Abstract
After ocular herpes simplex virus type 1 (HSV-1) infection, the virus travels up axons and establishes a lifelong latent infection in neurons of the trigeminal ganglia. LAT (latency-associated transcript), the only known viral gene abundantly transcribed during HSV-1 neuronal latency, is required for high levels of reactivation. The LAT function responsible for this reactivation phenotype is not known. Recently, we showed that LAT can block programmed cell death (apoptosis) in neurons of the trigeminal ganglion in vivo and in tissue culture cells in vitro (G.-C. Perng et al., Science 287:1500–1503, 2000; M. Inman et al., J. Virol. 75:3636–3646, 2001). Consequently, we proposed that this antiapoptosis function may be a key to the mechanism by which LAT enhances reactivation. To study this further, we constructed a recombinant HSV-1 virus in which the HSV-1 LAT gene was replaced by an alternate antiapoptosis gene. We used the bovine herpes virus 1 (BHV-1) latency-related (LR) gene, which was previously shown to have antiapoptosis activity, for this purpose. The resulting chimeric virus, designated CJLAT, contains two complete copies of the BHV-1 LR gene (one in each viral long repeat) in place of the normal two copies of the HSV-1 LAT, on an otherwise wild-type HSV-1 strain McKrae genomic background. We report here that in both rabbits and mice reactivation of CJLAT was significantly greater than the LAT null mutant dLAT2903 (P < 0.0004 and P = 0.001, respectively) and was at least as efficient as wild-type McKrae. This strongly suggests that a BHV-1 LR gene function was able to efficiently substitute for an HSV-1 LAT gene function involved in reactivation. Although replication of CJLAT in rabbits and mice was similar to that of wild-type McKrae, CJLAT killed more mice during acute infection and caused more corneal scarring in latently infected rabbits. This suggested that the BHV-1 LR gene and the HSV-1 LAT gene are not functionally identical. However, LR and LAT both have antiapoptosis activity. These studies therefore strongly support the hypothesis that replacing LAT with an antiapoptosis gene restores the wild-type reactivation phenotype to a LAT null mutant of HSV-1 McKrae.
Herpes simplex virus type 1 (HSV-1) is a large (152-kb) double-stranded DNA virus with neurotropic properties. HSV-1 establishes lifelong latent infections in host sensory neurons. This virus is wide spread in the general population. When the eye is infected, the virus travels up nerves and establishes latent infection in neurons of the trigeminal ganglia (TG). During neuronal latency, HSV-1 has no apparent impact on the infected individual. The latent virus can reactivate at various times throughout the life of the individual. This occurs through a mechanism(s) that is currently not completely understood. HSV-1 reactivation in the TG results in virus returning to the eye via the same route previously used to get from the eye to the TG. At the eye, reactivated HSV-1 can produce recurrent disease. Recurrent HSV-1 infection in the eye can cause corneal scarring leading to loss of vision. As a result, HSV-1 is one of the most common infectious causes of corneal blindness in the developed world.
During neuronal latency, LAT (latency-associated transcript) is the only abundantly transcribed viral gene (30, 37). The primary LAT transcript is ca. 8.3 kb long (7, 45) and overlaps two viral genes, ICP0 and ICP34.5, in an antisense direction (30, 37). A very stable intron, the 2-kb LAT is spliced from the primary transcript (9) and is the major LAT RNA detected during latency (7, 33, 36, 42–44).
LAT enhances the induced and spontaneous reactivation phenotypes in the rabbit ocular model (12, 21) and the induced reactivation phenotype in mice (1a, 6, 19, 26, 31, 35). The reduced reactivation phenotypes of LAT− mutants does not necessarily imply that LAT is directly involved in the molecular mechanism of HSV-1 reactivation from latency. LAT might enhance reactivation by increasing the initial amount of latency established and/or by maintaining a high level of latently infected neurons. The larger pool of latently infected neurons and/or the larger pool of neurons containing high copy numbers of the HSV-1 genome would be expected to increase reactivation. Several reports have, in fact, shown that in experimentally infected animals more neurons become latently infected with LAT+ viruses compared to LAT− viruses (29, 31, 40). In addition, we have recently shown that LAT has antiapoptosis activity that could protect acutely infected neurons from virus-induced programmed cell death and result in increased establishment of latency (15, 20).
Because of LAT’s antisense orientation to ICP0, it was originally proposed that LAT might function in the latency-reactivation cycle by regulating ICP0 expression via an antisense mechanism (30, 37). In addition, an apparent readthrough LAT transcript extends well into the region of another important immediate-early gene, ICP4, also in an antisense orientation. Thus, it has been proposed that LAT may suppress these immediate-early genes via an antisense mechanism and that this in turn results in virus shutdown and increased establishment of latency, subsequently resulting in increased reactivation (3, 10). However, the LAT function that enhances reactivation maps to within the first 1.5 kb of the primary LAT transcript (2, 24). This 1.5-kb region does not overlap ICP0 or ICP4, encompasses only the first 837 nucleotides of the stable 2-kb LAT, and does not retain the 2-kb LAT stability (24). Thus, LAT must enhance reactivation in the rabbit by a mechanism that does not involve antisense regulation of ICP0, does not require production of the entire 2-kb LAT, does not require splicing of the 2-kb LAT, and does not require the presence of a stable LAT RNA.
We recently showed that LAT has antiapoptosis activity in the absence of other viral genes, which correlates well with a LAT null mutant inducing high levels of virus-induced programmed cell death (apoptosis) in neurons of HSV-1-infected rabbit TG (20). LAT’s antiapoptosis activity has been independently confirmed in tissue culture and in the mouse model of ocular HSV-1 (1). Moreover, we have shown that this antiapoptosis activity maps to within the same 1.5-kb region to which enhancement of the spontaneous reactivation phenotype maps (15). This further suggests that LAT’s antiapoptosis activity is important in the latency-reactivation cycle.
To investigate this further, we constructed an HSV-1 mutant in which both copies of the region of LAT that enhances the reactivation phenotype were removed and replaced by a different antiapoptosis gene. If a recombinant virus containing an alternative antiapoptosis gene in place of LAT had a LAT null mutant-like reactivation rate, it would suggest that interfering with apoptosis is not the primary LAT function responsible for enhancing reactivation. In contrast, if an alternative antiapoptosis gene could effectively replace LAT and produce a virus with wild type-like reactivation, it would suggest that LAT’s antiapoptosis activity was key to LAT’s ability to enhance reactivation.
We report here on the construction of a chimeric recombinant HSV-1 virus, CJLAT, in which both copies of LAT were replaced with the bovine herpesvirus type 1 (BHV-1) latency-related (LR) gene. Like LAT, the LR gene can block apoptosis in transient-transfection assays in tissue culture (4). We show here that, compared to the LAT− mutant dLAT2903, CJLAT had significantly higher in vivo spontaneous reactivation in the rabbit (P = 0.0003 and P < 0.0001 in two independent experiments) and significantly higher in vitro-induced reactivation in the mouse (P = 0.001). These results support the hypothesis that LAT’s antiapoptosis activity is important for LAT’s ability to enhance reactivation.
