Abstract
Administration of either lamivudine (2′-deoxy-3′-thiacytidine) or l-FMAU (2′-fluoro-5-methyl-β-l-arabinofuranosyluracil) to woodchucks chronically infected with woodchuck hepatitis virus (WHV) induces a transient decline in virus titers. However, within 6 to 12 months, virus titers begin to increase towards pretreatment levels. This is associated with the emergence of virus strains with mutations of the B and C regions of the viral DNA polymerase (T. Zhou et al., Antimicrob. Agents Chemother. 43:1947–1954, 1999; Y. Zhu et al., J. Virol. 75:311–322, 2001). The present study was carried out to determine which of the mutants that we have identified conferred resistance to lamivudine and/or to l-FMAU. When inserted into a laboratory strain of WHV, each of the mutations, or combinations of mutations, of regions B and C produced a DNA replication-competent virus and typically conferred resistance to both nucleoside analogs in cell culture. Sequencing of the polymerase active site also occasionally revealed other mutations, but these did not appear to contribute to drug resistance. Moreover, in transfected cells, most of the mutants synthesized viral DNA nearly as efficiently as wild-type WHV. Computational models suggested that persistence of several of the WHV mutants as prevalent species in the serum and, by inference, liver for up to 6 months following drug withdrawal required a replication efficiency of at least 10 to 30% of that of the wild type. However, their delayed emergence during therapy suggested replication efficiency in the presence of the drug that was still well below that of wild-type WHV in the absence of the drug.
Hepatitis B virus (HBV) infection is a major public health problem, responsible for about 1.2 million deaths a year. With the availability of a vaccine and hepatitis B immunoglobulin preparations, it is possible to prevent nearly all HBV infections. However, universal vaccination has not yet been achieved, and there remain about 350 million HBV carriers worldwide. For carriers with active liver disease, alpha interferon therapy over a 4- to 6-month period has the potential to eliminate the virus, possibly by activating the host response to viral antigens. Unfortunately, this therapy induces virus clearance in only about 30% of these carriers. Thus, better treatments are needed.
Lamivudine is a cytosine analog that has been approved for clinical use against human immunodeficiency virus and HBV by the U.S. Food and Drug Administration. Lamivudine therapy can reduce the serum HBV titer, normalize serum alanine aminotransferase (ALT) levels, facilitate an increase in circulating, virus-reactive T cells, and improve liver histology, all without major side effects (5, 13, 16). Unfortunately, prolonged lamivudine treatment leads to the emergence of lamivudine-resistant mutants, generally followed by an increase in virus titers, elevations of ALTs beyond the normal range, and progressing liver pathology (13). Long-term virus suppression, without emergence of drug-resistant variants, has been seen in only about 30% of HBV carriers treated continuously for 3 years.
Five conserved regions, A to E, have been recognized in the active site of RNA-dependent DNA polymerases (4, 17), including the HBV polymerase. HBV resistance to lamivudine is always associated with a mutation of the YMDD motif in the C region of the viral polymerase, either to YIDD or YVDD (rtM204I/V; numbered according to reference 23). The latter mutation is always accompanied by a Leu-to-Met mutation upstream at amino acid rtL180 of the B region; this may also be found with the YIDD mutation (Fig. 1C) (2). Cell culture studies have revealed that these mutations could produce resistance to l-FMAU as well (9). The upstream mutation, by itself, is able to confer resistance to another anti-HBV nucleoside that has been employed in clinical trials, famciclovir, the prodrug of penciclovir (1, 3, 6, 9, 24).
FIG. 1.
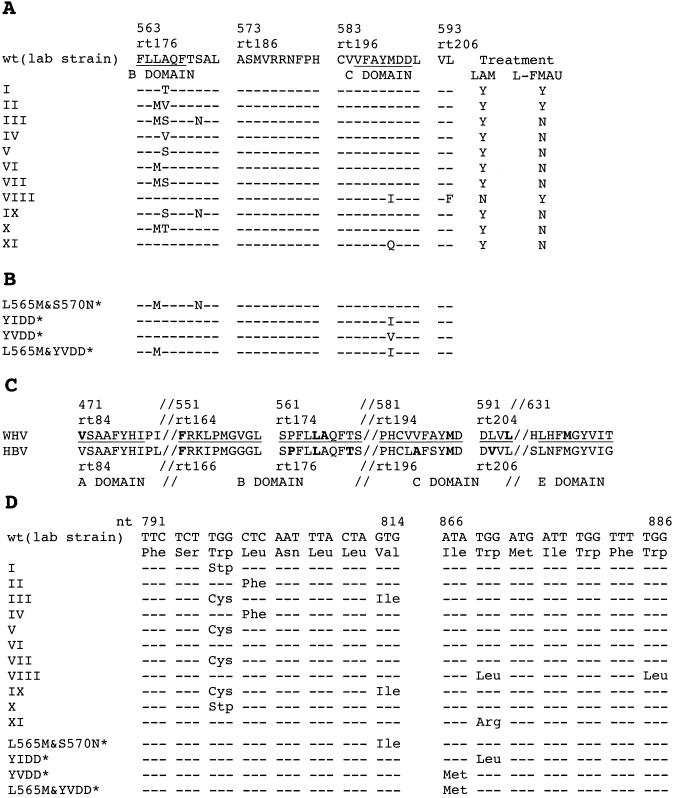
Mutations in the polymerase active site. (A) Eleven WHV variants (types I to XI) with mutations in the polymerase active site were detected in the sera of lamivudine- and/or l-FMAU-treated woodchucks. Two (VIII and XI) have mutations in the C region; the remainder have mutations only in the B region. The amino acid sequence of a cloned wild-type WHV polymerase (12, 27) is shown at the top. The columns to the right indicate which drug treatment was associated with the emergence of a variant. (B) Sequence of polymerase mutants of WHV produced for phenotypic analyses. These particular variants have not been found, so far, to occur in drug-treated woodchucks. The YIDD, YVDD, and L565M+YVDD mutants were analogous to HBV mutations that arise in patients subjected to lamivudine therapy. (C) Comparison of amino acid sequences in conserved regions of the HBV and WHV genomes. Amino acids known to be mutated in drug-resistant variants are shown in boldface. (D) Predicted mutations in the overlapping envelope protein due to drug resistance mutations of the WHV polymerase. Amino acid changes in the envelope were predicted for all but the type VI mutant. Among these, a tryptophan-to-stop codon change was predicted for type I and X mutants. In panels A and C, amino acid positions in the WHV polymerase ORF are numbered from the first AUG of polymerase and from the beginning of the reverse transcriptase region (23); only the latter convention is used for HBV.
