-EGFP resulted in increased [3H]InsP production within the cells. The InsP production in cells transfected with pcDNA3/R33 and pcDNA3/R33-EGFP only (4.0 μg/106 cells) was 3.2 times higher than that in cells transfected with an equal amount of pcDNA3 only. Interestingly, a significant difference in InsP levels was not observed between pcDNA3- and pcDNA3/EGFP-R33-transfected cells (data not shown). Taken together, these data indicate that (i) these proteins activate signaling pathways leading to increased InsP production rates and (ii) these proteins signal in an agonist-independent fashion.
FIG. 1.
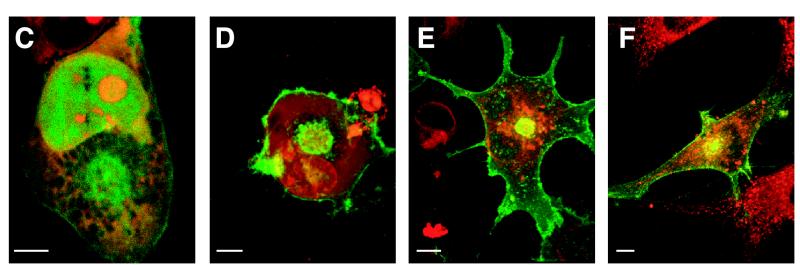
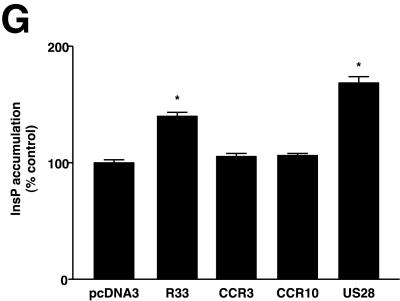
pR33- and pR33-EGFP-mediated induction of inositol phosphate (InsP) accumulation. (A) COS-7 cells (106) were transiently transfected with either pcDNA3, pcDNA3/R33, or pcDNA3/R33-EGFP. At 48 h after transfection, the InsP accumulation was measured. (B) Expression of pR33-EGFP. COS-7 cells were transiently transfected with various concentrations of pcDNA3/R33-EGFP. After permeabilization, the cells were immunologically stained using an anti-EGFP polyclonal antiserum. The staining was quantified by measuring the optical density at 490 nm (OD490) by spectrophotometry. (C and D) Cellular localization of EGFP (C) and pR33-EGFP (D). COS-7 cells were transiently transfected with either pcDNA3/EGFP or pcDNA3/R33-EGFP, fixed, stained using propidium iodide (to visualize nucleic acids), and subjected to confocal microscopy at 488 and 568 nm. (E) Cellular localization of pR33-EGFP and the ER. pcDNA3/R33-EGFP-transfected cells were stained using SERCA 2-specific antibodies. (F) Cellular localization of pR33-EGFP and the Golgi apparatus. pcDNA3/R33-EGFP-transfected cells were stained using a trans-Golgi marker. Bars = 5 μm. (G) Modulation of InsP accumulation by various receptors. The InsP accumulation was determined after transient transfection of COS-7 cells with either pcDNA3, pcDNA3/R33 (R33), pcDNA3/CCR33 (CCR3), pcDNA3/CCR10 (CCR10), or pcDNA3/US28 (US28) at 0.5 μg of DNA/106 cells. A representative experiment performed in triplicate is shown. Each experiment was repeated at least twice. The asterisks indicate a statistically significant difference (P < 0.05) with data from pcDNA3-transfected cells. Error bars, standard error of the mean.
To examine the expression levels as well as intracellular localization of the R33-encoded proteins, both ELISA and confocal microscopy were performed. Since appropriate anti-pR33 antibodies were unavailable, we could investigate only the expression of the EGFP fusion proteins by virtue of the presence of the EGFP tag in these proteins. The semiquantitative, ELISA-based monolayer assay showed that transfection of increasing amounts of pcDNA3/R33-EGFP resulted in increasing intracellular levels of pR33-EGFP (Fig. 1B). By contrast, expression of pEGFP-R33 was not detected (data not shown). These expression data are in agreement with the activity of these EGFP-tagged proteins in the InsP accumulation assay.
The cellular expression of pR33-EGFP was studied further by confocal microscopy. Figure 1D shows that the EGFP fluorescence colocalizes with intracellular, perinuclear vesicles as well as with the cell membrane of COS-7 cells transfected with pcDNA3/R33-EGFP. This expression pattern differs considerably from that seen in nonfused EGFP-expressing COS-7 cells, in which EGFP is seen dispersed throughout the nucleus and cytoplasm (Fig. 1C). These results strongly suggest that pR33-EGFP is properly expressed on the cell surface of transfected cells. In order to identify the intracellular vesicles to which pR33-EGFP localized, we stained the cells either with a trans-Golgi-specific marker (Bodipy TR ceramide) or with antibodies directed against ER-specific SERCA proteins. However, as shown in Fig. 1E and F, the EGFP signal does not colocalize with the signals that were generated with these compartment-specific markers. This indicated that pR33-EGFP localizes to intracellular compartments other than ER or trans-Golgi. Nevertheless, other attempts to identify these compartments have not yet been undertaken. Whether the presence of pR33-EGFP in these compartments is the consequence of high level of expression of this protein is not known.
To evaluate the specificity of the constitutive activity of pR33 in the InsP accumulation assay, we compared the activity of this protein with that of two mammalian β-chemokine receptors, CCR3 and CCR10, with which pR33 shares some homology, and a constitutively active viral GPCR, HCMV pUS28. As shown in Fig. 1G, cells transfected with either pcDNA3/R33 or pcDNA3/US28 produced significantly higher levels of InsP than cells transfected with either pcDNA3 (mock) or the mammalian GPCR expression constructs. These data show that constitutive activity of GPCRs in the InsP assay is not a general phenomenon but rather a characteristic of a subset of receptors, as was recognized previously (11).