MATERIALS AND METHODS
Virus and cells.
All parental and mutants viruses were triple plaque purified and passaged only one or two times in rabbit skin (RS) cells prior to use. Wild-type McKrae, the McKrae-derived LAT− mutant dLAT2903, and marker-rescued dLAT2903R have been previously described (21). RS and CV-1 cells were grown in Eagle minimal essential medium supplemented with 5% fetal calf serum.
Construction of CJLAT.
The plasmid p1658 contains two HSV-1 DNA restriction fragments. One restriction fragment (HpaI-EcoRV) corresponds to the region from 1800 to 161 nucleotides upstream of the LAT start site (LAT nucleotides −1800 to −161). The other restriction fragment (HpaI-MluI) corresponds to LAT nucleotides 1667 to 2850. A unique PacI restriction site was cloned between these restriction fragments (i.e., between LAT nucleotides −161 and +1667). PacI linkers were added to the complete BHV-1 LR gene DNA, which was then cloned into the above unique PacI restriction site of p1658. The LR gene was a 1.9-kb HindIII-SalI fragment derived from the HindIIID fragment of BHV-1 strain Cooper (BHV-1 nucleotides 1 to 1941) that contains the LR promoter and the known coding sequences (18). The resulting plasmid, CJLATP1658, was cotransfected with infectious dLAT2903 genomic DNA into RS cells as we previously described for construction of other HSV-1 mutants (21, 25). After homologous recombination between the HSV-1 DNA flanking the BHV-LR DNA in the plasmid and the genomic DNA flanking the LAT deletion in dLAT2903, the cotransfection mix was plated on RS cells, and viral plaques were isolated. Individual plaques were analyzed by restriction digestion and Southern analysis. In the final chimeric virus, CJLAT, the core LAT promoter and the region of the LAT gene corresponding to the first 1.7-kb of the primary LAT transcript of HSV-1 McKrae has been replaced by the BHV-1 LAT gene. CJLAT was triple plaque purified and its genomic structure was confirmed by additional restriction digestion-Southern analysis.
Rabbits.
Eight- to ten-week-old New Zealand White male rabbits (Irish Farms) were used. Rabbits were treated in accordance with ARVO (Association for Research in Vision and Ophthalmology), AALAC (American Association for Laboratory Animal Care), and National Institutes of Health guidelines. Rabbits were bilaterally infected without scarification or anesthesia by placing, as eye drops, 2 × 105 PFU of virus into the conjunctival cul-de-sac, closing the eye, and rubbing the lid gently against the eye for 30 s as we previously described (30). Analysis of virus replication in eyes was done as previously described (21)
Mice.
Swiss-Webster mice were used. Mice were ocularly infected as for rabbits except that the infectious dose was 2 × 106 PFU/eye. Analysis of virus replication in the eyes and TG was done as previously described (26).
Neutralization assay.
Serum-neutralizing antibody titers were determined by standard plaque reduction assays as previously described (28).
Northern blot hybridization.
Subconfluent cell monolayers were infected at a multiplicity of infection (MOI) of 5 PFU/cell, total RNA was isolated, and Northern blot analysis was performed according to standard protocols by using a 32P-labeled cloned restriction fragment specific for the detection of BHV LR-RNA or specific for the detection of HSV-1 LAT and ICP0.
RT-PCR.
For the reverse transcription (RT) step, 1 μg of total cell RNA (DNase treated) was mixed with 1 μg of random hexamer primers in a total volume of 4 μl, incubated at 65°C for 7.5 min, and chilled on ice (denaturation). Then, 16 μl of ice-cold RT mix (20 mM Tris-HCl [pH 8.3]), 50 mM KCl, 2.5 mM MgCl2, 100 μg of bovine serum albumin/ml, 1 mM dithiothreitol, a 0.5 mM concentration of each deoxynucleoside triphosphate, 10 U of RNasin, and 100 U of RNase H− reverse transcriptase [Stratagene]) was added. The reaction mixture was incubated for 10 min at 20.5°C and then for 90 min at 45°C. RT was terminated by incubation at 95°C for 5 min. As a control for DNA contamination in the RNA samples, 0.5 μg of RNA (DNase treated) was mixed with ice-cold RT mix lacking reverse transcriptase in a final volume of 10 μl.
An aliquot (1.5 μl) of RT reaction mixtures was used for each PCR. PCRs were carried out in 50 μl of a mixture containing 1× commercial PCR buffer, a 200 μM concentration of each deoxynucleoside triphosphate, 1.0 mM MgCl2, a 1 μM concentration of each LR primer, 10% glycerol, and 1 U of Taq polymerase. Amplification was carried out for 40 cycles of denaturing at 95°C for 1 min, annealing at 65°C for 1 min, and extending at 72°C for 2 min. After completion of the last cycle, reaction mixtures were further incubated at 72°C for 7 min to ensure complete extension of the amplified products. All PCRs were “hot started.” The use of RNase H− reverse transcriptase, a high temperature for RT, 10% glycerol, and a “hot start” in the PCRs allow amplification through RNA regions of complex secondary structure. Amplified products were visualized on agarose gels. The LR primers used—L3B upstream (nucleotides 1755 to 1775; 5′-TTCTCTGGGCTCGGGGCTGC-3′) and L3B downstream (nucleotides 1924 to 1947; 5′-AGAGGTCGACAAACACCCGCGGT-3′)—were described previously (13).
Statistical analysis.
Analyses were performed by using the personal computer program Prizm GraphPad, version 3.02.
RESULTS
Construction and genomic structure of CJLAT.