As with HBV carriers, emergence of drug-resistant variants occurs in woodchuck hepatitis virus (WHV)-infected woodchucks treated with lamivudine (15, 28). Resistance also develops in woodchucks treated with l-FMAU (29). Elevation of WHV titers towards pretreatment levels usually occurs within 6 to 12 months. Mutant virus is sometimes prevalent in the serum months before virus titers begin to significantly rebound, as was also found in HBV carriers (6). However, the pattern of mutations is different between the two viruses. Whereas mutation of the YMDD motif typifies HBV resistance, such mutations are not frequently found as either lamivudine or l-FMAU resistance develops in infected woodchucks. Rather, mutations typically map upstream, in the B region. In initial studies, three B region mutants were identified and shown to be lamivudine resistant (15, 28). Subsequent studies revealed six additional mutants in lamivudine-treated woodchucks, but their ability to cause drug resistance was not determined (29). In the current study, we show that these mutants, and two more that we have recently identified, are resistant to lamivudine and a second drug, l-FMAU. Mutations in six of these newly identified variants mapped to the B region and in only two cases mapped to the downstream YMDD motif. The mutations conferred drug resistance when substituted into a wild-type strain of WHV. Transfection assays suggested that some of the mutants replicated with efficiency comparable to that of the wild type. However, consideration of the hepadnavirus mutation rate (19), kinetics of emergence of the mutants during drug treatment, and their persistence following cessation of drug therapy, was more consistent with a reduced replication efficiency, though greater than 10% of that of the wild type.
MATERIALS AND METHODS
Viral DNA.
Virus was obtained from the sera of woodchucks chronically infected with WHV that had been administered either lamivudine or l-FMAU (15, 28, 29). To extract WHV DNA for sequencing, 50 μl of serum was layered onto a 10 to 20% sucrose step gradient containing 150 mM NaCl and 20 mM Tris-HCl (pH 7.4). Virus was pelleted by centrifugation for 3 h at 50,000 rpm, at 4°C in a Beckman SW60 rotor. The virus pellet was suspended in a mixture of 20 mM Tris-HCl (pH 7.5), 10 mM EDTA, 100 mM NaCl, 0.2% (wt/vol) sodium dodecyl sulfate (SDS), and 0.5 mg of Pronase per ml, and the mixture was incubated for 1 h at 37°C. Following two extractions with phenol-chloroform (1:1), virus DNA was precipitated by addition of 2 volumes of ethanol. The precipitate was collected by centrifugation, and the viral DNA was dissolved in 30 μl of water.
Genotyping of WHV.
Direct sequencing of PCR products spanning portions of the WHV polymerase gene encoding the DNA polymerase active site was carried out as previously described (28). For more detailed analyses, PCR products were cloned prior to sequence determinations. Five microliters of virus DNA solution, extracted from 50 μl of serum, was used as a template. PCR was carried out in 50-μl reaction mixtures containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTP), 0.4 μM sense primer (5′-AGATTGGTGGTGCACTTCTCTCAG-3′; WHV nucleotides [nt] 385 to 408) (12), 0.4 μM antisense primer (5′-CCACGGAATTGTCAGTGCCCAACA-3′; nt 1451 to 1474), and 1.25 U of AmpliTaq Gold (Perkin-Elmer, Santa Clara, Calif.). Following a denaturation step of 10 min at 94°C, reactions were carried through 30 cycles of 1 min at 94°C, 1 min at 57°C, and 1.25 min at 72°C. A 7-min incubation at 72°C was carried out after the last cycle. The PCR products were purified with a QIAquick PCR purification kit (Qiagen) and cloned with a TOPO PCR cloning kit (Invitrogen). The nucleotide sequence of the cloned DNA was then determined. Mutations in the polymerase active site were identified by comparison with the nucleotide sequence of the cloned WHV DNA used in this laboratory (28). Viral DNA synthesis in cells transfected with this clone is inhibited by lamivudine and l-FMAU (28, 29).
Analysis of viral DNA synthesis in HepG2 cells.
For transfection studies, we employed a plasmid in which WHV pregenomic RNA is transcribed from a cytomegalovirus (CMV) immediate-early (IE) promoter (20, 28). The CMV IE promoter was used because the endogenous promoters of WHV appear to function poorly in HepG2 cells (8, 21; Q. Di and W. S. Mason, unpublished observations). Mutations in the polymerase active site were introduced into this plasmid by site-directed mutagenesis, and the constructs were verified by DNA sequencing.
HepG2 cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium-F12 medium supplemented with 10% fetal bovine serum. A total of 1.5 × 106 cells were seeded onto 60-mm-diameter dishes 24 to 48 h before transfection. Growth medium with or without drug was replaced at least 6 h before transfection. Lamivudine and l-FMAU were both used at a concentration of 100 μM. Cells were transfected with 1.0 μg of a plasmid containing WHV DNA and 0.5 μg of a green fluorescent protein (GFP) expression vector by using Effectene transfection reagent (Qiagen) according to the manufacturer’s recommendations. With GFP-positive cells as a standard, transfection efficiencies between plates did not differ by more than ±25%. Cells transfected with 0.5 μg of the GFP expression vector were used as a negative control. Growth media were replaced daily. Five days posttransfection, the time of maximal accumulation of WHV DNA replication intermediates (with wild-type WHV), the monolayers were rinsed twice with phosphate-buffered saline and stored at −80°C for subsequent DNA extraction. All transfection studies were repeated at least twice with similar results.
Detection of viral DNA replication intermediates in transfected cells.
Monolayers were thawed and incubated for 10 min at 37°C in 1 ml of a lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1% Nonidet P-40, 50 mM NaCl, and 8% sucrose. Cell lysates were transferred into 1.5-ml centrifuge tubes and centrifuged at 11,000 × g for 2 min. The aqueous phase was collected and adjusted to contain 10 mM magnesium acetate, 100 μg of RNase A per ml, and 100 μg of DNase I per ml and incubated at 37°C for 30 min. The mixture was clarified by centrifugation for 4 min at 11,000 × g. The supernatant was adjusted to contain 17 mM EDTA. Two hundred-sixty microliters of 35% polyethylene glycol 8000, dissolved into 1.5 M NaCl, was next added. After 1 h at 4°C, virus nucleocapsids were collected by centrifugation for 5 min at 12,000 rpm and 4°C. The pellet was dissolved in 500 μl of a digestion buffer containing 25 mM Tris-HCl (pH 7.4), 10 mM EDTA, 0.1 M NaCl, 0.5% (wt/vol) SDS, and 0.5 mg of Pronase per ml, and incubated at 37°C for 1 h. After extraction with phenol and then phenol-chloroform (1:1), 50 μl of 3 M sodium acetate and 1 ml of ethanol were added to precipitate the DNA. DNA was collected by centrifugation for 15 min at 11,000 × g and 4°C. The DNA pellet was rinsed once with 70% ethanol and dissolved in 100 μl of a mixture containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
One-fourth of each DNA sample was subjected to electrophoresis into 1.5% agarose gels and then transferred to nitrocellulose membranes (Protran; Schleicher & Schuell, Keene, N.H.). Filters were hybridized with a 32P-labeled DNA probe representing the complete viral genome. After being washed in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS at 65°C for 1 h, signals on the filters were quantified with a Fujix BAS 1000 Bio-Imaging analyzer (Fuji Photo Film Co., Tokyo, Japan).
Cloning of the full-length WHV genome and subcloning of the polymerase gene.
Serum samples were from a lamivudine trial described in an earlier publication (28). Viruses from these samples were used as a template for amplification of full-length WHV genomes by PCR. PCR was performed as described above, except with 2 mM MgCl2, 5′-TTCACCTGTGCCTTGTTTTT-3′ (WHV nt 1941 to 1960) as the sense primer, 5′-GTGAAAAAGATACATGGTTAC-3′ (nt 1945 to 1925) as the antisense primer, 1.25 U of Takara EX Taq (Takara Shuzo Co., Ltd., Japan), initial denaturation for 4 min at 94°C, and a 4-min elongation step in each cycle at 72°C. PCR products were inserted into the pGEM-T vector (Promega Corporation, Madison, Wis.). The nucleotide sequence of the cloned DNA was determined by using the primers listed in Table 1.