In summary, we have found pR33-EGFP to colocalize with the cell membrane of transfected COS-7 cells and to induce signaling pathways in an expression level-dependent fashion, leading to the production of InsP. Since both pcDNA3/R33-EGFP- and pcDNA3/R33-transfected cells demonstrate similar activities in the InsP assay, we infer that pR33 is expressed on the surface of transfected cells in a similar fashion as as demonstrated for pR33-EGFP.
pR33 couples to G proteins of the Gq and Gi/0 classes.
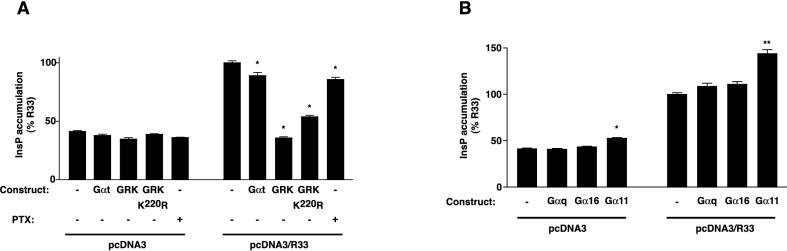
An intracellular rise in InsP levels is predominantly achieved following activation of PLC by G protein α subunits of the q class (Gαq). In addition, PLC activity can be stimulated by free G protein βγ subunits (16). To investigate whether the constitutive rise in InsP accumulation that is seen in cells expressing pR33 and pR33/EGFP is indeed mediated through the interaction of these proteins with heterotrimeric G proteins, COS-7 cells were transfected with both pcDNA3/R33 and constructs expressing either the Gα transducin subunit (Gαt), GRK2, or GRK2K220R. Gαt is known to sequester βγ subunits, whereas both GRK2 and GRK2K220R not only scavenge βγ subunits but also specifically inhibit Gαq activation (10, 20, 40). In addition, in contrast to GRK2K220R, GRK2 can phosphorylate GPCRs, leading to internalization and degradation of the receptors (18). As shown in Fig. 2A, cotransfection of pcDNA3/R33 and pcDNA3/Gαt resulted in a lower level of InsP accumulation (80%) than transfection of pcDNA3/R33 (basal) alone (100%). However, even lower levels of InsP were seen after cotransfection of pcDNA3/R33 and either pcDNA3/GRK2 or pcDNA3/GRK2K220R (25 and 40%, respectively). These InsP levels are similar to those measured in pcDNA3-transfected cells (30%). Together, these results suggest that the InsP production is mediated via coupling of pR33 to βγ as well as Gαq subunits. Since COS-7 cells endogenously express only two members of the Gq class, Gq and G11 (56), it is likely that the pR33-induced PLC activation is mediated by the α subunits of Gq and/or G11. In order to confirm this interaction of pR33 with the α subunits of Gq/11 and to investigate the putative interaction with another member of the Gαq family, Gα16, which is not expressed endogenously by COS-7 cells, cells were cotransfected with both pcDNA3/R33 and constructs expressing either Gαq, Gα11, or Gα16. As shown in Fig. 2B, cotransfection of pcDNA3/R33 with pCMV5/Gα11 resulted in a higher level of InsP production (145%) than transfection of pcDNA3/R33 alone (100%), whereas cotransfection of pcDNA3/R33 with either pcDNA3/Gαq or pC1H1/Gα16 did not change InsP production levels significantly compared to pcDNA3/R33 alone. These findings suggest that pR33 specifically activates G11 in a constitutive fashion but does not couple constitutively to Gq or G16.
FIG. 2.
(A) Effects of Gα transducin subunits, GRKs, and PTX on pR33-mediated InsP accumulation. COS-7 cells were transiently transfected with either pcDNA3 or pcDNA3/R33 in the presence of either pcDNA3/Gαt (Gαt), pcDNA3/GRK2 (GRK2), or pcDNA/GRK2K220R (GRK2K220R) (each construct at 2.0 μg of DNA/106 cells). The InsP accumulation was subsequently assessed either in the presence (+) or absence (−) of PTX in the culture medium. (B) Effects of Gαq, Gα16, and Gα11 on pR33-mediated InsP accumulation. COS-7 cells were transiently transfected with either pcDNA3 or pcDNA3/R33 in the presence of either pcDNA3/Gαq, pC1H1/Gα16, or pCMV5/Gα11, and the InsP accumulation was assessed after 48 h. Representative experiments performed in triplicate are shown. Each experiment was repeated at least three times. The asterisks indicate statistically significant differences (P < 0.05) between specific results and the result from pcDNA3/R33-transfected cells. Statistically significant differences (P < 0.05) were not found between results from the pcDNA3-transfected cells. Error bars, standard error of the mean.
Previously, it has been reported that PLC-activating βγ subunits can be released by G proteins of the Gi/0 class (16). To examine if these Gi/0 proteins are also involved in pR33-mediated constitutive PLC activation, we incubated pcDNA3- and pcDNA3/R33-transfected cells in the presence of a Gi/0 inhibitor (PTX), and measured the InsP accumulation. Figure 2A shows that the InsP accumulation level in pcDNA3/R33-transfected cells is significantly lower in the presence of PTX (75%) than in the absence of PTX (100%), indicating that at least part of the pR33-induced InsP production is mediated via Gi/0 proteins.
pR33-induced inhibition of CRE-mediated transcription.