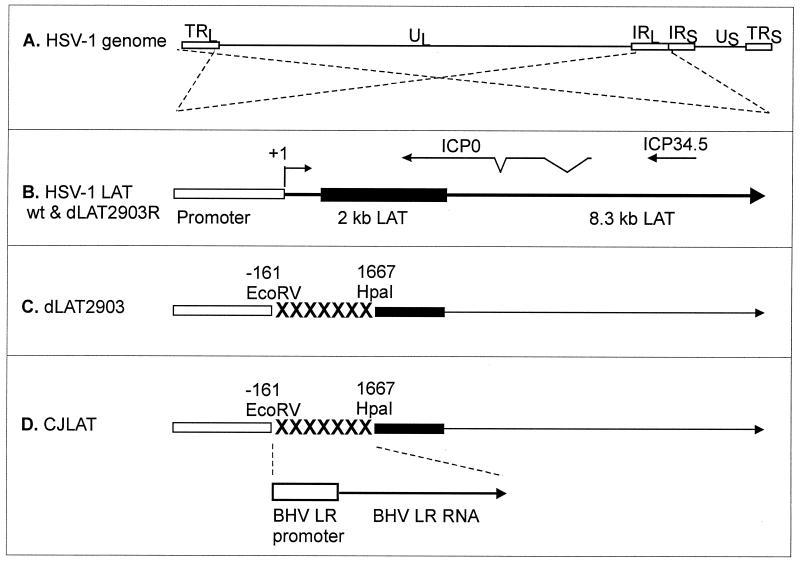
The genomic structures of CJLAT and other viruses used in this study are shown schematically in Fig. 1. All viruses were derived from HSV-1 strain McKrae. Figure 1A shows the prototypic structure of the wild-type HSV-1 McKrae genome. The viral long repeats are expanded (dashed lines) to show the relative location and status of the LAT gene in McKrae and marker-rescued dLAT2903R (Fig. 1B). The dashed lines cross to indicate that the repeats are inverted (i.e., the DNA sequences are in opposite orientations). The locations of the ICP0 and ICP34.5 genes are shown for reference. The primary LAT transcript in wild-type virus and marker-rescued dLAT2903R is ca. 8.3 kb (41, 42). A very stable and easily detected 2-kb LAT (solid rectangle) appears to be an intron derived by splicing of the primary LAT (9). The LAT region of dLAT2903 (Fig. 1C) contains a deletion in both copies of LAT from −161 to +1,677 relative to the start of the primary LAT transcript (indicated by “XXXXX”). This virus is missing key promoter elements, makes no LAT RNA, is a true LAT null mutant, and reactivates from latency inefficiently (21). The LAT region of CJLAT (Fig. 1D) contains the complete BHV-1 LR gene, including the LR promoter, inserted into the deletion in dLAT2903. CJLAT thus contains two complete copies of the BHV-1 LR gene (one in each viral long repeat) driven by the BHV-1 LR promoter. This virus is equivalent to marker-rescued dLAT2903R (Fig. 1B), except that the deletion in dLAT2903 was “rescued” by using BHV-1 DNA rather than HSV-1 DNA.
FIG. 1.
Schematic representation of mutant viruses. (A) Genomic structure of wild-type HSV-1 strain McKrae. TRL and IRL indicate the viral long repeats (terminal and internal, respectively). IRS and TRS indicate the viral short repeats. UL and US indicate the long and short unique regions. The dashed lines indicate that the region of the TRL and IRL are expanded below with the TRL inverted relative to the IRL so that both identical regions can be represented by a single image. (B) The LAT region of wild-type and marker-rescued dLAT2903R. The LAT promoter is represented by the open rectangle. The primary 8.3-kb LAT transcript is indicated by the large arrow. The solid rectangle indicates the stable 2-kb LAT intron. The start of LAT transcription is indicated by the arrow at +1 (LAT nucleotide 1). The relative locations of the ICP0 and ICP34.5 RNAs are shown for reference. (C) The deletion from LAT nucleotide −161 to +1667 in dLAT2903 is indicated by “XXXXXXX.” dLAT2903 is a true LAT null mutant that is missing primary LAT promoter elements between positions −161 and +1. dLAT2903 is also lacking a putative secondary LAT promoter, LAP2, located within the 5′ end of the primary LAT transcript prior to the start of the 2-kb LAT (11). This mutant therefore is not capable of expressing any LAT RNA (21). (D) The LAT region of CJLAT. The complete LR gene of BHV-1, including the promoter (a 1,941-bp HindIII-SalI fragment), was inserted into the deletion in dLAT2903. The resulting virus expresses the LR-RNA from the HSV-1 LAT region.
The construction of dLAT2903 and dLAT2903R have been previously described (21). Because the BHV-1 LR gene has no sequence homology to the HSV-1 LAT gene, construction of CJLAT by homologous recombination required that the BHV-1 LR gene was first cloned into a plasmid containing appropriate HSV-1 flanking sequences. This and additional details of the construction of CJLAT are given in Materials and Methods.
CJLAT expresses BHV LR-RNA but no HSV-1-LAT.
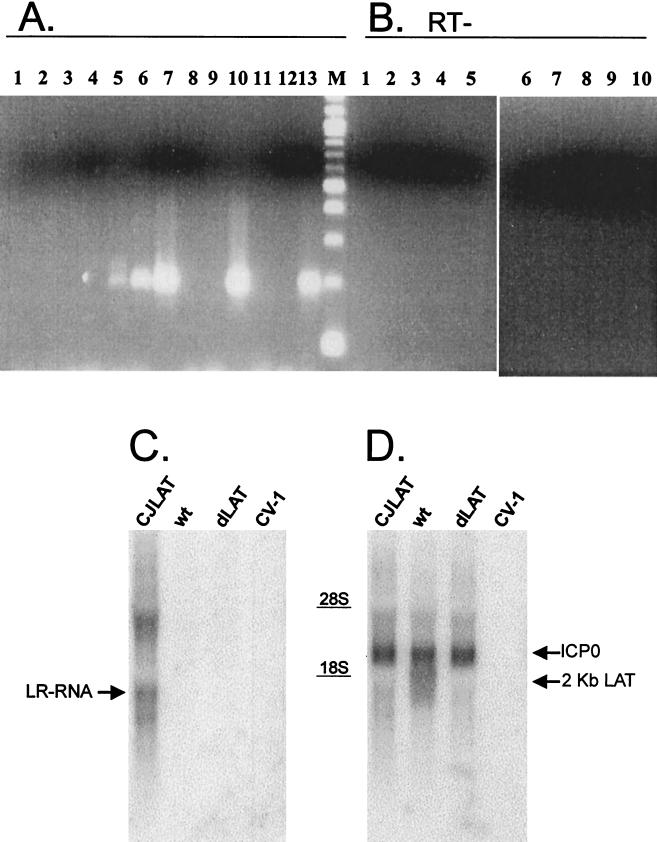
CV-1 cells were infected with CJLAT, dLAT2903, or wild-type McKrae at an MOI of 5. Total RNA was isolated at various times postinfection (p.i.). RT-PCR was performed by using the L3B primers specific for BHV-1 LR-RNA, and the RT-PCR products were analyzed by agarose gel electrophoresis as described in Materials and Methods (Fig. 2A). Lane 13 shows the PCR product generated from cloned LR DNA and acts as a size marker. An RT-PCR product of the same size was seen with RNA from cells infected with CJLAT. The amount of the RT-PCR product appeared to increase from 6 to 12 to 24 h p.i. (lanes 5, 6, and 7, respectively) and then remained constant from 24 to 48 h p.i. (compare lanes 7 and 10). This is consistent with expression of LR-RNA in BHV-1-infected cells (18) and for LAT in HSV-1-infected cells (34). As expected, no RT-PCR product was seen at any time p.i. with RNA from wild-type HSV-1 McKrae-infected cells (lanes 1, 2, and 8) or dLAT2903-infected cells (lanes 3, 4, and 9). Also, as expected, no RT-PCR product was seen with RNA from mock-infected cells (lane 11) or when RNA (lane 12) was left out of the reaction mix. In addition, no RT-PCR product was seen when reverse transcriptase was left out of the reaction mix (Fig. 2B, lanes 1 to 10).