TABLE 1.
Primers for WHV genome sequencing
| Primer name | Position no.a | Nucleotide sequence |
|---|---|---|
| 877 | 2400–2421 | GAACTCCAGCTCCATATAGACC |
| 879 | 2631–2654 | AACCAAGCTGCTTAGTTCAATCCG |
| 750 | 3066–3092 | AACCACTTATCCTCAGAATCAGTCAGT |
| 729 | 3195–3221 | CACTTGGCAAGGATTTCCTGTGGATCA |
| 738 | 8–31 | GACATACCACGTGGTTTAGTTCCG |
| 696 | 510–528 | GGATGTATCTGCGGCGTTT |
| 742 | 1030–1053 | GGTGTATTGCCACAAGACAAACAT |
| 744 | 1542–1565 | GACGTCCTTCTGCTACGTCCCTTC |
| 878 | 1874–1853 | TCTAGGATCAATGCTGCCCTCC |
| 743 | 1481–1458 | GACAACACCACGGAATTGTCAGTG |
| 741 | 990–967 | GCCCCACCATTTTGTTTTATTGAC |
| 739 | 473–450 | CAAGTGTTTGCAAGTTTGTAACTG |
| 811 | 10–1, 3320–3310 | GTCCCGAATTCCGGGAACTAT |
| 882 | 3217–3196 | CCACAGGAAATCCTTGCCAAGT |
| 749 | 2975–2952 | CTGTCTGTGTTCCCAAGAATATGG |
| 880 | 2654–2631 | CGGATTGAACTAAGCAGCTTGGTT |
| 907 | 2451–2432 | GGAAGAGTCGAGAGAATGGG |
Numbering system is according to reference 11.
Cloned WHV DNAs were then digested with NsiI and AflII, which cut in the precore and core regions, respectively, to release the entire polymerase gene of 884 amino acids. This DNA was then inserted into the homologous site of a laboratory strain of wild-type WHV. Transfection studies were carried out with these recombinant clones, as described above.
A computational model for evolution of WHV variants in a fully infected liver.
A Fortran program was created to simulate the effects of replication rate on enrichment of wild-type virus over a mutant virus, which has a lower replication rate constant, after withdrawal of lamivudine therapy. Executable code for the MacIntosh may be obtained from S. Litwin (s_litwin@fccc.edu). This Monte Carlo model of the liver assigned an initial viral covalently closed circular DNA (cccDNA) distribution to each of 10,000 simulated hepatocytes. The program was an extension of our previous work (29). Output was the ratio of the amounts of the two viruses at time t to their ratio at time 0 (i.e., enrichment).
Two different inputs were allowed for the cccDNA distribution at time 0. One is based on the assumption that the total cccDNA has a truncated Poisson distribution among hepatocytes, with a mean and range that can be specified. The mean of each cccDNA genotype in those hepatocytes in which it occurs was the same as its mean in the liver as a whole. The other assumes that each cccDNA genotype in a doubly infected hepatocyte has, on average, a 50:50 distribution with the other.
Two options were evaluated for cccDNA synthesis following hepatocyte division. Each assumed a binomial distribution of cccDNA to daughter cells. In the first, new cccDNA is produced from replicative intermediate DNA inherited from the parent, and therefore reflects the cccDNA distribution in the parent (see Fig. 10B). In the second, cccDNA is synthesized from newly made replicating DNA and thus reflects the inherited population of cccDNA (Fig. 10C). In both models, it is assumed that the rate of virus DNA synthesis greatly exceeds the rate of cell division.
FIG. 10.
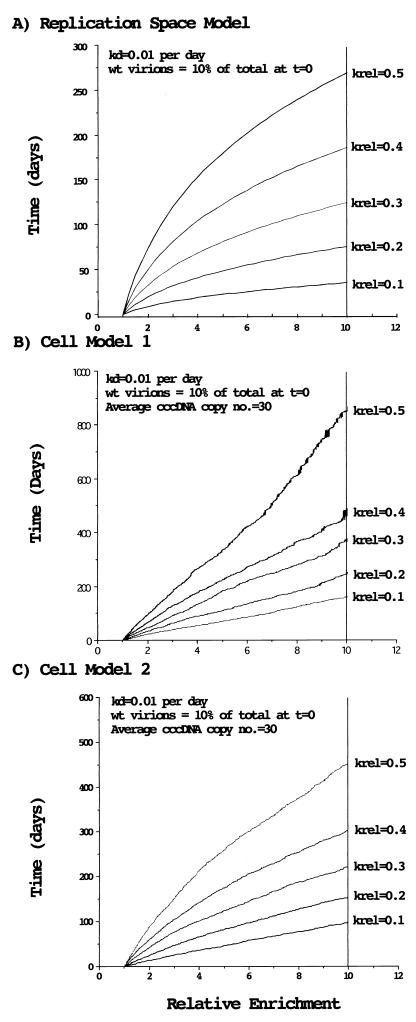
Models for the emergence of wild-type (wt) WHV after the termination of drug treatment. (A) Replication space model for the emergence of wild-type virus after termination of drug treatment. The curves were calculated from the formula of Zhang and Summers (26): t = (total liver regeneration)/(rate of hepatocyte death) = ln {Fwt · E^[1/(1 − krel)] + Fmut · E^[krel/(1 − krel)]}/(rate of cell death)Fwt and Fmut refer to the fraction of wild-type and mutant cccDNA at time 0, and E refers to the relative enrichment of wild type to mutant (that is, the ratio at time t divided by the ratio at time 0). krel is the ratio of the rate of replication of mutant to wild type, and is less than 1. kd is the rate constant (fraction of hepatocytes/day) for death of infected hepatocytes. Calculations were made assuming that wild-type virus was initially 10% of the total in the serum and that the daily rate of cell destruction was 1%. (B) Cellular model 1 for the emergence of wild-type virus following the termination of drug treatment. The model (sim4) is described in Materials and Methods. Calculations were made assuming that wild-type virus is initially 10% of the total in the serum and that cccDNA amplification following cell division occurs via reverse transcription of pregenomic RNA synthesized from inherited cccDNAs. (C) Cellular model 2 for the emergence of wild-type virus following the termination of drug treatment. In contrast to model 1, this model assumes that both cccDNA and replicative DNAs are inherited by daughter cells and that amplification of cccDNA in the daughter cells utilizes the inherited cytoplasmic DNAs.
RESULTS
Multidrug-resistant variants of WHV emerge during long-term treatment with lamivudine and l-FMAU.
During long-term treatment of WHV-infected woodchucks with either lamivudine or l-FMAU, rebound of virus titers often occurs within 6 to 12 months. Rebound is associated with the emergence of WHV strains with one or more mutations in the polymerase active site. These are typically found only in the B region of the polymerase, although mutation of the YMDD motif (C region) has been observed (15, 28, 29).