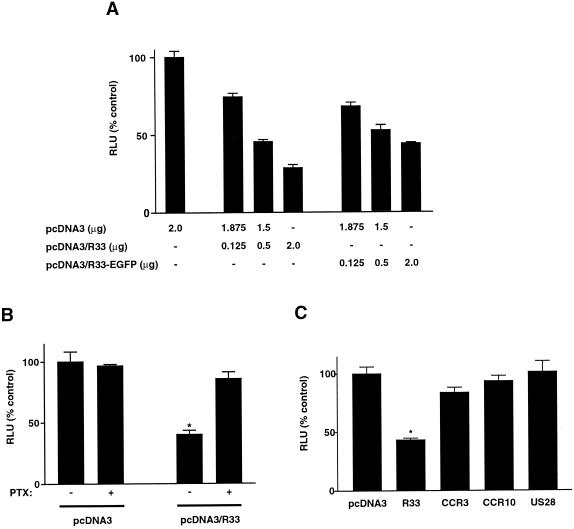
The interaction of pR33 with G proteins of the Gi/0 class was studied in more detail using the CRE reporter assay. This assay allows detection of the inhibitory effect of activated Gαi/0 subunits on adenylate cyclase (AC). The CRE assay is based on the artificial stimulation of AC with forskolin, which results in an increase of intracellular cAMP levels. The cAMP activates the CRE-binding protein (CREB), which can then bind and activate a CRE that controls transcription of a reporter gene (the luciferase gene). Hence, stimulation of cells carrying the CRE-luciferase gene with forskolin leads to increased production of luciferase transcripts and, subsequently, to higher levels of the luciferase protein, which can be quantified by measuring its activity. As described above, activated Gαi/0 subunits inhibit AC. Consequently, activation of these subunits by a GPCR in the CRE assay will result in lower luciferase levels and, thus, lower luciferase activity. To investigate whether pR33 can activate Gαi/0 subunits, we transfected COS-7 cells with the CRE-luciferase reporter construct and with either pcDNA3, pcDNA3/R33, or pcDNA3/R33-EGFP. Figure 3A shows a plasmid concentration-dependent inhibition of luciferase activity in both pcDNA3/R33- and pcDNA3/R33-EGFP-transfected cells, which indicates that pR33 can indeed constitutively activate Gαi/0 subunits. As expected, this activation can be inhibited by PTX, as shown in Fig. 3B. In order to study the specificity of this activity, we also tested constructs expressing either CCR3, CCR10, or US28 in the CRE assay. However, in contrast to pcDNA3/R33, these constructs did not induce an activity significantly different from that induced by pcDNA3 alone (Fig. 3C).
FIG. 3.
pR33-mediated inhibition of forskolin-induced, CRE-driven luciferase expression. (A) COS-7 cells (106) were transiently transfected with reporter plasmid pTLNC-21CRE (5.0 μg of DNA/106 cells) and with various amounts of either pcDNA3, pcDNA3/R33, or pcDNA3/R33-EGFP (in total, 2.0 μg of DNA/106 cells). When amounts of pcDNA3/R33 or pcDNA3/R33-EGFP lower than 2.0 μg were used, pcDNA3 was added to include a total of 2 μg of pcDNA3-derived plasmid per transfection. At 18 h after transfection, the cells were stimulated with forskolin. Six hours later, the CRE-driven luciferase expression and activity was quantified, and expressed as relative light units (RLU). (B) Influence of PTX on the pR33-mediated inhibition of CRE-driven expression. PTX (100 ng/ml) was added to the culture medium at 2 h after transfection. (C) Influence of various receptors on forskolin-stimulated, CRE-driven expression. The luciferase activity was determined similarly as described above, after transient transfection of COS-7 cells with either pcDNA3, pcDNA3/R33 (R33), pcDNA3/CCR3 (CCR3), pcDNA3/CCR10 (CCR10), or pcDNA3/US28 (US28) at 0.5 μg of DNA/106 cells. Data are presented as percentages of the result for the control (pcDNA3-transfected cells in the absence of PTX). Representative experiments performed in triplicate are shown, and each experiment was repeated at least three times. An asterisk indicates data that are statistically significantly different (P < 0.05) from that for the control. Error bars, standard error of the mean.
pR33-mediated activation of NF-κB- and SRE-driven transcription.
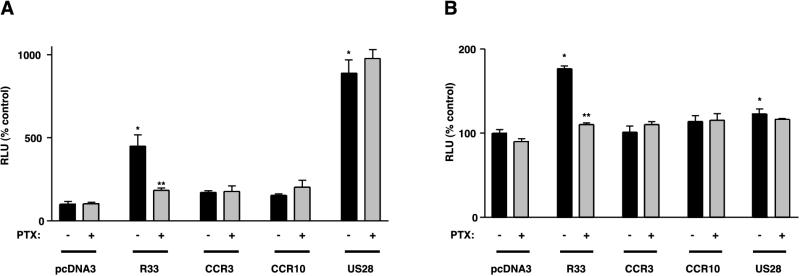
To assess the effect of constitutive pR33 signaling on the NF-κB promoter element and SRE, two additional reporter assays were used. In these assays, plasmid constructs are used in which the luciferase gene is placed under the control of either multiple SREs or the NF-κB promoter element. If receptor signaling induces upregulation of the intracellular concentration of activated NF-κB or serum response factor (SRF), an increase in activity will be measured in the NF-κB and SRE assay, respectively. Figure 4A shows that pcDNA3/R33-transfected cells have a significantly higher activity in the NF-κB assay than pcDNA3-transfected cells. As in the CRE assay, the pR33-induced activity was completely blocked in the presence of PTX (Fig. 4A). A similar observation was made for the activity of pR33-EGFP in the NF-κB assay (data not shown). We conclude that pR33 induces an upregulation of activated NF-κB that is mediated via a Gi/0 protein-dependent pathway(s). Of the other GPCRs that we tested in the NF-κB assay, only pUS28 was found to be active (Fig. 4A). As described previously, the activation of NF-κB by pUS28 is dependent not on Gi/0 protein-dependent pathways (since it is unaffected by the addition of PTX) but rather on Gq/11 protein-dependent pathways (11).
FIG. 4.
pR33-mediated NF-κB and SRE activation. COS-7 cells (106) were transiently transfected with either reporter plasmid pNF-κB-Luc (A) or pTLNC-3SRE (B) (5.0 μg DNA/106 cells) and with either pcDNA3, pcDNA3/R33 (R33), pcDNA3/CCR3 (CCR3), pcDNA3/CCR10 (CCR10), or pcDNA3/US28 (US28) at 0.5 μg of DNA/106 cells. At 48 h after transfection, luciferase activity was measured. The experiments were carried out either in the presence (+) or absence (−) of PTX in the culture medium. Representative experiments performed in triplicate are shown, and each experiment was repeated at least three times. Single asterisks indicate differences statistically significant (P < 0.05) from data from pcDNA3-transfected cells (in the absence of PTX). Double asterisks (**) indicate statistically significant differences (P < 0.05) between data from PTX-treated and -untreated cells that were transfected with the same expression construct. Error bars, standard error of the mean.
