Abstract
The hepatitis C virus (HCV) envelope protein E2 has been shown to accumulate in the lumen of the endoplasmic reticulum (ER) as a properly folded glycoprotein as well as large aggregates of misfolded proteins. In the present study, we have identified an additional unglycosylated species, with an apparent molecular mass of 38 kDa (E2-p38). In contrast to the glycosylated E2, E2-p38 is significantly less stable and is degraded through the proteasome pathway. Correspondingly, E2-p38 is found to be ubiquitinated. E2-p38 is localized mostly in the cytosol, in contrast to the glycosylated form, which is exclusively membrane associated. Alpha interferon (IFN-α) treatment or overexpression of the double-stranded RNA-activated protein kinase (PKR) significantly increased the stability of E2-p38, consistent with a previous report (D. R. Taylor, S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai, Science 285:107–110, 1999) that E2 interacts with PKR and inhibits its kinase activity. Direct interaction between PKR and E2-p38, but not the glycosylated form of E2, was also observed. These results show that E2-p38 is the form of E2 that interacts with PKR in the cytosol and may contribute to the resistance of HCV to IFN-α. Thus, an ER protein can exist in the cytosol as an unglycosylated species and impair cellular functions.
Hepatitis C virus (HCV) is the major etiological agent of non-A, non-B blood-borne hepatitis. An estimated 2% of the world population is chronically infected, and these individuals may develop chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (4, 12). The only available therapies are alpha interferon (IFN-α) in monotherapy or in combination with ribavirin; however, the efficacy of these treatments is low, particularly for some of the HCV genotypes (13). At least two HCV proteins, the envelope glycoprotein E2 and the nonstructural protein NS5A, have been reported as potential inhibitors of the IFN response (8, 23).
HCV is an enveloped positive-strand RNA virus which belongs to the Flaviviridae family. Its RNA genome encodes a polyprotein of about 3,020 amino acids which is processed by host and viral proteases to generate the structural and nonstructural proteins (19). HCV encodes two envelope proteins, E1 and E2, both of which are type I transmembrane glycoproteins. These two proteins are believed to assemble as a noncovalent heterodimer which is retained at the endoplasmic reticulum (ER) membrane (5, 7). Their N-terminal domains are highly N-glycosylated with unmodified glycans, suggesting that they both are retained at the ER membrane and do not reach the Golgi apparatus (7). They both contain a signal sequence and an ER retention signal in their C-terminal domains. Their ER retention signals are atypical, consisting of charged residues, which are conserved among the different viral genotypes, flanked by two stretches of hydrophobic residues (3). E1 has another ER retention signal in its juxtamembrane region (16). The folding of E1 and E2 requires chaperone proteins such as calreticulin and calnexin (1). The proper folding of E1 further depends on the interaction with E2 (1). Very little E1 or E2 protein is expressed on the cell surface. The ER localization of E1-E2 suggests that virus assembly takes place in the ER, but the fate of most E1-E2 proteins is not known (for a review, see reference 6).
The E2 protein has also been implicated in conferring resistance to IFN. E2 contains a region homologous to the double-stranded RNA-activated protein kinase (PKR) and its substrate, the subunit α of the translation initiation factor eIF2α (23). The activation of PKR and the resultant phosphorylation of eIF2α mediate the antiviral effect of IFN by inhibiting protein synthesis (10). E2 has been shown to bind to PKR and inhibit its kinase activity in vitro and in vivo (23, 24), presumably leading to the inhibition of the antiviral effect of IFN. The PKR-eIF2α phosphorylation site homology domain of E2 is required for the inhibition of PKR by E2 (23). However, since E2 is localized inside the lumen of the ER, whereas PKR is localized outside the ER, associated with ribosomes in the cytosol (27), it is difficult to understand how they interact with each other. The observed inhibition of PKR by E2 thus raises the possibility that E2 may have another subcellular localization besides the ER.
In the present study, we have demonstrated that a portion of E2 remains unglycosylated, is retained in the cytosol, and is degraded by the proteasome pathway after ubiquitination. Thus, this form is either retro-translocated from the ER to the cytosol or is retained in the cytosol after translation. Furthermore, this unglycosylated E2, but not the glycosylated form, interacts with PKR and is stabilized by IFN. These results demonstrate a novel form of E2 glycoprotein and elucidate how and where E2 and PKR can interact with each other. They also have a more general implication that a protein synthesized by membrane-bound ribosomes can be exposed to the cytosolic environment.
MATERIALS AND METHODS
Plasmids.
A fragment from nucleotide 1452 to 2579 of a cDNA clone of an HCV 1b isolate (25) (corresponding to amino acids 371 to 746 of the HCV polyprotein) was amplified by PCR and cloned into a modified pcDNA 3.1 plasmid (Invitrogen) in which the cytomegalovirus promoter had been replaced by the EF-1α promoter (pcDNAEF) (26); the resulting plasmid is named pcDNAEF-E2. The authenticity of the insert was confirmed by sequencing. The plasmid encoding the vesicular stomatitis virus G protein (VSV G) was a kind gift of Paula M. Cannon (University of Southern California). Plasmids encoding PKR and its catalytically inactivated form, PKR K296R, have already been described (23).