FIG. 2.
RT-PCR and Northern blot analysis of LR-RNA expressed in CJLAT. (A) CV-1 cells were infected with CJLAT, dLAT2903, or wild-type McKrae at an MOI of 5. Total RNA was isolated at various times p.i., and RT-PCR was performed as described in Materials and Methods. Lanes: 1, McKrae (6 h pi); 2, McKrae (24 h p.i.); 3, dLAT2903 (6 h p.i.); 4, dLAT2903 (24 h p.i.); 5, CJLAT (6 h p.i.); 6, CJLAT (12 h p.i.); 7, CJLAT (24 h p.i.); 8, McKrae (48 h p.i.); 9, dLAT2903 (48 h p.i.); 10, CJLAT (48 h p.i.); 11, mock infected; 12, no RNA; 13, plasmid containing LR gene; M, 100-bp marker ladder (the fastest-migrating band is 100 bp). The expected size of the RT-PCR product is 192 bp. (B) No-RT controls. Lanes: 1 to 10 are the same as in panel A, except that no reverse transcriptase was added to the reactions. (C) Total RNA from CV-1 cells infected as described above was isolated 12 h p.i. and analyzed by Northern blot with a HindIII-SalI restriction fragment probe corresponding to the entire BHV-1 LR gene as described in Materials and Methods. The arrow points to the LR-RNA band. (D) The membrane from panel C was stripped and reprobed with an HSV-1 HpaI-MluI DNA restriction fragment (LAT nucleotides 1667 to 2850) capable of hybridizing to both LAT and ICP0 (arrows). CJLAT, CJLAT-infected CV-1 cells; wt, wild-type McKrae-infected CV-1 cells; dLAT2903, dLAT2903-infected CV-1 cells; CV-1, uninfected CV-1 cells.
Transcription of BHV-1 LR-RNA was also examined by Northern blot analysis. CV-1 cells were infected at an MOI of 5 with CJLAT, wild-type McKrae, or dLAT2903. Total RNA was isolated 12 h p.i. and subjected to standard Northern blot analysis with a probe specific for BHV-1 LR-RNA (a HindIII-SalI DNA restriction fragment containing the entire LR gene). An RNA transcript that hybridized to the LR-RNA specific probe and that corresponded in size to LR-RNA (ca. 1.3 to 1.5 kb) was readily detected in the CJLAT lane (Fig. 2C, arrow) but not in the wild-type, dLAT2903, or uninfected CV-1 cell lanes. Additional LR-RNA bands representing alternative splicing products or readthrough transcription were also seen in the CJLAT lane. The Northern blot in Fig. 2C was stripped and reprobed with an HSV-1 HpaI-MluI DNA restriction fragment (LAT nucleotides 1667 to 2850) capable of hybridizing to both LAT and ICP0 (Fig. 2D). As expected, the 2-kb LAT RNA was detected in wild-type HSV-1 McKrae-infected cells (lower arrow) but not in mock-, CJLAT-, or dLAT2903-infected cells. ICP0 was detected with all three viruses (upper arrow) but not in uninfected cells, and the intensities of the ICP0 bands were similar with all three viruses. These results shown in Fig. 2 suggest that LR-RNA was expressed by CJLAT and that ICP0 expression was not perturbed in CJLAT.
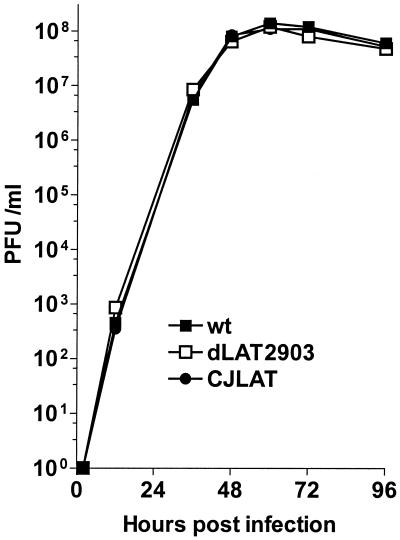
Replication of CJLAT in tissue culture.
CV-1 cells were infected at an MOI of 0.01 with CJLAT, dLAT2903 (the immediate parental virus), or wild-type McKrae (the parental virus for dLAT2903). Replication appeared similar for all three viruses (Fig. 3). Thus, insertion and expression of the BHV-1 LR gene in place of the HSV-1 LAT gene did not noticeably alter virus replication in tissue culture. This also supports the conclusion that the ICP0 gene is functioning correctly.
FIG. 3.
Replication of CJLAT in tissue culture. CV-1 cells were infected at an MOI of 0.01. The infected monolayers were harvested by freeze-thawing at the indicated times, and the amounts of infectious virus were determined by plaque assays as described in Materials and Methods.
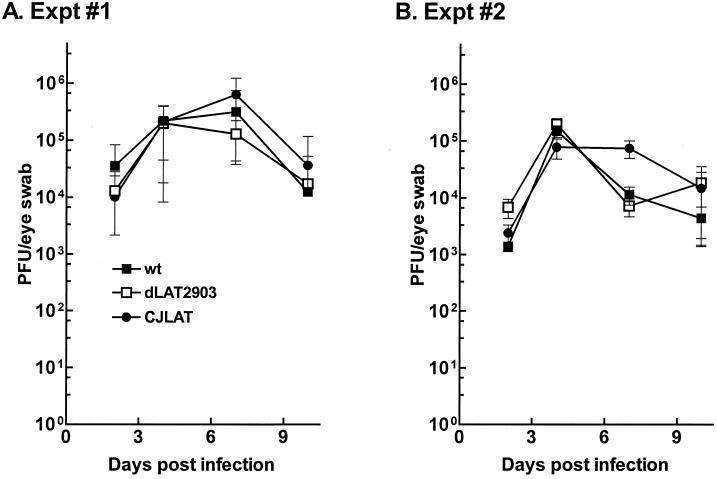
Replication of CJLAT in rabbit eyes.
Rabbits were bilaterally ocularly infected with 2 × 105 PFU of CJLAT, dLAT2903, or wild-type McKrae/eye as described in Materials and Methods. Tear films were collected from 10 eyes at various times, and the amount of virus in each tear film was determined by standard plaque assays on RS cells. Two independent experiments were performed (Fig. 4). Replication of CJLAT appeared similar to that of dLAT2903 and wild-type McKrae in both experiments.
FIG. 4.
Replication of CJLAT in rabbit eyes. Rabbits were infected as described in Materials and Methods. Tears were collected from 10 eyes/group on the days indicated and the amount of infectious virus was determined as described above. Panels A and B are from two independent experiments.