Three B region mutations found in an initial study (15), designated types I, II, and III (Fig. 1A), confer resistance to lamivudine when inserted into a wild-type WHV genome (28). Types I and II are also found in woodchucks that develop resistance to l-FMAU (29). An additional seven polymerase mutants were subsequently detected in woodchucks that had been treated with lamivudine (29; unpublished observations) (Table 2). One more mutant, not so far isolated from lamivudine-treated woodchucks, was found in a woodchuck that developed resistance to l-FMAU (29). These eight new mutants have been designated types IV to XI. Since the existence of these mutants was first demonstrable by sequencing of the PCR products encoding the polymerase active site that was obtained starting with total viral DNA from serum samples, all of the mutants may be inferred to have had an incidence of at least 10% in the sera in which they were detected. Our experiments also revealed that the number of polymerase mutants present during the emergence of lamivudine resistance was greater than we previously reported (27) (Table 2). Experiments involving site-specific mutagenesis of a wild-type laboratory clone of WHV were next carried out to determine if the mutations characterizing WHV types IV to XI mutations conferred resistance to lamivudine. l-FMAU resistance was also assessed for each mutant.
TABLE 2.
Multiple polymerase mutants found within treated individuals
| Woodchucka | Treatment | WHV variant (no. of clones)b | Previous report |
|---|---|---|---|
| 326 | 14 mo on, 4 mo off | V (5)c/wt (1) | ND |
| 14 mo on, 5 mo off | V (8)/wt (3) | ND | |
| 331 | 12 mo | IV (7)/I (3) | ND |
| 14 mo | I (7)/IV (3)d/II (1)d, VII (1), XI (1) | I/? | |
| 14 mo on, 2 mo off | I (8)/V (3)/II (1), IV (1), VII (1)e | ND | |
| 14 mo on, 5 mo off | V (6)/I (4)/II (1), IX (1), XI (1) | ND | |
| 335 | 12 mo | I (9)/IV (2)/V (1) | wt/I |
| 14 mo on, 1 mo off | IV (4), wt (4)f/I (2)/II (1), IX (1)g | ND | |
| 14 mo on, 4 mo off | I (5), IV (5)/wt (2)/II (1) | ND | |
| 14 mo on, 5 mo off | I (6)/IV (3)/wt (1), VII (1)h | ND | |
| 336 | 14 mo on, 1 mo off | II (10), VI (3)/I (1), VII (1) | ND |
| 14 mo on, 5 mo off | II (5)/VII (2), X (2)/III (1) | ND | |
| 337 | 12 mo | I (10)/VI (2)/wt (1), II (1), VII (1) | wt/I, II |
| 14 mo | I (10)/II (5)/VII (3)/VI (1) | ND | |
| 338 | 14 mo on, 5 mo off | I (6)/V (3) /wt (2) | ND |
| 342 | 14 mo on, 5 mo off | I (7)/wt (6) | ND |
Woodchucks were treated with lamivudine for ca 14 months (27), after which, lamivudine therapy was withdrawn (29). The nucleotide sequences of PCR products corresponding to nt 500 to 1050 of the WHV genome were determined. Aside from the identified mutations, individual clones contained less than 3 nt changes per 500 nt (on average, 0.105%). These mutations were thought to be spontaneous mutations or misamplification by Taq polymerase.
The active site of the pol gene was amplified from serum virus by PCR. The products were then cloned and sequenced. wt, wild type.
Type V produces an A566S amino acid change. One of the type V mutant clones also had a V471I amino acid change in the pol gene.
An F551L amino acid change in the pol gene was found in one of the type IV mutant clones and in the type II mutant clone.
An M635L amino acid change in the pol gene was found in the type VII clone.
One of the wild-type clones had an A566S amino acid change in the pol gene.
An M635L amino acid change in the pol gene was found in the type IX clone.
An M635L amino acid change in the pol gene was found in the type VII clone.
The B and C region mutations found in lamivudine- and l-FMAU-treated woodchucks conferred antiviral resistance when inserted into a wild-type clone of WHV.
To test for replication capacity and drug resistance, HepG2 cells were transfected with WHV expression vectors. Replication was assessed by measurement of intracellular accumulation of viral DNA replication intermediates.
As compared to wild-type WHV, all of the mutants were resistant to lamivudine and to l-FMAU (Fig. 2 and 3). Only with type IV (Fig. 1 to 3) was accumulation of replication intermediates much more strongly suppressed by lamivudine than by l-FMAU. This was surprising, since the natural variant homologous to the type IV construct arose in woodchucks treated with lamivudine (Table 2); as shown below, this greater than expected inhibition by lamivudine was not offset by the presence of additional polymerase mutations in natural isolates of type IV.
FIG. 2.
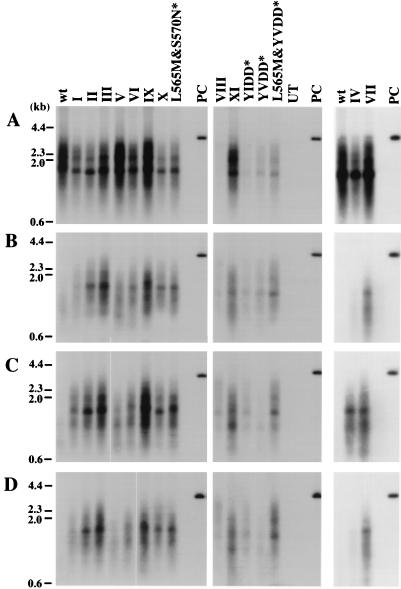
Replication of wild-type (wt) and mutant WHV in HepG2 cells. (A) Transfected cells maintained in drug-free medium. (B) Cells cultured with growth medium containing 100 μM lamivudine. (C) Cells cultured with growth medium containing 100 μM l-FMAU. (D) Cells cultured with growth medium containing 100 μM lamivudine and 100 μM l-FMAU. Molecular weight markers are shown as bars at the left. UT, DNA extracted from negative control; PC, 25 pg of linear, full-length WHV DNA. Amino acid sequences of the mutants are shown in Fig. 1. Asterisks indicate mutants that have not been found in woodchucks.
FIG. 3.
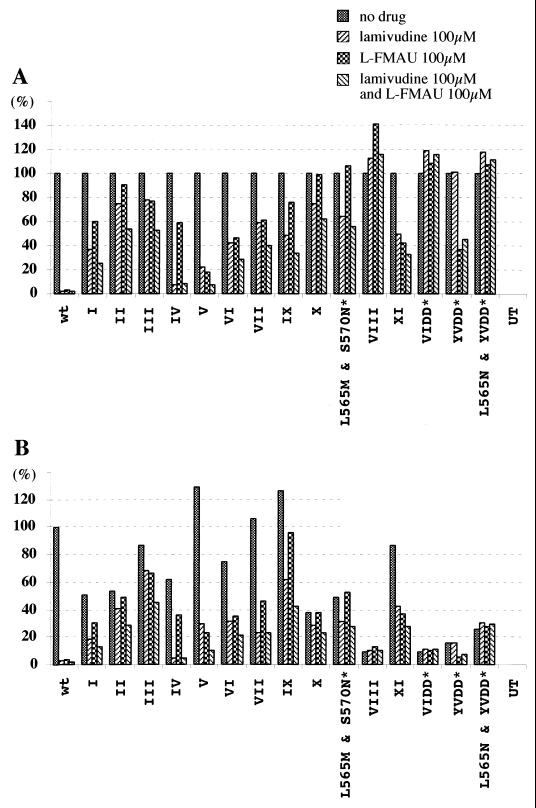
Quantitative summary of transfection results. Signal intensities for the Southern blots shown in Fig. 2 were quantified with a Fujix BAS 1000 Bio-imaging analyzer. A standard amount of cloned WHV DNA (25 pg), lane PC in Fig. 2, served as a hybridization control. (A) Inhibition of accumulation of virus DNA replication intermediates in the presence and absence of drug(s). (B) Accumulation of virus DNA replication intermediates in the presence and absence of drug(s), normalized to wild type (no drug). Asterisks indicate mutants that have not been found in woodchucks.