Figure 4B shows that pcDNA3/R33-transfected cells also have a significantly higher activity in the SRE assay than pcDNA3-transfected cells. Again, this activity as well as the activity of pcDNA3/R33-EGFP-transfected cells (data not shown) was blocked in the presence of PTX, indicating that pR33 induces an upregulation of activated SRF that is mediated via a Gi/0 protein-dependent pathway(s). A moderate but significant activity in the SRE assay was also measured in cells transfected with constructs expressing pUS28 (Fig. 4B). However, in contrast to the activity of pR33, the activity of pUS28 was not significantly affected by addition of PTX.
pR33 is unable to bind RANTES.
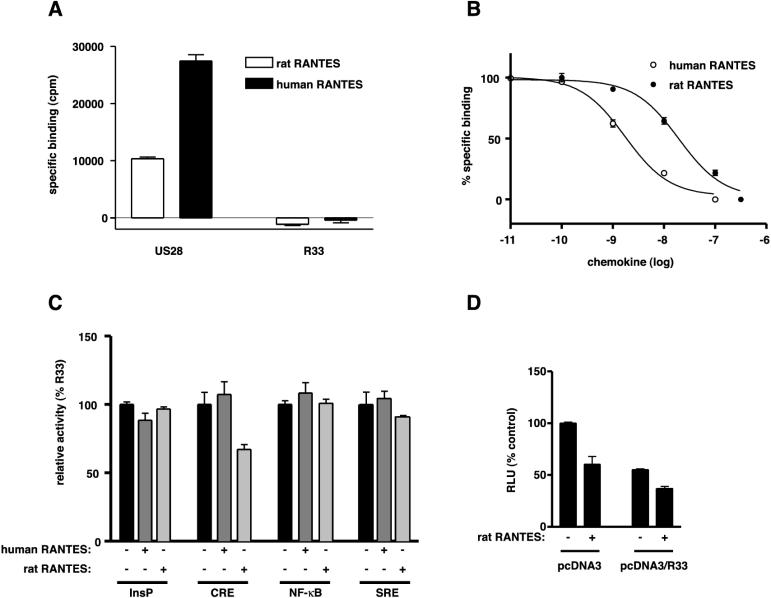
We previously reported that pR33 shows similarity with CC-chemokine-binding GPCRs (5, 54). In addition, the pR33 homolog from HHV-6B, pU12, was shown to bind CC-chemokine RANTES (24). To test whether RANTES could also bind to and/or modulate the constitutive activity of pR33, we transfected COS-7 cells with pcDNA3/R33 and, as a positive control, pcDNA3/pUS28 and monitored the binding of either 125I-labeled human or rat RANTES by the transfected cells. Although pUS28 was found to bind human and rat RANTES with a moderate affinity (Ki = 0.9 and 15.8 nM, respectively) (Fig. 5B), binding of these chemokines by pR33 was not detected (Fig. 5A). Furthermore, the addition of human or rat RANTES in the culture medium of pcDNA3/R33-transfected COS-7 cells did not modulate the activity of pR33 in the InsP production, NF-κB, and SRE assays (Fig. 5C). The addition of rat RANTES in the CRE assay resulted in a further inhibition of CRE-mediated transcription in pcDNA3/R33-transfected cells (Fig. 5C). However, a similar inhibitory effect of rat RANTES was seen in pcDNA3-transfected cells (Fig. 5D), indicating that this was not a specific pR33-mediated effect. Taken together, these findings suggest that, unlike HHV-6B pU12, RCMV pR33 does not bind and is not responsive to RANTES. In addition, several human chemokines of the CC (eotaxine, Teck, MCP-1), CX3C (fractalkine), and CXC class (IP-10, IP-9, GRO-α, and interleukin-8 [IL-8]) were not able to change the activity of pR33 significantly in any of the signaling assays used in this study (data not shown).
FIG. 5.
pR33 does not bind rat RANTES. (A) Binding of human and rat RANTES by pUS28 and pR33. COS-7 cells were transiently transfected with either pcDNA3, pcDNA3/US28 (US28), or pcDNA3/R33 (R33). Binding was performed 48 h after transfection on whole cells using 6 nM 125I-labeled human or rat RANTES. Nonspecific binding was determined in the presence of 0.1 μM unlabeled competitor (human or rat RANTES). (B) Human and rat RANTES competition curves on pUS28. COS-7 cells were transiently transfected with pcDNA3/US28 and incubated with 0.6 nM 125I-labeled human RANTES and various amounts of unlabeled human or rat RANTES. (C) Influence of human and rat RANTES on pR33-mediated constitutive signaling. COS-7 cells were transiently transfected with either pcDNA3 or pcDNA3/R33. Human or rat RANTES was added to the supernatants, both in the [3H]inositol phosphate production assay (InsP) and in the reporter gene assays (CRE, NF-κB, and SRE). Data are presented as percentages of pR33 activity (100%). (D) Influence of rat RANTES on pcDNA3- and pcDNA3/R33-transfected cells in the CRE assay. Error bars, standard error of the mean.
pR33- and pR33-EGFP-induced stimulation of SRE-mediated transcription in Rat2 cells.
In previous experiments, GPCR activity was measured in COS-7 cells, which are well characterized and highly suitable for signal transduction studies. However, since these cells are not likely to represent appropriate hosts for efficient replication of the species-specific RCMV, we set out to test the activity of pR33, pR33-EGFP, and pEGFP-R33 in rat cells (Rat2). These cells have previously been demonstrated to support full replication of RCMV (e.g., see reference 51). We first investigated the expression of pR33-EGFP in Rat2 cells by confocal microscopy and found the expression pattern of this protein to be similar to that in COS-7 cells (data not shown). This suggests that pR33-EGFP may be properly expressed as a functional GPCR in Rat2 cells.