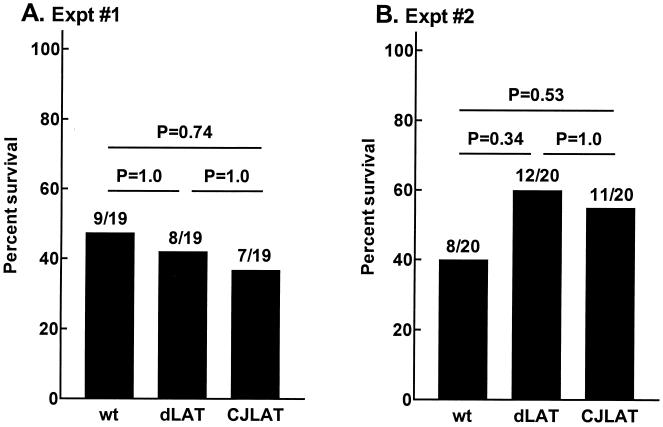
Rabbit survival.
In the above experiments rabbit survival (with death due to viral encephalitis) was determined on day 21 p.i. There was no significant difference in rabbit survival among the three viruses in either experiment (Fig. 5). Thus, after ocular infection of rabbits, CJLAT appeared to have wild-type virulence as measured by survival.
FIG. 5.
Rabbit survival. The numbers above each bar indicate the number of surviving rabbits on day 21 p.i. over the number of rabbits initially infected. P values were determined by chi-square analysis. Bars: wt, wild-type McKrae; dLAT, dLAT2903; CJLAT, CJLAT.
Spontaneous reactivation.
Beginning 30 days p.i., a time at which latency is well established, spontaneous reactivation can be determined by analyzing tears for the presence of infectious virus (i.e., virus shedding). However, if corneal scarring develops (<10% of wild type-infected eyes) detection of infectious virus in the tears of that eye becomes difficult, and these eyes are routinely excluded (29). In both sets of infected rabbits described above, ca. 80% of the eyes in the CJLAT group developed corneal scarring. Therefore, we were unable to use this approach to examine spontaneous reactivation of CJLAT.
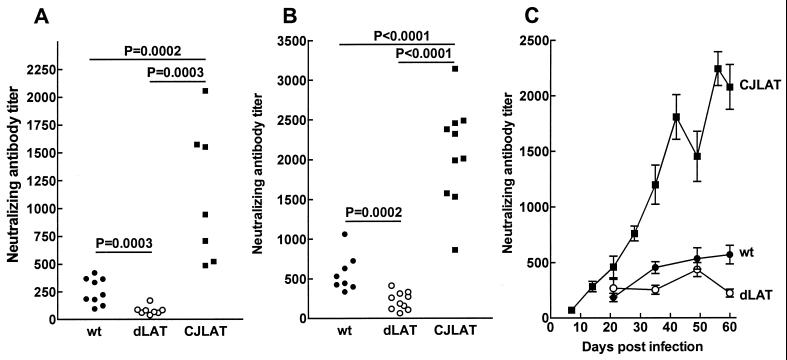
In rabbits infected with viruses showing efficient (LAT+ and wild type) or inefficient (LAT−) spontaneous reactivation, neutralizing antibody titers increase and are similar throughout the first 45 days p.i. (28). However, by day 59 p.i. (ca. 31 to 45 days after latency is established), the continued restimulation of the immune system in rabbits latently infected with efficiently reactivating viruses results in increased neutralizing antibody titers compared to rabbits infected with poorly reactivating viruses (28). This difference has been used to determine whether mutant viruses show wild-type LAT+-like or LAT−-like spontaneous reactivation (29). An increase in neutralizing antibody titers is also a sensitive measure of detecting dexamethasone-induced reactivation of calves latently infected with BHV-1 (17), demonstrating that an increase in virus-specific antibodies normally occurs after alphaherpesvirus reactivation. Thus, this approach was used to examine spontaneous reactivation of CJLAT.
Sera were collected on day 59 p.i. from each of the surviving rabbits shown in Fig. 5A (experiment 1) or from the 8 wild-type-infected surviving rabbits or the 10 dLAT2903- and CJLAT-infected surviving rabbits shown in Fig. 5B (experiment 2). Neutralizing antibody titers were determined on each serum, and the results were plotted as scattergrams (Fig. 6A and B). As expected, in both experiments the wild-type-infected rabbits had significantly higher neutralizing antibody titers than the dLAT2903-infected rabbits, a finding indicative of higher spontaneous reactivation with the wild-type virus. The CJLAT-infected rabbits also had significant higher average neutralizing antibody titers than the dLAT2903-infected rabbits in both experiments (Fig. 6A and B). Interestingly, the neutralizing antibody titers in CJLAT-infected rabbits were also significantly higher than in wild-type-infected rabbits. This suggests that CJLAT not only reactivated more efficiently than dLAT2903 but may also have reactivated more efficiently than wild-type McKrae.
FIG. 6.
Spontaneous reactivation. Rabbits were infected as described above, and individual sera were collected on day 59 p.i. (A), day 60 p.i. (B), or on the days indicated (C). In panels A and B, each datum point indicates the neutralizing antibody titer of one sera. In panel C, each datum point is the average of between 5 and 10 sera. Panels A and B are from separate experiments. Panels B and C represent sera from the same set of rabbits. wt, wild-type McKrae; dLAT, dLAT2903.
In experiment 2, neutralizing antibody titers were also determined for sera collected on additional days (Fig. 6C). The neutralizing antibody titers in the wild-type and CJLAT groups continued to rise throughout the entire study period, a result indicative of spontaneous reactivation. In contrast, in the dLAT2903 group the neutralizing titer declined after day 49. Consistent with the results for a single day shown in Fig. 6A and B, the CJLAT neutralizing antibody titers were higher and appeared to rise more sharply in the CJLAT group than in the wild-type group (Fig. 6C). Since the neutralizing antibody titers were determined by plaque reduction assays with wild-type HSV-1, it was very unlikely that the increased neutralizing antibody titers in the CJLAT group were due to anti-LR antibody somehow neutralizing wild-type HSV-1. Nonetheless, we confirmed that anti-LR protein antibody did not neutralize either wild-type HSV-1 or BHV-1 (not shown). These results therefore strongly suggest that spontaneous reactivation of CJLAT was at least as efficient as that of wild-type McKrae.