Cells and transfection.
Human embryonic kidney HEK-293T cells and HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS), 100 IU of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine. Transfections were performed with FuGENE6 (Roche) with a DNA/FuGENE reagent ratio of 1:3.
Metabolic labeling and immunoprecipitation.
Twenty-four hours after transfection, cells were starved for 30 min in DMEM without methionine and cysteine (Sigma) containing 2% FBS. Cells were then labeled for 5 or 15 min with 50 μCi of 35S-protein label mix (ICN) per ml. For pulse-chase experiments, cells were washed twice after labeling with 10× Met-Cys DMEM and then incubated for different lengths of time with 5× Met-Cys DMEM containing 2% FBS at 37°C. In experiments with tunicamycin, cells were treated with 10 μM tunicamycin (Biomol) 1 h prior to and throughout the pulse-chase period. In experiments with IFN-α treatment, recombinant IFN-α (Fitzgerald) was added 6 h after transfection, i.e., 18 h before harvesting, at a concentration of 1,000 IU/ml. The same concentration of IFN-α was maintained during starvation, labeling, and chase. After labeling or chase, cells were washed twice with ice-cold phosphate-buffered saline and lysed with TBS buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl) containing 0.5% Triton X-100. After one cycle of freeze-thaw, cell lysates were cleared by centrifugation in an Eppendorf model 5415C centrifuge at 1,200 rpm at 4°C for 5 min. Supernatants were used for immunoprecipitation. Briefly, lysates were first precleared with 50 μl of protein A-Sepharose (Zymed); antibodies were then added and incubated for at least 4 h at 4°C with rotation. Immune complexes were then incubated with 50 μl of protein A-Sepharose for an additional 1 h, collected by centrifugation, and washed twice with NET buffer (50 mM Tris-HCl [pH 7.4]-150 mM NaCl-0.5% NP-40-0.1% sodium dodecyl sulfate [SDS]-0.5% deoxycholic acid). For coimmunoprecipitation, the following buffer was used for the washing step: 50 mM Tris-HCl (pH 7.6)-150 mM NaCl-0.1% NP-40-0.03% SDS. For protein analysis, immune complexes were resuspended in 40 μl of 3× denaturing buffer and loaded on an SDS-12.5% polyacrylamide gel electrophoresis (PAGE) gel. After drying, gels were exposed with Kodak Biomax single emulsion film.
Antibodies.
The following antibodies were used (dilutions used for immunoprecipitation are indicated in parentheses): anti-E2 mouse monoclonal antibody H62 (1:1,000) (a kind gift of Jean Dubuisson, Institut Pasteur de Lille, Lille, France), anti-E2 mouse monoclonal antibody A11 (1:1,000) (a kind gift of Stephen Feinstone, FDA), anti-E2 monoclonal antibody BDI715 (1:50) (Biodesign International), anti-VSV G mouse monoclonal antibody P5D4 (1:500) (Sigma), anti-PKR mouse monoclonal antibody B-10 (1:300) (Santa Cruz Biotechnology), anti-ubiquitin mouse monoclonal antibody (1:500) (Chemicon International), and anti-ubiquitin rabbit polyclonal FL-76 (1:500) (Santa Cruz Biotechnology).
Membrane flotation analysis.
HEK-293T cells were transfected with 10 μg of pcDNAEF-E2 per 10-cm plate. Twenty-four hours posttransfection, cells were labeled for 15 min and harvested as previously described except that the lysis buffer used contained only 0.2% Triton X-100. Cellular extracts were subjected to fractionation by the membrane flotation method as described previously (14). Briefly, cellular extracts (≈0.7 ml) were diluted into 2 ml of 75% (wt/wt) sucrose in low-salt buffer (LSB) (50 mM Tris-HCl [pH 7.4], 25 mM KCl, 5 mM MgCl2) and overlaid with 2 ml of 55% (wt/wt) sucrose in LSB and 0.3 ml of 10% (wt/wt) sucrose in LSB. Sucrose gradients were then centrifuged in a Beckman SWTi rotor at 4°C for 14 h at 38,000 rpm. After centrifugation, 1-ml fractions were collected from the top, diluted in 2.5 ml of LSB and 0.82 ml of 5× TBS-2.5% Triton X-100, and incubated for 2 h at 4°C with rotation. Each fraction was then subjected to immunoprecipitation as described previously (anti-E2 monoclonal antibody H62 diluted 1:4,000).
Immunoblotting.
After SDS-PAGE separation, proteins were transferred to a nitrocellulose membrane (Hybond-C extra; Amersham Life Science) using a Trans-blot apparatus (Bio-Rad). E2 detection was performed with the anti-E2 antibody H62 (dilution, 1:1,000), followed by a goat anti-mouse immunoglobulin conjugated to horseradish peroxidase (dilution, 1:10,000; American Qualex). Immunocomplexes were detected using an enhanced chemiluminescence detection kit (ECL+Plus; Amersham).
RESULTS
Detection of a novel unglycosylated form of the E2 glycoprotein in mammalian cells.