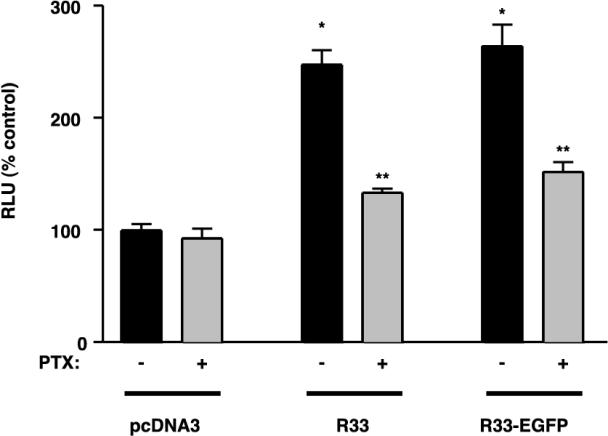
To test whether pR33-EGFP and pR33 are capable of signaling in Rat2 cells, we transfected these cells with either pcDNA3/R33 or pcDNA3/R33-EGFP and tested their activity in the SRE assay. Indeed, both pR33 and pR33-EGFP were found to signal in an agonist-independent, constitutive fashion in Rat2 cells (Fig. 6), leading to a PTX-sensitive activation of SRE-driven transcription. Thus, pR33 and pR33-EGFP induce an upregulation of SRF that is mediated via a Gi/0 protein-dependent pathway(s) in both COS-7 and Rat2 cells. As of yet, we have not been able to detect pR33 activity in Rat2 cells in any of the other reported assays.
FIG. 6.
pR33- and pR33-EGFP-mediated SRE activation in Rat2 cells. Rat2 cells (106) were transiently transfected with reporter plasmid pTLNC-3SRE (5.0 μg of DNA/106 cells) and with either pcDNA3, pcDNA3/R33 (R33), or pcDNA3/R33-EGFP (R33-EGFP) at 0.5 μg of DNA/106 cells. At 48 h after transfection, luciferase activity was measured. The experiment was done in six replicates, either in the presence (+) or absence (−) of PTX in the culture medium. Single asterisks indicate differences statistically significant (P < 0.05) from data from pcDNA3-transfected cells (in the absence of PTX). Double asterisks indicate statistically significant differences (P < 0.05) between data from PTX-treated and -untreated cells that were transfected with the same expression construct. Error bars, standard error of the mean.
DISCUSSION
The biological significance of the RCMV R33-like genes in the pathogenesis of CMV infections has previously been demonstrated in studies with recombinant viruses (5, 17). In contrast to their WT counterparts, RCMV and MCMV mutant viruses lacking a functional R33 or M33 gene, respectively, were unable to replicate in the salivary glands and displayed lower mortality rates in infected animals (5, 17). Although these findings illustrated the importance of the R33-like genes, not much was known about the biochemical or pharmacological properties of the proteins encoded by these genes. Here, we report that R33 encodes a membrane-associated protein that functions as a GPCR. Furthermore, we demonstrate for the first time that a member of the R33-like gene family encodes a constitutively active receptor that activates multiple signaling pathways in both COS-7 and Rat2 cells.
Currently, the concept of constitutive signaling by GPCRs is well accepted. It has been demonstrated for multiple endogenously expressed GPCRs, such as the H3 receptor in the rat brain (36). Although the biological relevance is still poorly understood, it is clear that constitutive activity can be of major pathophysiological significance. Various diseases, including proliferative disorders, have been described that are caused by GPCRs that are constitutively active due to naturally occurring mutations in these receptors (42, 43, 46, 48).
Previously, two other virus-encoded GPCRs were demonstrated to be constitutively active. The Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF74-encoded GPCR (KSHV-GPCR) and HCMV pUS28 both signal in an agonist-independent fashion, resulting in constitutive activation of various signaling pathways (2, 3, 11, 35, 45, 47). KSHV is thought to be the causative agent of AIDS- and non-AIDS-related Kaposi’s sarcoma as well as Castleman’s disease (12), and the KSHV-GPCR has been proposed as a key mediator of both the proliferative and angiogenic properties of KSHV (2, 3, 12, 28, 35, 45, 47, 57). As described in the introduction, several interesting pUS28 activities have been suggested to be of importance in the pathogenesis of HCMV infections. However, it is still unclear which role constitutive signaling by pUS28 plays in these activities. Similarly, the physiological importance of constitutive pR33 signaling remains speculative.
Both RCMV pR33 and HCMV pUS28 were found to constitutively induce activation of PLC. Upon ligand binding, chemokine receptors also activate PLC, and this is predominantly achieved by release of βγ subunits from activated, PTX-sensitive Gi proteins, which stimulate the PLC isoform PLC-β2 (30, 50). In addition, some chemokine receptors can activate PLC by direct interaction with the α subunits of Gq class proteins like Gαq, Gα11, Gα14, Gα15, or Gα16 (1, 30, 50). Similarly, the constitutive pR33-induced PLC activation involves Gq as well as Gi/0 classes of G proteins, whereas constitutive pUS28-mediated PLC activation is solely dependent on Gq class proteins (11). Additionally, pR33 shows a remarkable level of selectivity for Gα11 over Gαq while pUS28 interacts both with Gαq and Gα11 to enhance PLC activity in COS-7 cells. A wide variety of GPCRs, including chemokine receptors, can couple to PLC via activation of Gα16, a hematopoesis-specific member of the Gq class (34). Even receptors that selectively couple to Gi/0 proteins, like the muscarinic acetylcholine M2 receptor, induce InsP production in the presence of PTX when Gα16 is present (34). Surprisingly, pR33-mediated constitutive PLC activation is not enhanced by expression of Gα16. Since GPCR-induced PLC activation via Gα16 has been demonstrated as an agonist-mediated response, it is possible that pR33 might interact with Gα16 only upon ligand binding and not in a ligand-independent fashion. On the other hand, pR33 might not be capable of Gα16 activation at all, as has been shown previously for certain CC-chemokine receptors (30). Nevertheless, we were not able to investigate these notions because at present no ligands for pR33 have been identified.