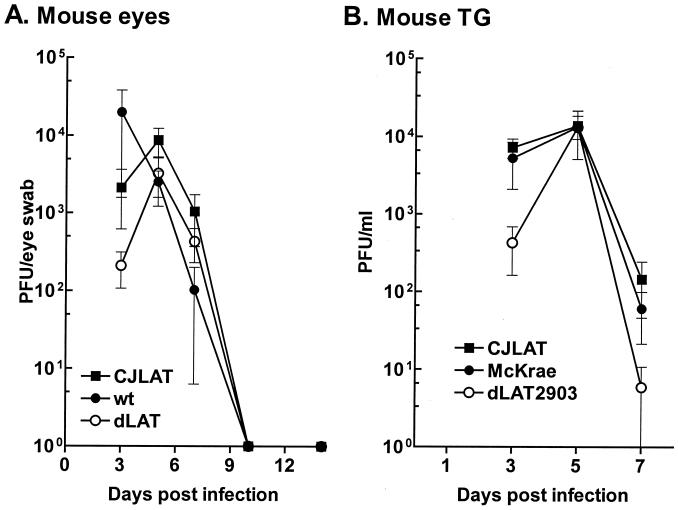
Replication of CJLAT in mouse eyes and TG.
To determine whether CJLAT also had LAT+ reactivation in a second animal model, studies were performed in mice. Mice were infected with 2 × 106 PFU of CJLAT, dLAT2903, or wild-type McKrae/eye without corneal scarification as described in Materials and Methods. Eye swabs were collected from 10 eyes/group at various times, and the amount of infectious virus was determined (Fig. 7A). All three viruses appeared to replicate with similar kinetics and to a similar peak titer. Mice were euthanized at various times, TG were removed and homogenized, and total infectious virus was determined for each TG. All three viruses appeared to replicate with similar kinetics and to a similar peak titer (Fig. 7B). Thus, CJLAT was not impaired for replication in mouse eyes or TG.
FIG. 7.
Replication of CJLAT in mouse eyes and TG. Mice were ocularly infected as described in Materials and Methods. (A) Tears were collected from 10 eyes/group on the indicated days, and the amounts of infectious virus were determined by plaque assay. (B) Mice were infected as in panel A. Five mice/group were euthanatized on the days indicated, and individual TG (10/group/time point) were homogenized, and the amounts of infectious virus were determined by plaque assay.
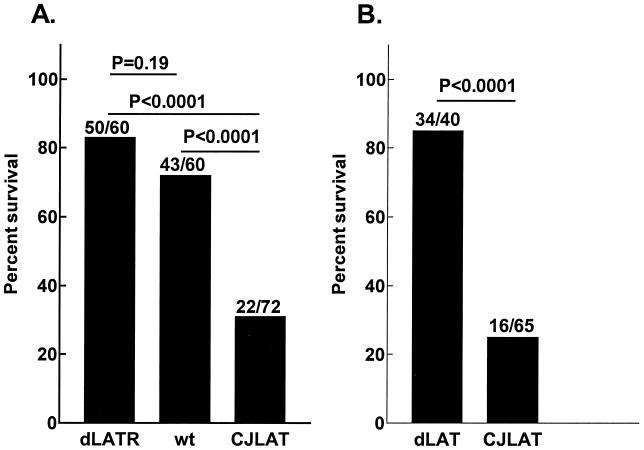
Mouse survival.
Mice were ocularly infected with CJLAT, wild-type McKrae, or dLAT2903R (marker-rescued dLAT2903), and death due to viral encephalitis was monitored for up to 21 days. Significantly fewer CJLAT-infected mice survived compared to wild-type- or dLAT2903R-infected mice (Fig. 8A). Survival was similar for dLAT2903R- and wild type-infected mice. In a second experiment, mice were infected as described above with CJLAT or dLAT2903. Again, significantly fewer CJLAT-infected mice survived (Fig. 8B). The symptoms demonstrated by the mice that died from CJLAT infection were similar to those that died from wild-type McKrae, dLAT2903, or dLAT2903R infection, suggesting that the CJLAT-infected mice also died from encephalitis. As judged by death, CJLAT appeared to have increased virulence in mice.
FIG. 8.
Mouse survival. Mice were ocularly infected with 2 × 106 PFU of virus/eye, and survival was determined on day 21 p.i. Panels A and B are from separate experiments. wt, wild-type McKrae; dLATR, dLAT2903R-infected mice.
Mouse explant cultivation reactivation.
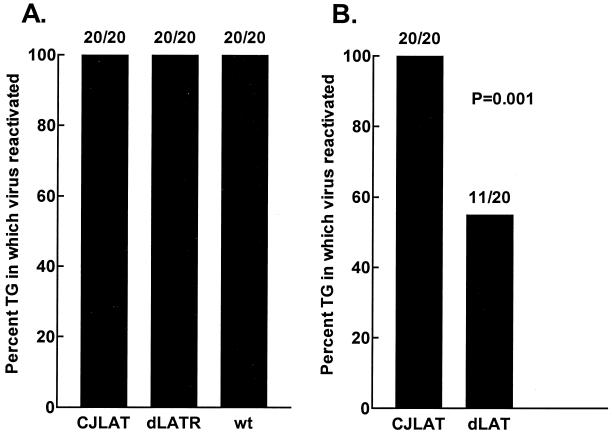
To examine the reactivation of CJLAT compared to wild-type viruses in the mouse model, 10 mice/group were randomly selected from the surviving mice shown in Fig. 8A. On day 30 p.i., the mice were sacrificed, and individual TG were cultured in tissue culture media. Aliquots of media were removed from each culture daily for up to 21 days and plated on indicator cells (RS cells) to look for the presence of reactivated virus. Reactivated virus was detected from all 20 CJLAT, wild-type McKrae, and dLAT2903R TG (Fig. 9A). To examine reactivation of CJLAT compared to dLAT2903, TG from surviving mice shown in Fig. 8B were similarly analyzed. Again, all 20 TG from mice latently infected with CJLAT reactivated. In contrast, significantly fewer TG from dLAT2903 latently infected mice appeared to reactivate (11 of 20, P = 0.001). Thus, in this standard mouse reactivation system, CJLAT appeared to have wild-type reactivation that was significantly greater than that of LAT− virus and at least as efficient as that of wild-type McKrae.
FIG. 9.
Explant reactivation. Ten mice from each of the groups shown in Fig. 8 were selected randomly, and the TG were removed on day 30 p.i. and individually incubated in tissue culture media at 37°C. Aliquots of the tissue culture media were removed daily for up to 21 days and inoculated onto RS cell monolayers to look for the presence of reactivated virus.
Since in the above studies media from the explanted TG cultures were plated daily, the time at which reactivated virus first appeared in the explanted TG cultures could be determined. No cultures contained any detectable infectious virus during the first 3 days of explant. This finding confirmed that none of the TG contained cell-free infectious virus at the time of explant. In the experiment comparing CJLAT to wild-type viruses (Fig. 9A), 80% of the CJLAT TG cultures had detectable reactivated virus on day 4 after explant. In contrast, 80% reactivation was not seen in the wild-type and dLAT2903R cultures until day 6. This was significantly different by survival curve analysis (P = 0.0002 for CJLAT compared to dLAT2903R; P = 0.03 for CJLAT compared to wild type). Consistent with the serum neutralizing antibody titer results, this apparent higher reactivation of CJLAT suggests that CJLAT may have a more efficient reactivation phenotype than did the wild-type virus.