Only two of the mutants had changes in the C region. Type VIII, with a YIDD rather than YMDD motif and a downstream L594F amino acid change, replicated poorly in transfected cells (Fig. 3B). The same was true for mutant constructs containing a YIDD motif, YVDD motif, and YVDD plus L565M motif (28) (Fig. 3B), which was also found in lamivudine-resistant isolates of HBV. Interestingly, these four WHV mutants were more resistant to lamivudine than any of the others, being essentially unaffected, and all but the YVDD mutant were also highly resistant to l-FMAU. The inefficient selection of these four mutants in lamivudine-treated woodchucks may be due to some effect of the polymerase mutations on the function or immunogenicity of the overlapping S open reading frame (ORF) product (Fig. 1D).
Lamivudine, a deoxycytidine analog, functions as a chain terminator to inhibit synthesis of HBV and HIV DNAs (22). In contrast, l-FMAU, a thymidine analog, represses replication of Epstein-Barr virus, and by inference, WHV, without incorporation into viral DNA (25). Nonetheless, in combination, total inhibition was generally that of the stronger inhibitor (Fig. 2D and 3A). This fits with the observations that a single set of mutants produces resistance to both agents.
Lack of effect of additional mutations in the A, B, and E regions of the active site on production of a drug-resistant WHV polymerase.
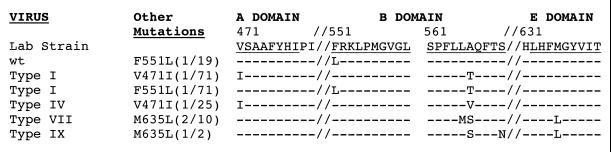
All of the clones listed in Table 2 were sequenced through a 550-nt region of the polymerase active site that included not only the B and C regions, but also the conserved A, D, and E regions. As summarized in Fig. 4, three additional mutations were found in the A, B, and E regions, respectively, of a fraction of the type I, IV, VII, IX, and wild-type clones. Two mutations, at amino acids V471 and F551, alter sites that are conserved among >70% of viruses that have an RNA-dependent polymerase (17), which suggested that the respective mutations might have significant effects on virus drug resistance.
FIG. 4.
Additional mutations in the polymerase A, B, and E regions. Three additional amino acid changes were predicted for some of the mutants summarized in Table 2, based on the sequencing of the polymerase active site. Their frequency and coding capacity are summarized here in comparison to the sequence of a laboratory strain of wild-type (wt) WHV. The mutation V471I in the A region of polymerase would be accompanied by an M281I change in the overlapping S-ORF. The F551L change in the B region does not change the amino acid sequence of the overlapping S gene product. The E region does not overlap S.
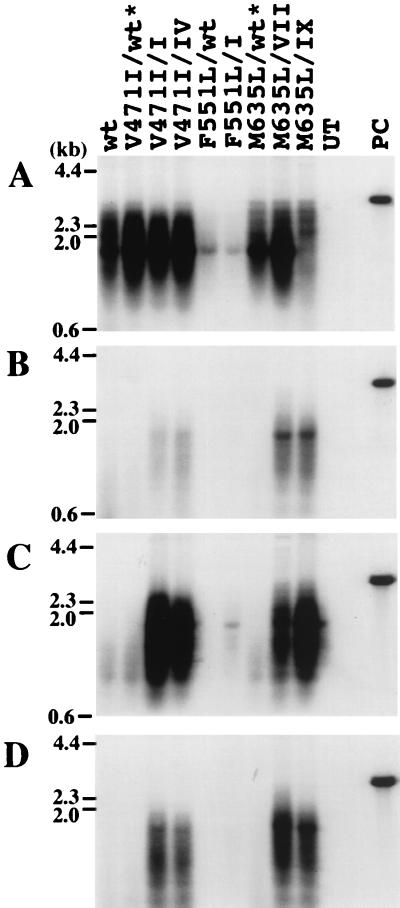
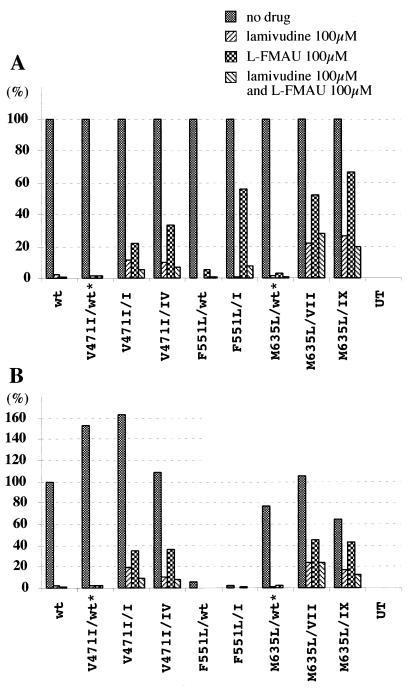
As shown in Fig. 5 and quantified in Fig. 6, V471I and M635L did not confer drug resistance. Within the limits of the assays, V471I in combination with the type I and IV mutations, and M635L in combination with the type VII and IX mutations did not enhance the resistance to lamivudine and l-FMAU. F551L was also without apparent effect, although the assay was of limited sensitivity, because this mutation strongly inhibited viral DNA synthesis. This F551L change in the B region is also found in combination with the YIDD mutation of HBV and again appeared to inhibit HBV replication (14). Direct sequencing of the PCR products of serum virus collected for up to 6 months after lamivudine withdrawal (29) did not detect emergence of any these mutations in the prevalent viral strains (T. Yamamoto, data not shown). However, this may be because the mutations were still relatively rare (<10%) and the follow-up time was relatively short.
FIG. 5.
Effect of the V471I, F551L, and M635L mutations on WHV DNA synthesis in HepG2 cells. Southern blots of core DNA from transfected cells maintained in drug-free medium (A), medium containing 100 μM lamivudine (B), 100 μM l-FMAU (C), or 100 μM lamivudine plus 100 μM l-FMAU (D). Hybridization signals were quantified, with the results summarized in Fig. 6. wt, wild type. UT, DNA extracted from negative control; PC, 25 pg of linear full-length WHV DNA. Asterisks indicate mutants that have not been found in woodchucks.
FIG. 6.
Quantitative summary of analyses of V471I, F551L, and L565M mutations. (A) Inhibition of accumulation of virus DNA replication intermediates in the presence and absence of drug(s). (B) Accumulation of virus DNA replication intermediates in the presence and absence of drug(s), normalized to wild type (no drug). wt, wild type. Asterisks indicate mutants that have not been found in woodchucks.
Lack of effect of polymerase mutations outside the active site region.