As indicated above, pR33 constitutively couples to Gi proteins, and this interaction stimulates the pR33-induced PLC activity. Like PLC stimulation by Gi-coupled chemokine receptors, this effect might be mediated by the release of Gi βγ subunits, because the pR33-induced PLC activity in the presence of PTX was similar to that seen in the presence of βγ scavengers. Since COS-7 cells lack the PLC isoform that is most sensitive to stimulation by Gi-derived βγ subunits (PLC-β2), it is likely that the pR33-induced βγ-mediated signal activates other PLC isoforms expressed in COS-7 cells, i.e., PLC-β1 and PLC-β3 (16, 27, 56).
In addition to PLC activation, both pR33 and pUS28 also induce NF-κB-mediated transcription in a constitutive fashion. In the case of pUS28, this constitutive effect is due to the activation of Gq proteins via the release of Gq βγ subunits (11). Interestingly, pR33-mediated NF-κB transcription is completely inhibited by PTX, indicating that this activity of pR33 is mainly due to the activation of Gi/0 proteins. Similarly, pR33 induces SRE-mediated transcription and inhibits CRE-mediated transcription in a PTX-sensitive, constitutive fashion. These two signaling parameters, however, are not affected by pUS28. Together with the PLC data, these findings illustrate that although pR33 and pUS28 are both virus-encoded receptors which signal constitutively, they are completely different with regard to their respective signaling profiles.
It is likely that the PTX-sensitive, pR33-induced signaling to CRE, SRE, and NF-κB, may be mediated by the Gi2 protein, since COS-7 cells do not endogenously express any other PTX-sensitive G proteins apart from Gi2 (27). Previously, signaling by Gi2 has been demonstrated to be linked to downregulation of cytokine expression (32). Additionally, PTX-sensitive NF-κB-mediated transcription has been shown to influence the expression of cytokines, chemokines, growth factors, and cell adhesion molecules as well as some acute-phase proteins (15). The induction of NF-κB-mediated transcription has also been linked to inflammatory events associated with various disease states, such as autoimmune diseases and atherosclerosis (15). Whether or not (the constitutive activity of) pR33 plays a role in any of these processes during RCMV infection in vivo will have to be addressed in future studies.
A factor that may influence the effect of (constitutive) pR33 signaling in vivo is the presence of endogenous ligands. Interaction of pR33 with endogenous ligands may modulate constitutive activity as well as (de)activate additional signaling pathways. Previously, modulation of constitutive activity by agonists and inverse agonists has been demonstrated for KSHV-GPCR and pUS28 in vitro. The constitutive activity of the KSHV-GPCR is modulated positively by GRO-α and negatively by IP-10 and SDF-1α, which are thus acting as agonist and inverse agonists, respectively (23, 35, 44, 45). Nevertheless, not all ligands that bind to a certain GPCR also affect the constitutive activity of that GPCR. This was previously shown for the KSHV-GPCR ligand IL-8 (23, 44). Similarly, pUS28 binds and responds to various CC-chemokines, whereas none of these CC-chemokines have been reported to modulate the constitutive activity of pUS28. By contrast, fractalkine was identified as a partial inverse agonist for pUS28, inhibiting both constitutive PLC activation and NF-κB-mediated transcription (11).
The identification of agonists and, in particular, inverse agonists of constitutive viral GPCR signaling might provide valuable tools for elucidation of the role of this constitutive activity in the pathogenesis of viral infection. In addition, inverse agonists may be regarded as putative leads for antiviral drug design. In order to identify potential (inverse) agonists of pR33, we tested several chemokines for modulation of pR33-mediated constitutive activity. However, neither the human chemokines of the CC (RANTES, eotaxine, Teck, MCP-1), CX3C (fractalkine) or CXC class (IP-10, IP-9, GRO-α, IL-8) nor the CC-chemokine rat RANTES was able to change the activity of pR33 significantly in any of the signaling assays used in this study. In addition, pR33 was unable to bind RANTES, in contrast to its homolog from HHV-6B (pU12). The latter protein was shown to bind RANTES, MCP-1, and MIP-1α as well as MIP-1β, resulting in an intracellular calcium flux (25). These findings illustrate that individual members of the R33 family can have significantly different ligand-binding characteristics.
Taken together, we found pR33 to activate and inhibit various signaling pathways in the absence of exogenously added ligands. However, it cannot be ruled out that the activity of pR33 is the result of binding of yet-unidentified, endogenously produced ligands. In this respect it is interesting that a novel CC-chemokine was identified, CCL28, which is a ligand for both CCR10 and CCR3 (39, 55). This chemokine, also known as mucosa-associated epithelial chemokine, is expressed in human and murine epithelial cells of diverse mucosal tissues and directs chemotaxis of CCR10-expressing lymphoid cells. Interestingly, CCL28 transcripts are most abundant in the salivary glands (39). With respect to the sequence similarity between CCR10, CCR3, and pR33, we hypothesized that the ability of RCMV to establish a successful infection within the salivary glands could be dependent on the interaction of a CCL28-like chemokine with pR33 (54). This putative interaction of pR33 with the rat homolog of CCL28 is currently under investigation.
Acknowledgments
Y.K.G. and P.C. contributed equally to this work.
We are grateful to C. Gerard and D. Bota for providing us with plasmid pcDNA3/CCR10. We thank Patrick Beisser for critically reading the manuscript and Erik Beuken for DNA sequencing.
Y.K.G. is supported by a grant (901-02-224) from The Netherlands Organization for Scientific Research (NWO, medical sciences). C.V. and M.J.S. are supported by a fellowship of the Royal Netherlands Academy of Arts and Sciences (KNAW).