DISCUSSION
Since its discovery in 1987 (30, 37), LAT, the only HSV-1 gene abundantly transcribed during neuronal latency, has been thought to be involved in the viral latency-reactivation cycle. This concept is supported by numerous reports showing that in mice and rabbits infected with LAT− viruses the amount of reactivation is significantly reduced compared to that seen in wild type-infected or marker-rescued virus-infected animals (1a, 6, 12, 19, 21, 26, 31, 35). This reduced reactivation phenotype may be the result of LAT playing a direct role in the reactivation process and/or the result of LAT influencing the amount of latency that is established after acute infection. Although LAT reduces expression of immediate-early viral genes (3, 10) and inhibits virus-induced apoptosis (15, 20) (1), the mechanism by which LAT enhances the reactivation phenotype remains unclear. We report here that replacing the HSV-1 LAT gene with an alternative gene known to have antiapoptosis activity resulted in a recombinant virus with a reactivation phenotype at least equivalent to that of the wild-type virus. This supports the hypothesis that LAT’s antiapoptosis activity is a key LAT function involved in enhancing spontaneous reactivation.
Mice and rabbits infected with LAT− virus have ca. 50% fewer TG neurons in which latency is established (29, 31). It has been proposed that this may completely account for the reduced reactivation phenotype of LAT− mutants. However, in LAT− mutants, reactivation is often reduced by 3- to 10-fold (21, 24, 27). This suggests that LAT also influences the reactivation stage or that a LAT null mutant does not establish latency in specific neuronal populations that are capable of supporting reactivation. In addition, in at least one report, infection of rabbits with a virus containing a mutation in an HSV-1 gene other than LAT resulted in a >5-fold reduction in the establishment of latency, yet the spontaneous reactivation phenotype was indistinguishable from that of wild-type virus (23). This suggests that LAT enhances establishment in certain neuronal populations and reactivation of latency by different mechanisms.
The primary LAT transcript overlaps the important immediate-early gene ICP0 in an antisense orientation, suggesting that repression of ICP0 expression by an antisense mechanism is crucial for regulating latency (30, 37). In this regard, LAT has been shown to down regulate immediate-early gene expression in the TG of acutely and latently infected mice (3, 10). However, it has been shown that mutants capable of expressing only the first 1.5 kb of LAT, a region that does not overlap either ICP0 or ICP4, have normal reactivation levels in the rabbit model (2, 24). Thus, if LAT antisense suppression of ICP0 and ICP4 occurs, it is not a critical factor in the reactivation phenotype as measured in rabbits. This does not rule out the possibility that LAT suppresses ICP0 and/or ICP4 by an antisense-independent mechanism and that this may play an important role in the reactivation phenotype.
We recently reported that the HSV-1 LAT can prevent neurons in the TG of infected rabbits from death by blocking virus-induced apoptosis (20). We also reported that, in a transient-expression assay in tissue culture, a plasmid expressing LAT blocks apoptosis induced by a variety of chemicals (15, 20). We proposed that this LAT antiapoptosis activity is an important factor in the ability of LAT to enhance reactivation. Our finding that LAT protects neurons of the TG from viral induced death was confirmed (39), however, because these authors could not detect TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining of neurons, they contended that LAT did not block apoptosis. Recently, our original report has been independently confirmed and extended by showing that plasmids expressing LAT can block apoptosis induced by anti-Fas antibody, and by showing that in TG of mice infected with LAT− virus TUNEL staining and staining for HSV-1 antigens colocalized, while this did not occur in TG from wild type-infected mice (1).
Like LAT, the BHV-1 LR gene is the only viral gene abundantly transcribed during neuronal latency. Like LAT, the LR-RNA overlaps and is antisense to the BHV-1 homologue of the HSV-1 ICP0 gene. In addition, a recent study demonstrated that a BHV-1 mutant containing stop codons near the beginning of the LR-RNA reactivates poorly compared to wild-type BHV-1 (14). Finally, both the BHV-1 LR-RNA gene (4) and the HSV-1 LAT (15, 20) have been shown to have antiapoptosis activity. Although the LR gene of BHV-1 is clearly the homologue of the HSV-1 LAT gene, there are some important differences. For example, the primary LAT transcript is 8.3 kb (45), whereas the primary LR transcript is only 1.5 kb (18), and there is no obvious sequence homology between LR-RNA and LAT. While the region of LAT to which the spontaneous reactivation phenotype maps does not appear to encode a protein involved in the latency-reactivation cycle (8), the BHV-1 LR gene does encode such a protein (16). Although an open reading frame in the stable 2-kb LAT may encode a protein (5, 38), this protein is unlikely to play an important role in reactivation since it is located outside the region of LAT to which the reactivation function maps in rabbits (2, 24) and mice (unpublished results). Expression of an LR protein appears to play a role in protecting cells from death (4). Finally, the LR gene has also been shown to inhibit cell cycle progression (32) and in the context of the viral genome plays an important role in virus shedding in the eyes of infected calves (14). Thus, structurally, the LR and LAT genes and RNAs are very different. As a first step toward testing the hypothesis that LAT’s antiapoptosis activity is responsible for LAT’s ability to increase reactivation, we therefore inserted the antiapoptosis BHV-1 LR gene into HSV-1 in place of LAT.
Replication of CJLAT in tissue culture, rabbit eyes, mouse eyes, and mouse TG was indistinguishable from that of wild-type McKrae. Surprisingly, although CJLAT appeared to have wild-type McKrae-like virulence in rabbits, as judged by survival after ocular infection, in mice CJLAT appeared to be significantly more virulent than McKrae. Although the reason for this remains unknown, we have previously shown that several mutants lacking portions of LAT have altered virulence and that with some of these mutants the virulence is asymmetric, or dissimilar, in rabbits compared to mice (22).
One unexpected result in these studies was that CJLAT caused severe corneal scarring in most of the infected rabbits. The corneal scarring was not apparent until after day 30 p.i., a time well after all primary infection had subsided and latency was established. It is unclear whether the corneal scarring represents recurrent disease or if it is an aftermath of the primary infection. We previously reported that viruses expressing enhanced green fluorescent protein driven by the LAT promoter also produced severe corneal scarring in most rabbit eyes after day 30 p.i. (29). Thus, regardless of whether the corneal scarring is due to acute or recurrent infection, we speculate that it may be due to high-level long-term expression of a foreign protein in the eye (LR protein with CJLAT). This does not occur in wild type-infected rabbits because LAT does not appear to encode a protein. Alternatively, the increased corneal scarring with CJLAT in rabbits could be the result of increased reactivation, more virus in the eye following reactivation, or infection of different cell types. Interestingly, in mice CJLAT did not cause any significant increase in corneal scarring above that seen with wild-type HSV-1 or dLAT2903. That spontaneous reactivation occurs in rabbits but not in mice suggests that the corneal scarring seen in rabbits is due to recurrent disease following spontaneous reactivation.