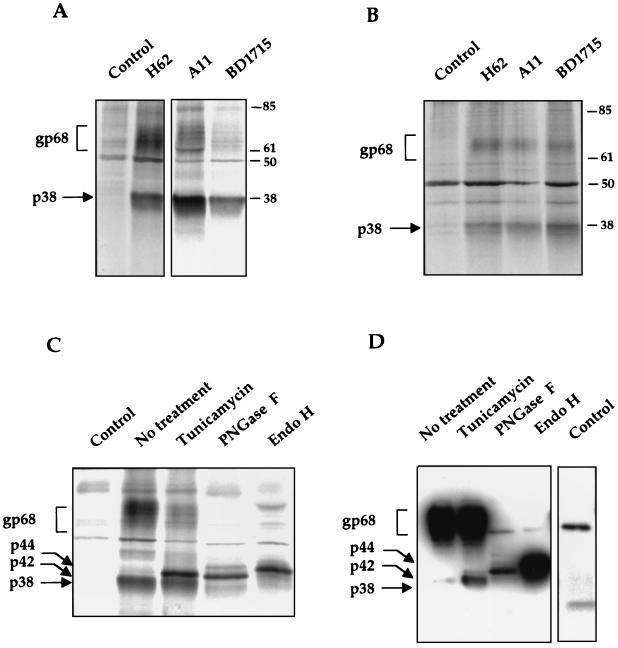
To study the processing of HCV E2 protein, we cloned the full-length E2 of genotype 1b and its signal sequence, corresponding to the last 20 amino acids of E1, under the EF-1α promoter (pcDNAEF-E2). Human embryonic kidney cells (293T cells) were transfected with pCDNAEF-E2, labeled with [35S]methionine and -cysteine, and immunoprecipitated using three different anti-E2 monoclonal antibodies of different origins. Two major HCV-specific products were immunoprecipitated with all three antibodies used (Fig. 1A). One (≈68 kDa) corresponds to the glycosylated form of E2, with various bands representing different degrees of glycosylation (7), and the other, with an apparent molecular mass of 38 kDa, corresponds in size to that predicted for the unglycosylated E2. The same results were obtained in HeLa cells, although the level of E2 expression in this cell line was lower (Fig. 1B). To confirm that the low-molecular-weight product corresponds to an unglycosylated form of E2, transfected cells were labeled in the presence of tunicamycin, a glycosylation inhibitor, and immunoprecipitated with an anti-E2 antibody, or alternatively, the immunoprecipitated E2 was digested with glycosidases: peptide N-glycosidase F (PNGase F) or endoglycosidase H (Endo H) (Fig. 1C). Interestingly, after tunicamycin treatment, two distinct bands, of 38 and 42 kDa, were found. The lower band corresponds to p38, indicating that p38 is likely an unglycosylated form of E2 (E2-p38). The nature of p42 is not known, but it has also been previously described by Grakoui et al. (9). One possible interpretation is that p42 represents the product of another posttranslational modification of E2 that is not inhibited by tunicamycin. Complete removal of carbohydrates by PNGase F yielded the same two bands of 38 and 42 kDa as with the tunicamycin treatment, consistent with the interpretation that E2 may have another posttranslational modification in addition to N-glycosylation. The intensity of the band corresponding to E2-p38 was reduced after PNGase F treatment, probably due to dilution of the sample after digestion. Since E2-p42 was observed only after treatments that inhibit glycosylation or remove glycans and its presence corresponds to a decrease of E2-gp68, these results suggest that E2-p42 is the true deglycosylated form of the mature E2, E2-gp68. The absence of E2-p42 in untreated cells suggests either that E2-p42 is a transient conformation that is rapidly glycosylated or, more likely, that the putative additional posttranslational modification of E2-p42 occurs simultaneously with glycosylation. Endo H digestion yielded a third product, of 44 kDa, which probably represents a partially deglycosylated E2, since Endo H cleaves after the first N-acetylglucosamine of glycans but does not remove them completely. The same results were obtained in HeLa cells (data not shown). As an ultimate confirmation, all the protein bands described, p38, p42, and p44, were detected by immunoblot with an anti-E2 antibody (Fig. 1D), indicating that they were all derived from E2. The reactivity of the different bands with the antibody varied, probably reflecting differences in their conformation. Furthermore, three anti-E2 antibodies obtained from independent sources could detect all these bands in immunoblots (data not shown). These results combined indicate that p38 is most likely an unglycosylated form of E2. However, since treatments with an inhibitor of glycosylation or glycosidase digestions did not increase the amount of E2-p38, the latter is probably not a direct precursor of E2-gp68 and may have a different fate in the cells.
FIG. 1.
Detection of an unglycosylated form of E2 in mammalian cells. (A) Detection of E2 in 293T cells transfected with pcDNAEF-E2. Cells were radiolabeled with [35S]methionine and -cysteine and immunoprecipitated with different anti-E2 monoclonal antibodies. The control was cells transfected with the empty vector pcDNAEF and immunoprecipitated with H62. (B) Same as for panel A but in HeLa cells. (C) Immunoprecipitation of radiolabeled E2 after tunicamycin treatment or digestion with PNGase F or Endo H glycosidases. (D) Detection by immunoblotting, using H62 monoclonal antibody, of immunoprecipitated E2 after tunicamycin treatment or glycosidase digestions. Control lane was cells transfected with an empty vector; the two bands correspond to the light (≈25 kDa) and heavy (≈55 kDa) chains of the antibody used for immunoprecipitation. Exposure time for the control lane was longer to identify their positions. Markers are indicated in kilodaltons. The glycosylated form of E2 is termed gp68, the unglycosylated forms of E2 are termed p38 and p42, and the partially deglycosylated E2 is termed p44.