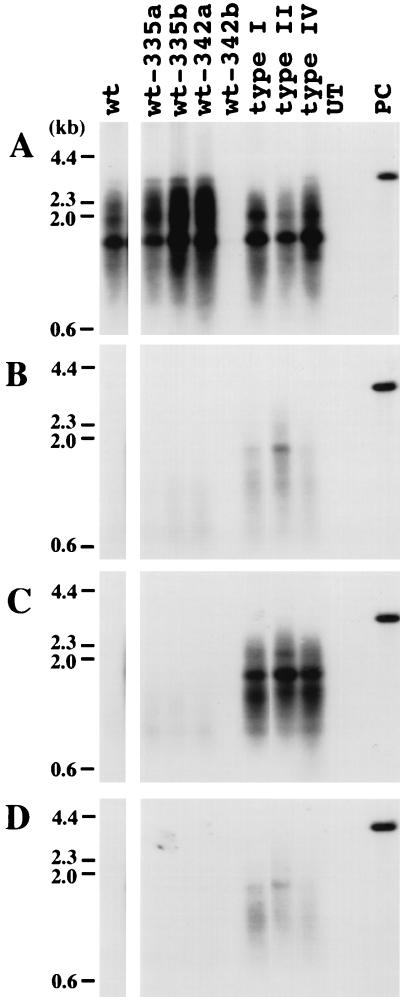
Most of the mutants analyzed above were inhibited two- to fourfold by drug treatment. To see if mutations outside of the conserved polymerase regions contributed a more efficient drug resistance, the complete wild-type, type I, type II, and type IV polymerase genes of WHV were cloned from serum virus and substituted for the wild-type gene present in the laboratory strain of WHV. (Although each contained polymerase ORF mutations outside the DNA polymerase active site, none contained mutations of the conserved regions A to E.) DNA replication was then assessed in the presence and absence of drug, as shown in Fig. 7. As summarized in Fig. 8, all three retained partial sensitivity to inhibition by lamivudine and l-FMAU. Thus, the mutations shown in Fig. 1 appear to be the only determinants of drug resistance of the WHV polymerase gene.
FIG. 7.
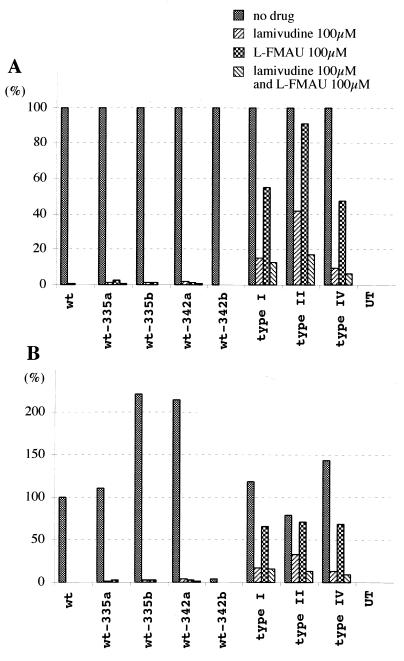
Replication of WHV genomes containing the complete polymerase gene of serum-derived virus from drug-treated woodchucks. WHV genomes were amplified by PCR with primers located in WHV nt 1941 to 1960 and 1945 to 1925. PCR products were cloned, and their nucleotide sequence was determined (accession no. AF410859 [wt-335a], AF410860 [wt-335b], AF410861 [wt-342a], AF410858 [342b], AF410857 [type I], AF410855 [type II], AF410856 [type IV]). Clones with the indicated mutations in the polymerase active site were cleaved upstream at AflII at nt 2303 and downstream at the NsiI site at nt 1914, releasing the complete WHV polymerase gene. This fragment was then substituted for the homologous region of an expression plasmid containing the laboratory strain of WHV. Accumulation of virus DNA was measured (A) in drug-free medium, (B) in medium containing 100 μM lamivudine, (C) in medium containing 100 μM l-FMAU, and (D) in medium containing 100 μM lamivudine and 100 μM l-FMAU. One of the wild-type clones, 342b, did not replicate; sequencing revealed a 6-nt (2 amino acid) deletion in the spacer region of the polymerase ORF. UT, DNA extracted from a negative control; PC, 25 pg of linear full-length WHV DNA. wt-335a, wt-335b, and type I polymerase genes were cloned from woodchuck 335 (Table 2) serum collected 2 months after termination of lamivudine therapy; wt-342a and wt-342b polymerase genes were cloned from woodchuck 342 serum collected 2 months after termination of lamivudine therapy. The type II and IV polymerase genes were cloned from woodchuck 331 serum collected upon termination of lamivudine therapy.
FIG. 8.
Quantitative summary of transfection assays in Fig. 7. (A) Inhibition of accumulation of virus DNA replication intermediates in the presence and absence of drug(s). (B) Accumulation of virus DNA replication intermediates in the presence and absence of drug(s), normalized to wild type (wt [no drug]).
Replication efficiency and the lack of reemergence of wild-type WHV as the predominant species following lamivudine withdrawal.
In a previous study (29), we observed that several of the WHV mutants characterized here remained more abundant than the wild type in woodchuck sera for up to 6 months after the termination of lamivudine treatment (summarized in Fig. 9). This was surprising, since the wild type was often detected in serum at the end of therapy, or shortly thereafter, implying an abundance of at least 10% of total serum virus. Since it was expected that the wild type would replicate more efficiently, it was anticipated that it would quickly reemerge as the most abundant species. However, factors that could contribute to a slow evolution of a virus population, and particularly a shift back to a presumably well-adapted wild-type virus, might include a high efficiency of mutant virus replication combined with a low rate of hepatocyte turnover.
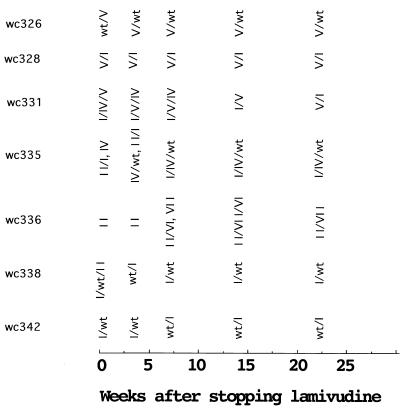
FIG. 9.
Summary of data on persistence of lamivudine-resistant WHV in the serum following cessation of drug treatment. Results are summarized from reference 29. The order in which the variants are listed reflects their relative abundance in the serum.
In a recent study, Zhang and Summers (26) showed that the emergence of a more replication-efficient strain of DHBV in chronically infected ducks depended on liver cell turnover to create virus replication space (26). Thus, when the death rate for infected hepatocytes was low, virus populations evolved only slowly. Although designed to show the dependence of virus evolution on replication space in a situation in which virus fills new space essentially instantaneously (virus replication much faster than liver regeneration), this model can also be used to show the relationship between time and virus evolution. This is done by taking advantage of the fact that the rate of creation of replication space is determined by the rate of cell death in an adult liver. In Fig. 10A, we have used the replication space model to estimate the time of enrichment of a virus (here denoted as wild type) that replicates more efficiently than competing viruses. A hepatocyte turnover rate of 1% per day is assumed, which probably represents a reasonable estimate for chronically infected woodchucks (15, 27, 29). The model predicts that a wild-type virus that was initially somewhat infrequent (i.e., 10% of the total) would achieve only a fivefold enrichment over the next 6 months (180 days) when competing against mutants that replicated ∼50% as well (krel = 0.5; Fig. 9). On the other hand, it would become predominant if it replicated at least threefold more efficiently. Thus, this analysis suggests that some of the mutants (types I, IV, and V) replicated at close to wild-type efficiency in vivo (that is, with >33% of wild-type efficiency).