REFERENCES
- 1.Arai, H., and I. F. Charo. 1996. Differential regulation of G-protein-mediated signaling by chemokine receptors. J. Biol. Chem. 271:21814–21819. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershenghorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347–350. [DOI] [PubMed] [Google Scholar]
- 3.Bais, C., B. Santamasso, O. Coso, L. Arvanitakis, E. Geras-Raaka, J. Silvio Gutkind, A. S. Asch, E. Cesarman, M. C. Gershenghorn, and E. A. Mesri. 1998. G-protein-coupled receptor of Kaposi’s sarcoma-associated is a viral oncogene and angiogenesis activator. Nature 391:86–89. (Erratum, 392:210.) [DOI] [PubMed] [Google Scholar]
- 4.Beisser, P. S., G. E. L. M. Grauls, C. A. Bruggeman, and C. Vink. 1999. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J. Virol. 73:7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisser, P. S., C. Vink, J. G. van Dam, G. E. L. M. Grauls, S. J. V. Vanherle, and C. A. Bruggeman. 1998. The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection. J. Virol. 72:2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, G. L., and R. Horuk. 1997. Iodination of chemokines for use in receptor binding analysis. Methods Enzymol. 288:134–148. [DOI] [PubMed] [Google Scholar]
- 7.Billstrom, M. A., G. L. Johnson, N. J. Avdi, and G. S. Worthen. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 72:5535–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodaghi, B., T. R. Jones, D. Zipeto, C. Vita, L. Sun, L. Laurent, F. Arenzana-Seisdedos, J. Virelizier, and S. Michelson. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 188:855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broers, J. L., B. M. Machiels, G. J. van Eys, H. J. Kuijpers, E. M. Manders, van R. Driel, and F. C. Ramaekers. 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 112:3463–3475. [DOI] [PubMed] [Google Scholar]
- 10.Carman, C. V., J. L. Parent, P. W. Day, A. N. Pronin, P. M. Sternweis, P. B. Wedegaertner, A. G. Gilman, J. L. Benovic, and T. Kozasa. 1999. Selective regulation of Gα(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J. Biol. Chem. 274:34483–34492. [DOI] [PubMed] [Google Scholar]
- 11.Casarosa, P., R. A. Bakker, D. Verzijl, M. Navis, H. Timmerman, R. Leurs, and M. J. Smit. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 276:1133–1137. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi’s sarcoma-associated herpesvirus contains G-protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J. Virol. 70:8218–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerni, T. Horsnell, C. A. Hutchison, I. I. I., T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 14.Chee, M. S., S. C. Satchwell, E. Preddie, K. M. Weston, and B. G. Barell. 1990. Hum. cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 344:774–777. [DOI] [PubMed] [Google Scholar]
- 15.Chen, F., V. Castranova, X. Shi, and L. M. Demers. 1999. New insights into the role of nuclear factor-κB, a ubiquitous transcription factor in the initiation of diseases. Clin. Chem. 45:7–17. [PubMed] [Google Scholar]
- 16.Clapham, D. E., and E. J. Neer. 2000. G protein βγ subunits. Annu. Rev. Pharmacol. Toxicol. 37:167–203. [DOI] [PubMed] [Google Scholar]
- 17.Davis-Poynter, N. J., D. M. Lynch, H. Vally, G. R. Shellam, W. D. Rawlinson, B. G. Barell, and H. E. Farrell. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 71:1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diviani, D., A. L. Lattion, N. Larbi, P. Kunapuli, A. Pronin, J. L. Benovic, and S. Cotecchia. 1996. Effect of different G protein-coupled receptor kinases on phosphorylation and desensitization of the α1B-adrenergic receptor. J. Biol. Chem. 271:5049–5058. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez, G., T. R Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federman, A. D., B. R. Conklin, K. A. Schrader, R. R. Reed, and H. R. Bourne. 1992. Hormonal stimulation of adenylyl cyclase through Gi-protein βγ subunits. Nature 356:159–161. [DOI] [PubMed] [Google Scholar]
- 21.Fluhmann, B., U. Zimmermann, R. Muff, G. Bilbe, J. A. Fischer, and W. Born. 1998. Parathyroid hormone responses of cyclic AMP-, serum- and phorbol ester-responsive reporter genes in osteoblast-like UMR-106 cells. Mol. Cell. Endocrinol. 139:89–98. [DOI] [PubMed] [Google Scholar]
- 22.Gao, J., and P. M. Murphy. 1994. Human cytomegalovirus open reading frame US28 encodes a functional β chemokine receptor. J. Biol. Chem. 269:28539–28542. [PubMed] [Google Scholar]
- 23.Geras-Raaka, E., A. Varma, H. Ho, I. Clark-Lewis, and M. C. Gershenghorn. 1998. Human interferon-γ-inducible protein 10 (IP-10) inhibits constitutive signaling of Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J. Exp. Med. 188:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51. [DOI] [PubMed] [Google Scholar]
- 25.Isegawa, Y., Z. Ping, K. Nakano, N. Sugimoto, and K. Yamanishi. 1998. Human herpesvirus 6 open reading frame U12 encodes a functional β-chemokine receptor. J. Virol. 72:6104–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmin, D. I., M. Rits, D. Bota, N. P. Gerard, G. J. Graham, I. Clark-Lewis, and C. Gerard. 2000. Cutting edge: identification of the orphan receptor G-protein-coupled receptor 2 as CCR10, a specific receptor for the chemokine ESkine. J. Immunol. 164:3460–3464. [DOI] [PubMed] [Google Scholar]
- 27.Katz, A., D. Wu, and M. I. Simon. 1992. Subunits beta gamma of heterotrimeric G protein activate beta 2 isoform of phospholipase C. Nature 360:686–689. [DOI] [PubMed] [Google Scholar]
- 28.Kirshner, J. R., K. Staskus, A. Haase, M. Lagunoff, and D. Ganem. 1999. Expression of the open reading frame of Kaposi’s sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73:6006–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kledal, T. N., M. M. Rosenkilde, and T. W. Schwartz. 1998. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett. 441:209–214. [DOI] [PubMed] [Google Scholar]