The unglycosylated form of E2 is more rapidly degraded than its glycosylated form.
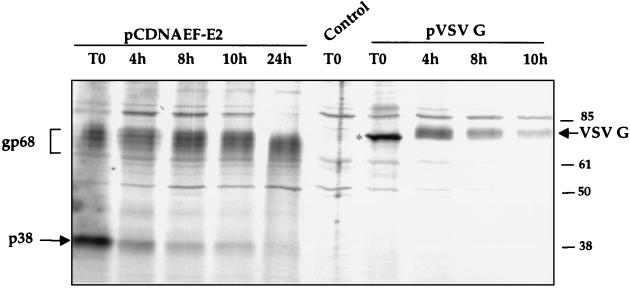
To study the processing of these two forms of E2, pulse-chase experiments were performed (Fig. 2). After a pulse-labeling of 5 min, a large proportion of E2 was glycosylated, suggesting that glycosylation occurs very rapidly after synthesis (in less than 5 min). Nevertheless, a sizable proportion of E2 remained as the unglycosylated form E2-p38. The dichotomy of rapid glycosylation of E2 and yet the presence of a large amount of the unglycosylated form suggests that the majority of E2-p38 may not be the precursor to E2-gp68. During the chase period, no significant increase of E2-gp68 was noted, whereas the amount of E2-p38 dropped rapidly, further suggesting that most of the E2-p38 was not chased into E2-gp68. In comparison, VSV G was found only in an unglycosylated form after 5 min of pulse-labeling, indicating that glycosylation of VSV G takes longer than that of E2. However, during the chase period, all of the VSV G protein existed as glycosylated species; no unglycosylated form remained. Once glycosylated, E2 became very stable, whereas VSV G protein was comparatively less stable, with a shorter half-life than the glycosylated E2. In contrast to the glycosylated form of E2, the E2-p38 form was very unstable. The rate of degradation of E2-p38 was almost equivalent to that of VSV G protein. These results combined suggest that the glycosylated and unglycosylated forms of E2 have different rates of degradation and different fates.
FIG. 2.
Kinetics of degradation of E2 and VSV G proteins. Cells were pulse-labeled with [35S]methionine and -cysteine for 5 min and chased for various lengths of time as indicated in hours. Immunoprecipitations of E2 and VSV G were performed with their respective monoclonal antibodies H62 and P5D4. Control cells were transfected with pcDNAEF and immunoprecipitated with P5D4. Marker is indicated in kilodaltons. VSV G is indicated by an arrow and its unglycosylated form is marked by an asterisk.
E2-p38 is degraded via the proteasome pathway and is ubiquitinated.
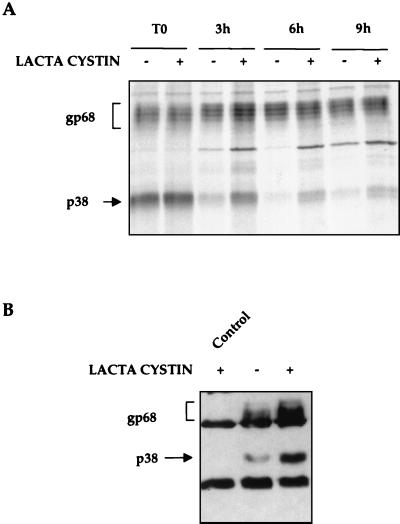
To test whether E2-p38 goes into the degradation pathway, the stability of E2-p38 was studied in the presence of lactacystin, a proteasome inhibitor. After pulse-labeling, roughly similar amounts of E2-p38 were synthesized, either in the absence or the presence of lactacystin. However, during chase, E2-p38 was more slowly degraded in the presence of lactacystin than in its absence (Fig. 3A). Since E2-gp68 was already a very stable protein, treatment with lactacystin did not make any significant difference on its stability during the chase period studied here. These results established that E2-p38 is not as stable as E2-gp68 because E2-p38 is degraded through the proteasome pathway.
FIG. 3.
Degradation of E2. (A) Stability of E2 in the presence of a proteasome inhibitor. Cells were pulse-labeled for 15 min (T0), chased with (+) or without (−) lactacystin for various lengths of time in hours, and immunoprecipitated with anti-E2 antibody. (B) Transfected cells were treated with (+) or without (−) 20 μM lactacystin for 1 h. Cell lysates were immunoprecipitated with a mixture of anti-ubiquitin mouse monoclonal (1:500) and anti-ubiquitin rabbit polyclonal FL-76 (1:500) antibodies. Proteins were immunoblotted for E2. The control was cells transfected with the empty vector and treated with lactacystin.