A feature of the replication space model is that it provides a minimum estimate of enrichment time (assuming that immune selection is not involved) because it treats all replication space as accessible to all viruses. However, it is possible that in a fully infected liver, restriction to hepatocytes, which may be superinfection resistant, slows the emergence of wild-type virus, particularly if the liver regenerates from mature, infected hepatocytes. To determine the effects of restricting replication space availability, we have constructed “cellular” models (see Materials and Methods) in which different virus strains are distributed among hepatocytes in a random fashion at the time that lamivudine therapy ends. This randomization, with mixed cellular infections, is presumed to be the result of selective pressures that exist during drug therapy. As shown in Fig. 10B and C, if superinfections do not occur after withdrawal of lamivudine therapy and hepatocytes are replaced via division of infected cells, enrichment takes two to three times as long as in the replication space model. The actual time depends upon how new cccDNA is produced following cell division. In the model shown in Fig. 10B, cccDNA synthesis following hepatocyte division employs newly synthesized DNA and therefore reflects the distribution of inherited cccDNA. As modeled in Fig. 10C, new cccDNA is made from replicating DNA that was inherited from the parental cell and therefore reflects the population of cccDNA in the parental cell. Three to 6 months would be just sufficient (Fig. 10C) for a wild-type WHV that replicated 10 times more efficiently to become more abundant in the serum. Thus, if new replication space is restricted, mutants would need to replicate with >10 to 30% of wild-type efficiency to remain predominant for at least 6 months. Factors such as liver regeneration from uninfected hepatocyte progenitors or poor superinfection resistance would accelerate wild-type selection back towards that modeled in Fig. 10A.
In brief, the data summarized in Fig. 9 together with the analyses in Fig. 10 suggest that three of the drug-resistant mutants we have characterized (types I, IV, and V, each with only a single base mutation in the B region) replicate with at least 10 to 30% of wild-type efficiency in vivo (in the absence of drug). Types I and V may actually replicate their genomes more efficiently than the wild type, because both gained predominance over the wild type during the course of observation in woodchucks 326 and 338, respectively. Type I has a stop codon in the overlapping S gene as a result of the polymerase mutation and would presumably be incapable of spreading except in cells that were coinfected with a helper virus. This does not appear to be true for type V, which was previously observed as the predominant virus in a woodchuck that had not received any drug treatment (wc4961 of reference 15).
Replication efficiency and the emergence of mutant WHV during drug treatment.
Pult et al. (19) have shown that single nucleotide substitutions in duck hepatitus B virus (DHBV) were generated at a frequency of 10−4 to 10−5 per infecting virus during the infection of naive ducklings. If this mutation frequency also applies to WHV, four of the mutants, types I, IV, V, and VI, may be present in the liver at about this frequency in untreated animals. This possibility, combined with estimates of the mutant virus replication efficiency must, however, be reconciled with the observation that total cccDNA levels can greatly decline and some hepatocytes become WHV free during antiviral therapy (10, 29). For instance, in a study with l-FMAU, >80% of hepatocytes appeared to be virus free after 30 weeks of therapy, with resistant strains of WHV emerging thereafter (29). If, however, the mutant virus had the replication rate constant of the wild type (under drug-free conditions), which is sufficient for even a small amount of virus to fully infect the liver in 1 to 2 months (11), a large population of virus-free hepatocytes should never have appeared in these experiments. It therefore seems likely that the drug-resistant mutants have lower replication rate constants in the presence of l-FMAU than the wild-type does in its absence.
DISCUSSION
Several conclusions may be derived from the current study. (i) Resistance of WHV to lamivudine, and possibly l-FMAU as well, is associated primarily with mutations in the B region. This contrasts with HBV, where resistance to lamivudine and l-FMAU (in cell culture) (9) results primarily from mutations of the YMDD motif in the C region. B region mutations of HBV appear to enhance the replication competence of the C region mutants. Thus, the pattern of WHV polymerase mutations is not a perfect match to mutations developing in HBV patients, although the woodchuck remains a useful model for characterizing the biology of emergence of drug-resistant variants. (ii) The molecular basis of the resistance was due to mutations in three codons in the B region (L565, A566, and N570) and one codon (M589) of the C region of the WHV DNA polymerase. A second site in the C region, L594, was found to be mutated, but a role for this mutation in drug resistance was not apparent (Fig. 3). Eight of the 11 mutants had mutations of A566. Comparison of these mutants as aligned in Fig. 1A suggests that type X may have evolved from type I, type II from type IV, and types VII, IX, and III from type V. (iii) Polymerase ORF mutations outside of the B and C regions did not appear to play a role in drug resistance. (iv) Transfection assays suggested that most of the drug-resistant variants might replicate at ≥50% of the wild-type rate in the absence of drug. (The major exception was type VIII [∼10%], which was found in an l-FMAU-treated woodchuck.) As revealed by the computational models in Fig. 10, a high replication efficiency of several of the mutants (10% or more of the wild type), predicted from the cell culture studies, is also consistent with a 6-month follow-up study of woodchucks that were removed from lamivudine therapy (29). In only one of seven woodchucks did wild-type WHV reemerge as the most abundant species. Consideration of hepadnavirus mutation rates and the time to emergence of mutants in l-FMAU-treated woodchucks suggests, however, that the mutants all replicate less efficiently than the wild type.
While computational models can predict the effects of replication rates on the evolution of virus populations, it should be noted that the predictions are dependent upon initial conditions as well as specific assumptions about the virus infections. For the models we have illustrated (Fig. 10B and C), key assumptions were that the liver was fully infected and regenerated from mature, infected hepatocytes that were superinfection resistant. (An additional assumption was that virus replication rates significantly exceeded liver replication rates, an assumption that appears valid in the situation under consideration [11].) In the cell models, susceptibility to superinfection, which is probably low (18), but nonetheless may occur, as well as liver regeneration from uninfected progenitor cells, which then became susceptible, would each accelerate the selection of the virus with the higher replication rate. That is, the predicted outcome would approach that of the replication space model (26). The cell models also make a prediction that is not inherent in the replication space model. Mixed infections of cells with two or more viruses will ultimately segregate, as a result of random fluctuations, into pure infections, with each hepatocyte carrying only a single WHV variant. The segregation will be accelerated by differences in replication rate constants. The effect of this is to limit the ultimate enrichment of one virus over another. With the parameters we used here, this limit to wild-type enrichment was about 15-fold for the model in Fig. 10B and 40-fold for that in Fig. 10C.
The ability of B region mutations of WHV to cause lamivudine resistance remains obscure. Structural models of the active site of the HBV DNA polymerase suggest that mutations of the YMDD motif interfere with lamivudine binding (2, 7) and are responsible for drug resistance. It has been proposed, but not yet established, that the B region mutation of HBV (rtL180M), found together with the YMDD mutations, could either interfere with nucleotide binding directly or perturb the nucleotide-binding region in the YMDD motif. However, B region mutations of HBV have not been found by themselves to produce lamivudine resistance in patients, and their primary function appears to be to enhance the efficiency of viral DNA synthesis, which is depressed by the YMDD alterations. Presumably the B region mutations of WHV are better positioned to perturb the binding or positioning of lamivudine without concurrent mutations of other regions of the active site. Interestingly, some of the most prevalent WHV mutants result from a single nucleotide change in the B region. This, together with the expected mutation rate, as determined for DHBV (19), supports the possibility that these mutations are common even prior to drug treatment and may explain the observation that drug-resistant mutants of WHV typically emerge after only 6 to 12 months of drug therapy (15, 27–29). In contrast, and for reasons that are obscure, 30% of HBV patients still do not experience breakthrough even after 3 years of lamivudine treatment. Perhaps with these patients, the HBV genotype precludes the development of a resistant mutant via a single nucleotide change (e.g., the YIDD mutation).