- 30.Kuang, Y., Y. Wu, H. Jiang, and D. Wu. 1996. Selective G protein coupling by C-C chemokine receptors. J. Biol. Chem. 271:3975–3978. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn, D. E., C. J. Beall, and P. E. Kollattukudy. 1995. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem. Biophys. Res. Commun. 211:325–330. [DOI] [PubMed] [Google Scholar]
- 32.Luther, S. A., and J. G. Cyster. 2001. Chemokines as regulators of T cell differentiation. Nat. Immunol. 2:102–107. [DOI] [PubMed] [Google Scholar]
- 33.Margulies, B. J., H. Browne, and W. Gibson. 1996. Identification of the human cytomegalovirus G-protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milligan, G., F. Marshall, and S. Rees. 1996. G16 as a universal G protein adapter: implications for agonist screening strategies. Trends Pharmacol. Sci. 17:235–237. [DOI] [PubMed] [Google Scholar]
- 35.Montaner, S., A. Sodhi, S. Pece, E. A. Mesri, and J. S. Gutkind. 2001. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor promotes endothelial cell survival through the activation of AKT/protein kinase B. Cancer Res. 61:2641–2648. [PubMed] [Google Scholar]
- 36.Morisset, S., A. Rouleau, X. Ligneau, F. Gbahou, J. Tardivel-Lacombe, H. Stark, W. Schunack, C. R. Ganellin, J. C. Schwartz, and J. M. Arrang. 2000. High constitutive activity of native H3 receptors regulates histamine neurons in brain. Nature 408:860–864. [DOI] [PubMed] [Google Scholar]
- 37.Neote, K., D. DiGregorio, J. Y. Mak, R. Horuk, and T. J. Schall. 1993. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 72:415–425. [DOI] [PubMed] [Google Scholar]
- 38.Nicholas, J. 1996. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J. Virol. 70:5975–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, J., E. J. Kunkel, U. Gosslar, N. Lazarus, P. Langdon, K. Broadwell, M. A. Vierra, M. C. Genovese, E. C. Butcher, and D. Soler. 2000. A novel chemokine ligand for CCR10 and CCR3 expressed by epithelial cells in mucosal tissues. J. Immunol. 165:2943–2949. [DOI] [PubMed] [Google Scholar]
- 40.Premont, R. T., J. Inglese, and R. J. Lefkowitz. 1995. Protein kinases that phosphorylate activated G protein-coupled receptors. FASEB J. 9:175–182. [DOI] [PubMed] [Google Scholar]
- 41.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins, L. S., J. H. Nadeau, K. R. Johnson, M. A. Kelly, L. Roselli-Rehfuss, E. Baack, K. G. Mountjoy, and R. D. Cone. 1993. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 72:827–834. [DOI] [PubMed] [Google Scholar]
- 43.Robinson, P. R., G. B. Cohen, E. A. Zhukovsky, and D. D. Oprian. 1992. Constitutively active mutants of rhodopsin. Neuron 9:719–725. [DOI] [PubMed] [Google Scholar]
- 44.Rosenkilde, M. M., T. N. Kledal, H. Brauner-Osborne, and T. W. Schwartz. 1999. Agonists and inverse agonists for the herpesvirus 8-encoded constitutively active seven-transmembrane oncogene product. J. Biol. Chem. 274:956–961. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz, M., and P. M. Murphy. 2001. Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor constitutively activates NF-κB and induces proinflammatory cytokine and chemokine production via a C-terminal signaling determinant. J. Immunol. 167:505–513. [DOI] [PubMed] [Google Scholar]
- 46.Shenker, A., L. Laue, S. Kosugi, J. J. Merendino, Jr., T. Minegishi, and G. B. Cutler, Jr. 1993. A constitutively activating mutation of the luteinizing hormone receptor in familial male precocious puberty. Nature 365:652–654. [DOI] [PubMed] [Google Scholar]
- 47.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor upregulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 60:4873–4880. [PubMed] [Google Scholar]
- 48.Spiegel, A. M. 1996. Defects in G protein-coupled signal transduction in human disease. Annu. Rev. Physiol. 58:143–170. [DOI] [PubMed] [Google Scholar]
- 49.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511–520. [DOI] [PubMed] [Google Scholar]
- 50.Thelen, M. 2001. Dancing to the tune of chemokines. Nat. Immunol. 2:129–134. [DOI] [PubMed] [Google Scholar]
- 51.Vink, C., E. Beuken, and C. A. Bruggeman. 1997. Cloning and functional characterization of the origin of lytic-phase DNA replication of rat cytomegalovirus. J. Gen. Virol. 78:2963–2973. [DOI] [PubMed] [Google Scholar]
- 52.Vink, C., P. S. Beisser, and C. A. Bruggeman. 1999. Molecular mimicry by cytomegaloviruses. Function of cytomegalovirus-encoded homologues of G protein-coupled receptors, MHC class I heavy chains and chemokines. Intervirology 42:342–349. [DOI] [PubMed] [Google Scholar]
- 53.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vink, C., M. J. Smit, R. Leurs, and C. A. Bruggeman. 2001. The role of cytomegalovirus-encoded homologs of G protein-coupled receptors and chemokines in manipulation of and evasion from the immune system. J. Clin. Virol. 23:43–55. [DOI] [PubMed] [Google Scholar]
- 55.Wang, W., H. Soto, E. R. Oldham, M. E. Buchanan, B. Homey, D. Catron, N. Jenkins, N. G. Copeland, D. J. Gilbert, N. Nguyen, J. Abrams, D. Kershenovich, K. Smith, T. McClanahan, A. P. Vicari, and A. Zlotnik. 2000. Identification of a novel chemokine (CCL28), which binds CCR10 (GPR2). J. Biol. Chem. 275:22313–22323. [DOI] [PubMed] [Google Scholar]
- 56.Wu, D., A. Katz, and M. I. Simon. 1993. Activation of phospholipase C beta 2 by the alpha and beta gamma subunits of trimeric GTP-binding protein. Proc. Natl. Acad. Sci. USA 90:5297–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, T. Y., S. C. Chen, M. W. Leach, D. Manfra, B. Momey, M. Wiekowski, L. Sullivan, C. H. Jenh, S. K. Narula, S. W. Chensue, and S. A. Lira. 2000. Transgenic expression of the chemokine receptor encoded by human herpesvirus 8 induces an angioproliferative disease resembling Kaposi’s sarcoma. J. Exp. Med. 191:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]