The degradation of E2-p38 through the proteasome degradation pathway implies that E2-p38 may be ubiquitinated to be recognized by the 26S proteasome (11). To examine this possibility, 293T cells were transfected with pCDNAEF-E2 and cellular extracts were immunoprecipitated with a mixture of two anti-ubiquitin antibodies. Since ubiquitinated proteins are rapidly degraded, some samples were treated with lactacystin to allow accumulation of ubiquitinated products. After immunoprecipitation, proteins were immunoblotted with an anti-E2 monoclonal antibody. In untreated cells, the anti-ubiquitin antibodies precipitated small amounts of both E2-p38 and E2-gp68, which significantly increased in lactacystin-treated cells (Fig. 3B). These results combined demonstrate that E2 is ubiquitinated and that the unglycosylated form of E2 is degraded by the proteasome pathway. Although the glycosylated form of E2 is also ubiquitinated, it is very stable, suggesting that the degradation of this form is less efficient.
E2-p38 and E2-gp68 are localized in different subcellular compartments.
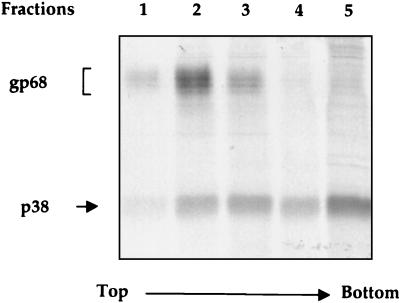
Since protein degradation through the proteasome pathway occurs outside the ER, it is expected that E2-p38 should be in the cytosol, in contrast to the known localization of E2 inside the lumen of the ER (7). Therefore, subcellular localization of E2-p38 was analyzed by a membrane flotation gradient assay. In this procedure, membrane-associated proteins float on top of the gradient after equilibrium centrifugation, whereas soluble proteins remain in the dense sucrose fractions (22). Cells transfected with pcDNAEF-E2 were pulse-labeled for 15 min. After very mild lysis, cellular extracts were used for flotation analysis. Gradients were collected from the top, and each fraction was immunoprecipitated with an anti-E2 antibody and analyzed by SDS-PAGE. Almost all of the E2-gp68 was found near the top of the gradient (fractions 1 to 3), with the largest amount being in fraction 2, corresponding to the membrane-associated proteins (Fig. 4). In contrast, E2-p38 was found in every fraction, with the predominant portion being in fraction 5, which reflects the distribution of the soluble proteins. Compared to the glycosylated form, a significantly smaller portion of E2-p38 was membrane associated. This result shows that a substantial fraction of the unglycosylated E2 is not associated with membranes and is likely localized outside the ER. This finding is consistent with the propensity of E2-p38 to be degraded by the proteasome.
FIG. 4.
Analysis by membrane flotation gradients of the subcellular distribution of E2. Transfected 293T cells were radiolabeled for 15 min, and cellular extracts were subjected to centrifugation on sucrose gradients (10 to 75% [wt/wt] sucrose) at 38,000 rpm for 14 h. Fractions were collected from the top and each fraction was immunoprecipitated using anti-E2 antibody H62.
E2-p38 stability is modulated by IFN treatment through PKR.
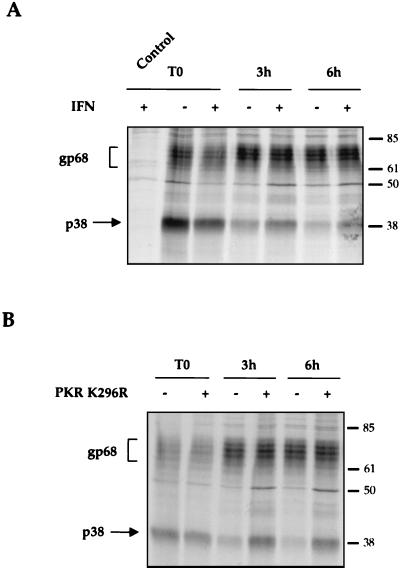
It has previously been shown that E2 plays a role in conferring IFN resistance by interacting with PKR and inhibiting its kinase function (23). Our present finding that E2-p38 is found in the cytosol raises the possibility that this is the E2 form that interacts with PKR. To address this possibility, the stability of E2-p38 was studied in the absence or presence of IFN. Cells transfected with pcDNAEF-E2 were treated with 1,000 IU of IFN-α per ml for 18 h and pulse-chase experiments were then performed with the same concentration of IFN. This length of treatment allowed PKR to be induced. After pulse-labeling, the overall amounts of proteins labeled in the presence of IFN were slightly smaller than in the absence of IFN, suggesting that IFN caused some translation shutoff (Fig. 5A, T0). However, after 3 and 6 h of chase, larger amounts of E2-p38 were found in IFN-treated cells than in untreated cells. In comparison, the amount of E2-gp68 was not affected by the IFN treatment. This result showed that E2-p38 stability was increased after IFN treatment, suggesting that certain IFN-induced proteins may modulate the stability of E2-p38.
FIG. 5.
Stability of E2 after IFN-α treatment or cotransfection with PKR K296R. (A) IFN treatment. Transfected cells were treated with or without 1,000 IU of IFN-α per ml for 18 h, pulse-labeled for 15 min (T0), and then chased for various lengths of times as indicated. Control with empty vector pcDNAEF is included. (B) PKR K296R. Cells were transfected with E2 alone (−) or cotransfected with PKR K296R (+) (ratio, 1:1). A pulse-chase experiment was done as for panel A.