Our neutralizing antibody titer results in rabbits and our explant TG reactivation results in mice each strongly suggested that the BHV-1 LR gene supported HSV-1 reactivation at least as efficiently as the HSV-1 LAT gene. Thus, it appeared that the HSV-1 LAT activity responsible for enhancing reactivation in the rabbit and the mouse could be replaced by an activity supplied by the BHV-1 LR gene. Since the BHV-1 LR gene can block apoptosis (4), it is tempting to conclude that replacing the HSV-1 LAT with an antiapoptosis gene allows for efficient LAT+-like reactivation. It would then be further tempting to conclude that the CJLAT results reported here prove that LAT’s antiapoptosis activity is responsible for LAT’s ability to enhance reactivation. However, it remains possible that an as-yet-unknown additional activity common to both the BHV-1 LR gene and the HSV-1 LAT gene are responsible for enhancing reactivation. Experiments are under way to determine whether replacing LAT with other known antiapoptosis genes also produces LAT+-like reactivation.
Acknowledgments
This work was supported by Public Health Service grants EY07566, EY11629, EY12823, and P20RR15635; USDA grant 2000-02060; the Discovery Fund for Eye Research; and the Skirball Program in Molecular Ophthalmology.
REFERENCES
- 1.Ahmed, M., M. Lock, C. G. Miller, and N. W. Fraser. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76:717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.ock, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618–630. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, D. C., J. M. Hill, G. Devi-Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, S. H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin, R. S., S. K. Thomas, D. P. Thomas, and D. S. Latchman. 1998. The herpes simplex virus 2-kb latency associated transcript (LAT) leader sequence allows efficient expression of downstream proteins which is enhanced in neuronal cells: possible function of LAT ORFs. J. Gen. Virol. 79:3019–3026. [DOI] [PubMed] [Google Scholar]
- 6.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson, A. T., F. Sederati, G. Devi-Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63:3844–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drolet, B. S., G. C. Perng, J. Cohen, S. M. Slanina, A. Yukht, A. B. Nesburn, and S. L. Wechsler. 1998. The region of the herpes simplex virus type 1 LAT gene involved in spontaneous reactivation does not encode a functional protein. Virology 242:221–232. [DOI] [PubMed] [Google Scholar]
- 9.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goins, W. F., L. R. Sternberg, K. D. Croen, P. R. Krause, R. L. Hendricks, D. J. Fink, S. E. Straus, M. Levine, and J. C. Glorioso. 1994. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J. Virol. 68:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117–125. [DOI] [PubMed] [Google Scholar]
- 13.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507–8515. [DOI] [PMC free article] [PubMed]
- 15.Inman, M., G. Perng, G. Henderson, H. Ghiasi, A. Nesburn, S. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133–8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, C., T. J. Newby, T. Holt, A. Doster, M. Stone, J. Ciacci-Zanella, C. J. Webster, and M. W. Jackwood. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185–3195. [DOI] [PubMed] [Google Scholar]
- 18.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leib, D. A., C. L. Bogard, M. Kosz-Vnenchak, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perng, G., C. Jones, H. Ciacci-Zanella, G. Henderson, A. Yukht, S. Slanina, F. Hofman, H. Ghiasi, A. Nesburn, and S. Wechsler. 2000. Virus induced neuronal apoptosis blocked by the herpes simplex virus latency associated transcript (LAT). Science 287:1500–1503. [DOI] [PubMed] [Google Scholar]
- 21.Perng, G. C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perng, G. C., D. Esmaili, S. M. Slanina, A. Yukht, H. Ghiasi, N. Osorio, K. R. Mott, B. Maguen, L. Jin, A. B. Nesburn, and S. L. Wechsler. 2001. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J. Virol. 75:9018–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. High-dose ocular infection with a herpes simplex virus type 1 ICP34.5 deletion mutant produces no corneal disease or neurovirulence yet results in wild-type levels of spontaneous reactivation. J. Virol. 70:2883–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perng, G. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, and S. L. Wechsler. 1996. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J. Virol. 70:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perng, G. C., S. M. Slanina, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1996. A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. J. Virol. 70:2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perng, G. C., S. M. Slanina, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2001. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. J. Gen. Virol. 82:1117–1122. [DOI] [PubMed] [Google Scholar]
- 27.Perng, G. C., S. M. Slanina, A. Yuhkt, B. S. Drolet, W. J. Keleher, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1999. A herpes simplex virus type 1 latency associated transcript (LAT) mutant with increased virulence and reduced spontaneous reactivation. J. Virol. 73:920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perng, G. C., S. M. Slanina, A. Yuhkt, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1999. Herpes simplex virus type 1 serum neutralizing antibody titers increase during latency in rabbits latently infected with LAT positive but not LAT negative viruses. J. Virol. 73:9669–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perng, G. C., S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J. Virol. 74:1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rock, D. L., A. B. Nesburn, H. Ghiasi, J. Ong, T. L. Lewis, J. R. Lokensgard, and S. L. Wechsler. 1987. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J. Virol. 61:3820–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawtell, N. M., and R. L. Thompson. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J. Virol. 66:2157–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70:3807–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spivack, J. G., and N. W. Fraser. 1988. Expression of herpes simplex virus type 1 (HSV-1) latency-associated transcripts and transcripts affected by the deletion in avirulent mutant HFEM: evidence for a new class of HSV-1 genes. J. Virol. 62:3281–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiner, I., J. G. Spivack, R. P. Lirette, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, and N. W. Fraser. 1989. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 8:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, J. G. 1990. Transcripts associated with herpes simplex virus latency. Adv. Exp. Med. Biol. 278:199–204. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, J. G., E. K. Wagner, G. B. Devi-Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, S. K., G. Gough, D. S. Latchman, and R. S. Coffin. 1999. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. J. Virol. 73:6618–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, R. L., and N. M. Sawtell. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J. Virol. 71:5432–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, E. K., G. Devi-Rao, L. T. Feldman, A. T. Dobson, Y. F. Zhang, W. M. Flanagan, and J. G. Stevens. 1988. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J. Virol. 62:1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wechsler, S. L., A. B. Nesburn, R. Watson, S. M. Slanina, and H. Ghiasi. 1988. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J. Virol. 62:4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wechsler, S. L., A. B. Nesburn, J. Zwaagstra, and H. Ghiasi. 1989. Sequence of the latency-related gene of herpes simplex virus type 1. Virology 168:168–172. [DOI] [PubMed] [Google Scholar]
- 44.Zwaagstra, J., H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1989. In vitro promoter activity associated with the latency-associated transcript gene of herpes simplex virus type 1. J. Gen. Virol. 70:2163–2169. [DOI] [PubMed] [Google Scholar]
- 45.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]