Acknowledgments
We are grateful to J. Summers (University of New Mexico, Albuquerque) for advice on computational models for the evolution of virus populations and for sharing unpublished data. We thank J. Taylor, C. Seeger, and J. Summers for a critical reading of the manuscript and acknowledge the DNA sequencing, oligonucleotide synthesis, and cell culture facilities of the Fox Chase Cancer Center for technical support.
This work was supported by USPHS grants AI-18641, 3P01-CA-4073711S1, and CA-06927 from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Ahmed, S. N. S., D. Tavan, C. Pichoud, F. Berby, L. Stuyver, M. Johnson, P. Merle, H. Abidi, C. Trepo, and F. Zoulim. 2000. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in patients treated with lamivudine for chronic hepatitis B. Hepatology 32:1078–1088. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K.-A. Walters, D. L. J. Tyrrell, N. Brown, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670–1677. [DOI] [PubMed] [Google Scholar]
- 3.Aye, T. T., A. Bartholomeusz, T. Shaw, S. Bowden, A. Breschkin, J. McMillan, P. Angus, and S. Locarnini. 1997. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J. Hepatol. 26:1148–1153. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomeusz, A., L. C. Groenen, and S. A. Locarnini. 1997. Clinical experience with famciclovir against hepatitis B virus and development of resistance. Intervirology 40:337–342. [DOI] [PubMed] [Google Scholar]
- 5.Boni, C., A. Bertoletti, A. Penna, A. Cavalli, M. Pilli, S. Urbani, P. Scognamiglio, R. Boehme, R. Panebianco, F. Fiaccadori, and C. Ferrari. 1998. Lamivudine treatment can restore T cell responsiveness in chronic hepatitis B. J. Clin. Investig. 102:968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chayama, K., Y. Suzuki, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711–1716. [DOI] [PubMed] [Google Scholar]
- 7.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di, Q., J. Summers, J. B. Burch, and W. S. Mason. 1997. Major differences between WHV and HBV in the regulation of transcription. Virology 229:25–35. [DOI] [PubMed] [Google Scholar]
- 9.Fu, L., S. H. Liu, and Y. C. Cheng. 1999. Sensitivity of l-(−)2,3-dideoxythiacytidine resistant hepatitis B virus to other antiviral nucleoside analogues. Biochem. Pharmacol. 57:1351–1359. [DOI] [PubMed] [Google Scholar]
- 10.Genovesi, E. V., L. Lamb, I. Medina, D. Taylor, M. Seifer, S. Innaimo, R. J. Colonno, D. N. Standring, and J. M. Clark. 1998. Efficacy of the carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob. Agents Chemother. 42:3209–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jilbert, A. R., D. S. Miller, C. A. Scougall, H. Turnbull, and C. J. Burrell. 1996. Kinetics of duck hepatitis B virus infection following low dose virus inoculation: one virus DNA genome is infectious in neonatal ducks. Virology 226:338–345. [DOI] [PubMed] [Google Scholar]
- 12.Kodama, K., N. Ogasawara, H. Yoshikawa, and S. Murakami. 1985. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J. Virol. 56:978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau, D. T.-Y., M. F. Khokhar, E. Doo, M. G. Ghany, D. Herion, Y. Park, D. E. Kleiner, P. Schmid, L. D. Condreay, J. Gauthier, M. C. Kuhns, J. K. Liang, and J. H. Hoofnagle. 2000. Long-term therapy of chronic hepatitis B virus with lamivudine. Hepatology 32:828–834. [DOI] [PubMed] [Google Scholar]
- 14.Ling, R., D. Mutimer, M. Ahmed, E. H. Boxall, E. Elias, G. M. Dusheiko, and T. J. Harrison. 1996. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology 24:711–713. [DOI] [PubMed] [Google Scholar]
- 15.Mason, W. S., J. Cullen, G. Moraleda, J. Saputelli, C. E. Aldrich, D. S. Miller, B. Tennant, L. Frick, D. Averett, L. D. Condreay, and A. R. Jilbert. 1998. Lamivudine therapy of WHV-infected woodchucks. Virology 245:18–32. [DOI] [PubMed] [Google Scholar]
- 16.Nowak, M. A., S. Bonhoeffer, A. M. Hill, R. Boehme, H. C. Thomas, and H. McDade. 1996. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 93:4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Protzer, U., M. Nassal, P. W. Chiang, M. Kirschfink, and H. Schaller. 1999. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc. Natl. Acad. Sci. USA 96:10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pult, I., N. Abbott, Y.-Y. Zhang, and J. Summers. 2001. Frequency of spontaneous mutations in an avian hepadnavirus infection. J. Virol. 75:9623–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeger, C., B. Baldwin, and B. C. Tennant. 1989. Expression of infectious woodchuck hepatitis virus in murine and avian fibroblasts. J. Virol. 63:4665–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeger, C., and J. Maragos. 1989. Molecular analysis of the function of direct repeats and a polypurine tract for plus-strand DNA priming in woodchuck hepatitis virus. J. Virol. 63:1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severini, A., X. Y. Liu, J. S. Wilson, and D. L. J. Tyrrell. 1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 39:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuyver, L. J., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751–757. [DOI] [PubMed] [Google Scholar]
- 24.Tillmann, H. L., C. Trautwein, T. Bock, K. H. Boker, E. Jackel, M. Glowienka, K. Oldhafer, I. Bruns, J. Gauthier, L. D. Condreay, H. R. Raab, and M. P. Manns. 1999. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology 30:244–256. [DOI] [PubMed] [Google Scholar]
- 25.Yao, G. Q., S. H. Liu, E. Chou, M. Kukhanova, C. K. Chu, and Y. C. Cheng. 1996. Inhibition of Epstein-Barr virus replication by a novel l-nucleoside, 2′-fluoro-5-methyl-β-l-arabinofuranosyluracil. Biochem. Pharmacol. 51:941–947. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y.-Y., and J. Summers. 2000. Low dynamic state of a viral competition in a chronic avian hepadnavirus infection. J. Virol. 74:5257–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, T., J.-T. Guo, F. A. Nunes, K. L. Molnar-Kimber, J. M. Wilson, C. E. Aldrich, J. Saputelli, S. Litwin, L. D. Condreay, C. Seeger, and W. S. Mason. 2000. Combination therapy with lamivudine and adenovirus causes transient suppression of chronic woodchuck hepatitis virus infections. J. Virol. 74:11754–11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, T., J. Saputelli, C. E. Aldrich, M. Deslauriers, L. D. Condreay, and W. S. Mason. 1999. Emergence of drug-resistant populations of woodchuck hepatitis virus in woodchucks treated with the antiviral nucleoside lamivudine. Antimicrob. Agents Chemother. 43:1947–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]