To directly examine the possibility that PKR is involved in the IFN-induced increase of E2-p38 stability, pulse-chase experiments were performed in cells expressing E2 alone or cotransfected with PKR K296R. This mutant has an amino acid substitution in the kinase catalytic site and is enzymatically inactive. Since the E2/PKR ratio is critical for PKR inhibition, this mutant was used instead of wild-type PKR to eliminate the possible confounding effects caused by the PKR activities, so that the stability of E2-p38 could be unequivocally measured. The results showed that, after pulse-labeling, similar amounts of E2-p38 were labeled in the presence or absence of the overexpressed PKR mutant (Fig. 5B, T0). However, after 3 and 6 h of chase, larger amounts of E2-p38 were found in cells cotransfected with E2 and PKR than in cells transfected with E2 alone, suggesting that PKR stabilized E2-p38 and retarded its degradation. No effect on the stability of E2-gp68 was noted. These results are consistent with those observed in IFN-treated cells and confirm that the increase of E2-p38 stability after IFN treatment is mediated by PKR. These results suggest that E2-p38 interacts with PKR and may play a role in IFN resistance by interfering with the biological function of PKR.
E2-p38, but not E2-gp68, interacts with PKR.
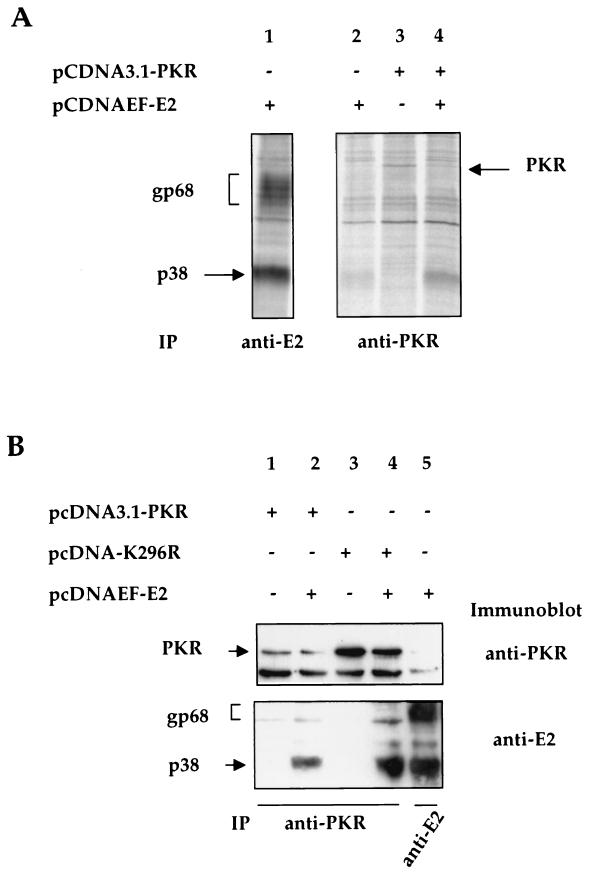
To further confirm that E2-p38 interacts directly with PKR, coimmunoprecipitation of E2 and PKR was performed. Because the endogenous PKR is expressed at a very low level in nonactivated cells, we overexpressed exogenous PKR to facilitate its detection. At 24 h posttransfection, cells were pulse-labeled for 15 min and cell lysates were used for immunoprecipitation with a PKR-specific monoclonal antibody. In cells transfected with PKR only, a band corresponding to the predicted size of PKR was detected (Fig. 6A, lane 3). In cells cotransfected with E2 and PKR, E2-p38 was coimmunoprecipitated with PKR (Fig. 6A, lane 4). In contrast, E2-gp68 was not coimmunoprecipitated with PKR. In cells transfected with E2 only, a small amount of E2-p38 was precipitated by the anti-PKR antibody (Fig. 6A, lane 2). This was probably as a result of coprecipitation with the endogenous PKR. Again, no E2-gp68 was precipitated.
FIG. 6.
Coimmunoprecipitation of E2-p38 with PKR. (A) Cells transfected with different plasmids were radiolabeled and precipitated with the anti-E2 (lane 1) or anti-PKR (lanes 2 to 4) antibodies. (B) Protein extracts of cells transfected with different plasmids were first immunoprecipitated with an anti-PKR antibody (lanes 1 to 4) or with an anti-E2 antibody (lane 5). After SDS-PAGE analysis, proteins were immunoblotted with an anti-PKR antibody (upper panel) or an anti-E2 antibody (lower panel). In the top panel, the lower band corresponds to the heavy chain (≈55 kDa) of the antibody used and the upper one corresponds to PKR.
Since multiple background bands were also detected in this coimmunoprecipitation experiment, a similar analysis was performed, but proteins were detected by immunoblotting after immunoprecipitation to confirm the identity of PKR and E2. Both the wild type and the mutant PKR K296R were used for transfection either alone or together with E2. At 48 h posttransfection, cells were harvested and lysates were used for immunoprecipitation with a specific anti-PKR monoclonal antibody. After SDS-PAGE analysis, proteins were immunoblotted with an anti-PKR monoclonal antibody (Fig. 6B, upper panel) or an anti-E2 monoclonal antibody (Fig. 6B, lower panel). A band corresponding to the predicted size of PKR (≈68 kDa) was detected (Fig. 6B, upper panel, top band, lanes 1 to 4) in the transfected cells, confirming the expression of PKR. As expected, the bands corresponding to PKR K296R, which does not inhibit translation, had a higher intensity than those of wild-type PKR (20). In cells cotransfected with E2 and PKR or PKR K296R, a band corresponding to E2-p38 was coimmunoprecipitated with PKR and revealed by the anti-E2 antibody by immunoblot (Fig. 6B, lower panel, lower band, lanes 2 and 4). Intensity of the E2-p38 band was higher when E2 was cotransfected with PKR K296R than when cotransfected with wild-type PKR, indicating that the quantity of E2 coprecipitated is proportional to the amount of PKR. In contrast, E2-gp68 was not coimmunoprecipitated with PKR in either case. As a control of E2 expression, cells were transfected with E2 alone (Fig. 6B, lane 5), and lysate was immunoprecipitated with an anti-E2 antibody (H62) and subjected to immunoblot with a different anti-E2 antibody (Biodesign). Both gp68 and p38 were detected (Fig. 6B, lower panel, lane 5). However, we could not detect any endogenous PKR coprecipitated by E2 (Fig. 6B, upper panel, lane 5), probably because of its low level of expression in nonactivated cells. These results establish that PKR interacts with the unglycosylated p38 form of E2 only. Moreover, this interaction probably occurs in the cytosol.
DISCUSSION
In the present study, we have identified a new unglycosylated form of the HCV envelope protein E2 and shown that it interacts with PKR. This novel form may thus account for the previously described effects of E2 in inhibiting PKR (23), which likely contributes to the resistance of HCV to IFN. The HCV E2 protein has previously been reported as a glycoprotein with an exclusive ER subcellular localization (2). This picture is probably an oversimplification of what happens in cells; proteins do not exist only under one static form but more likely maintain a dynamic equilibrium between different states. In this study, in which expression of E2 was mediated by a cellular promoter, an unglycosylated form of E2 (E2-p38) was observed in two different cell types and detected by three different antibodies. It is not clear why previous studies did not detect such an unglycosylated species. It is conceivable that the vaccinia virus expression system or strong viral promoters used by others may have interfered with its detection. It is interesting that E2 was glycosylated very rapidly (within 5 min of pulse-labeling), and yet a large amount of E2-p38 remained unglycosylated for several hours, suggesting that it represents a different pool of protein products which is separated from those being processed through the conventional glycosylation pathway in the ER. Indeed, our studies suggest that the majority of E2-p38 is degraded through the proteasome pathway. We have detected another product of 42 kDa in the presence of tunicamycin or after PNGase F treatment, suggesting that E2 may have another type of posttranslational modification. Among the various possibilities of protein modifications, we found a potential site of acylation in the C-terminal region of E2 based on sequence analysis. However, we did not find any incorporation of palmitic acid into E2 by metabolic labeling (data not shown). We have also tested if E2 could be phosphorylated, since E2 interacts with PKR, but no incorporation of [32P]orthophosphate was observed (data not shown). Thus, it is not clear whether E2 undergoes other posttranslational modifications or exists in different protein conformations.
The lactacystin experiment showed that E2-p38 was rapidly degraded by proteasome, and correspondingly, is ubiquitinated. The lactacystin treatment increased the accumulation of the ubiquitinated E2-p38. Since E2 has a strong tendency to misfold, it is not surprising that the cellular quality control mechanisms detect this improperly folded protein and degrade it. Surprisingly, our data showed that the glycosylated E2 is very stable despite the fact that it is also ubiquitinated. It is conceivable that large aggregates of glycosylated E2 may not be able to translocate to the cytosol to be degraded. It has been reported that, in some cases, ubiquitination is a posttranslational modification that does not lead to protein degradation. In fact, it has been shown that other viral proteins, such as the p6gag of human immunodeficiency virus type 1 and simian immunodeficiency virus, are ubiquitinated within viral particles (17). The significance of ubiquitinated viral proteins remains unclear, but it may have a specific role in viral pathogenesis through the host cell ubiquitin-proteasome pathway. In cells, ubiquitination can also play a role in the endocytosis of cell surface receptor as an internalization signal and lysosomal trafficking signal (18, 21). The rapid disappearance of E2-p38 through degradation by proteasome thus suggests that this unglycosylated form is largely present in the cytosol. Indeed, membrane flotation analysis showed that a significant amount of E2-p38 was in the cytosol, although it was also associated with the membrane. In contrast, E2-gp68 was found exclusively in membrane-bound fractions, as would be expected. How E2-p38 remains in the cytosol is an interesting question. E2 is translated by membrane-bound polysomes and is expected to be translocated into the lumen of the ER because of its leader sequence present in the C terminus of the preceding protein, E1 (7). One possibility is that E2 enters the ER and is immediately retro-transported back to the cytosol, where it is degraded. Another interesting possibility is that E2 is trapped near the ribosome or in the space between the ribosome and the outer layer of the ER membrane immediately after translation. For example, the ribosome-bound PKR could potentially trap the unglycosylated E2 in such a space. Indeed, recent structural analysis of the ribosome-ER pore channel complex suggests that such a space exists where the newly synthesized proteins can exit the ribosome-ER pore channel and be translocated to the cytosol (15).
In this perspective, we found that IFN treatment or cotransfection with a catalytically inactivated PKR enhanced E2-p38 stability. Since IFN up-regulates PKR, this effect was most likely due to the interaction of E2-p38 with PKR, which, as a result, prevents its degradation by the proteasome. Indeed, we found that PKR binds to the unglycosylated but not the glycosylated form of E2. These results solved the puzzling question of how E2, which was thought to reside exclusively in the lumen of the ER, could interact with PKR, which is in the cytosol as well as associated with ribosomes (27). Since E2 degradation was delayed after IFN treatment or PKR K296R cotransfection, this interaction likely occurs between the intact E2-p38 and PKR, but not with peptides derived from the degradation of E2-p38.
In conclusion, we have shown that E2, which is presumably an ER-resident viral glycoprotein, is also found in large amounts as an unglycosylated protein in the cytosol. This new finding helps us to understand how E2 interacts with PKR, which may lead to the impairment of the IFN response. This interaction could also contribute to the escape of HCV-infected cells from the immune response, thus leading to the chronic infection observed in 80% of HCV infections. Furthermore, it shows that viral glycoproteins can be found in various forms and thus have other unexpected functions in the cells than just being on the virion surface to interact with cellular receptors.
Acknowledgments
We are grateful to Jean Dubuisson and Stephen Feinstone for their kind gifts of the anti-E2 monoclonal antibodies. We thank Paula M. Cannon for providing the plasmid encoding the VSV G protein.
This project was supported by NIH research grant AI 40038. N.P. is a Research Associate and M.M.C.L. is an Investigator of the Howard Hughes Medical Institute. D.R.T. was supported by a postdoctoral fellowship from the National Institute of Allergy and Infectious Diseases, NIH.
REFERENCES
- 1.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo, M. 1998. The role of hepatitis C virus in hepatocellular carcinoma. Recent Results Cancer Res. 154:337–344. [DOI] [PubMed] [Google Scholar]
- 5.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135–148. [DOI] [PubMed] [Google Scholar]
- 7.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217–227. [DOI] [PubMed] [Google Scholar]
- 9.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershey, J. W. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717–755. [DOI] [PubMed] [Google Scholar]
- 11.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405–439. [DOI] [PubMed] [Google Scholar]
- 12.Kiyosawa, K., E. Tanaka, and T. Sodeyama. 1998. Hepatitis C virus and hepatocellular carcinoma. Curr. Stud. Hematol. Blood Transfus. 62:161–180. [DOI] [PubMed] [Google Scholar]
- 13.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Int. Med. 132:296–305. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto, M., T. Y. Hsieh, N. Zhu, T. VanArsdale, S. B. Hwang, K. S. Jeng, A. E. Gorbalenya, S. Y. Lo, J. H. Ou, C. F. Ware, and M. M. Lai. 1997. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J. Virol. 71:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menetret, J., A. Neuhof, D. G. Morgan, K. Plath, M. Radermacher, T. A. Rapoport, and C. W. Akey. 2000. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell 6:1219–1232. [DOI] [PubMed] [Google Scholar]
- 16.Mottola, G., N. Jourdan, G. Castaldo, N. Malagolini, A. Lahm, F. Serafini-Cessi, G. Migliaccio, and S. Bonatti. 2000. A new determinant of endoplasmic reticulum localization is contained in the juxtamembrane region of the ectodomain of hepatitis C virus glycoprotein E1. J. Biol. Chem. 275:24070–24079. [DOI] [PubMed] [Google Scholar]
- 17.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, 2nd, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paolini, R., and J. P. Kinet. 1993. Cell surface control of the multiubiquitination and deubiquitination of high-affinity immunoglobulin E receptors. EMBO J. 12:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55–84. [DOI] [PubMed] [Google Scholar]
- 20.Romano, P. R., M. T. Garcia-Barrio, X. Zhang, Q. Wang, D. R. Taylor, F. Zhang, C. Herring, M. B. Mathews, J. Qin, and A. G. Hinnebusch. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2 alpha kinases PKR and GCN2. Mol. Cell. Biol. 18:2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth, A. F., and N. G. Davis. 1996. Ubiquitination of the yeast a-factor receptor. J. Cell Biol. 134:661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanderson, C. M., H. H. Wu, and D. P. Nayak. 1994. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J. Virol. 68:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107–110. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, D. R., B. Tian, P. R. Romano, A. G. Hinnebusch, M. M. Lai, and M. B. Mathews. 2001. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. J. Virol. 75:1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161–172. [DOI] [PubMed] [Google Scholar]
- 26.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, S., P. R. Romano, and R. C. Wek. 1997. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J. Biol. Chem. 272:14434–14441. [DOI] [PubMed] [Google Scholar]