Abstract
Recent studies demonstrate that virus-cellular receptor interactions are not the sole determinants of adenovirus (Ad) tropism. It has been shown that the fiber shaft length, which ranges from 6 to 23 β-repeats in human Ads, also influences viral tropism. However, there is no report that investigates whether artificial extension of the shaft alters the infectivity profile of Ad. Therefore, we constructed Ad serotype 5 (Ad5) capsid-based longer-shafted Ad vectors by incorporating Ad2 shaft fragments of different lengths into the Ad5 shaft. We show that “longer-shafted” Ad vectors (up to 32 β-repeats) could be rescued. We also show that longer-shafted Ad vectors had no impact on knob-CAR (coxsackievirus and Ad receptor) interaction compared to wild-type Ad. Nevertheless, gene transfer efficiencies of longer-shafted Ad vectors were lower in CAR-positive cell lines compared to wild-type Ad. We suggest that artificial extension of the shaft can inhibit infectivity in the context of CAR-positive cell lines without modification of knob-CAR interaction.
The utility of adenovirus (Ad) for the delivery and expression genes both in vitro and in vivo is based on the widespread host range and the remarkable efficiency of cellular entry processes allowing high-level gene expression, together with their relative ease of preparation and purification (28). To achieve effective infection, two virus-cellular receptor interactions are required. The adsorption of Ad to target cell receptors, which have been identified as coxsackievirus and Ad receptor (CAR), histocompatibility class I molecule, or sialoglycoproteins, is initiated via the knob portion of the fiber (13, 20, 28). Subsequently, the interaction of the penton base with αV integrins facilitates the entry of Ad via clathrin-mediated endocytosis (1, 11, 22, 28). Ad can infect a wide range of cell types because the primary cellular receptors for Ad are expressed on most cells (28). However, specific gene delivery to a target cell is precluded due to the ubiquitous expression of CAR. On the other hand, Ad gene transfer is poor to some cell types due to a low level of Ad receptors on these cells (23, 28). It is thus apparent that receptor recognition is one of the key factors involved in viral tropism. To develop effective and specific gene delivery to target cells (resulting in increased safety in gene therapy applications), a number of recombinant Ad vectors have been constructed. Major attention has been given to modification of virus-cellular receptor interactions, including bispecific conjugates (6, 10, 28, 36), genetic modification of the fiber knob (5, 17, 28, 35, 37), modification of the hexon (3) or penton (34), ablation of the binding site on the knob (25, 28), and construction of chimeric vectors (9, 18, 28). However, it has become clear that virus-cellular receptor interactions (knob-CAR and penton base-αV integrins) are not the sole determinants of viral tropism.
In this regard, Roelvink et al. reported that Ad type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but that the shorter fiber of Ad9 permitted fiber-independent binding of Ad9 penton base to αV integrins (24). Moreover, they showed that Ads belonging to subgroups A, C, D, E, and F all bind to the same cellular fiber receptor CAR and suggested that differences in subgroup tropism is significantly influenced by the length of the fiber shaft (26). Additional data regarding the importance of shaft length were obtained by Ad5 capsid-based vectors with chimeric fibers (15, 30). These observations led to a more systematic analysis of the role of fiber shaft length in Ad infectivity. Shayakhmetov et al. generated viruses with chimeric fibers containing short shafts (Ad9 or Ad35) or long shafts (Ad5) in combination with CAR (Ad5 and Ad9)- or non-CAR (Ad35)-recognizing knob domains (29). For Ad5 or Ad9 knob-possessing vectors, long shafts were critical for efficient infection compared with the weak attachment of the engineered short-shafted vectors. In contrast, for the Ad35 knob-possessing vectors, which infected cells by a CAR-independent pathway, fiber shaft length had no significant influence on infection. This study clearly demonstrated that the length of the fiber shaft influenced CAR- and αV integrin-mediated infections.
Taken together, these reports have established shaft length as a key parameter whose modulation might allow tropism alternations. In this regard, the long-shafted Ad uses its fiber interaction (higher affinity than penton base-αV integrin interaction) exclusively for attachment to cell surface proteins and for charge-dependent repulsion (24, 26, 29), followed by internalization by the αV integrins. In contrast, for short-shafted Ad, including artificial fiberless Ad (19, 33), direct binding to the αV integrins (lower affinity than knob-CAR interaction) becomes dominant. In this instance, infectivity is decreased. Indeed, when the short-shafted fiber of Ad9 was engineered into the Ad5 capsid, a significant decrease in infectivity was observed compared to wild-type Ad5 (29). However, there is no significant difference in infection between wild-type Ad2 and wild-type Ad9, which expresses its short-shafted fiber but also the natural Ad9 capsid proteins, including the penton base (24). It is expected that Ad9 capsid does not require long-shafted fibers for efficient infection. On this basis, we hypothesized that the shaft length provides optimal spatial proportion between knob-CAR interaction and penton base-αV integrin interaction, especially when Ad uses CAR-integrin pathway for infection. Therefore, the length of shaft is limited from 6 β-repeats to 23 β-repeats in the process of evolution. Paradoxically, these observations provided us with the basis to explore whether artificial extension of the shaft alters the infectivity profile of Ad. We focused on rescuing recombinant Ad vectors, which express artificial longer shaft over 23 β-repeats.
The primary sequence of the fiber shaft consists of 15-residue pseudorepeats. The length of the shaft is determined by the number of β-repeats, which ranges from 6 (Ad3, Ad11, and Ad35) β-repeats to 23 (Ad12) β-repeats in human Ads (2). Green et al. predicted that these repeats contained two β-strands and two turns (the cross-β model) (12). Stouten et al. subsequently proposed a triple β-helical model, taking into account length measurements from electron microscopy and fiber diffraction patterns (31). Recently, van Raaij et al. revealed a novel structural motif for the shaft as a triple β-spiral model based on the crystallographic data (32). According to this model, we attempted to make Ad5 capsid-based longer-shafted Ad vectors by incorporating Ad2 shaft fragments of different lengths into Ad5 shaft and investigated whether extension of the shaft over 23 β-repeats altered the infectivity profiles of human Ads.
MATERIALS AND METHODS
Cells and cell culture.
Human embryonal kidney (293) cells transformed with Ad5-E1 DNA were purchased from Microbix (Toronto, Ontario, Canada) and cultured in Dulbecco modified Eagle medium-Ham’s F-12 medium (DMEM-F12) containing 10% fetal calf serum (FCS) and 2 mM glutamine. Human cervix carcinoma (HeLa) cells, human umbilical vein endothelial cells (HUVEC), and human embryonal rhabdomyosarcoma (RD) cells were obtained from the American Type Culture Collection (Rockville, Md.). HeLa cells, HUVEC, and RD cells were cultured in DMEM-F12 with 10% FCS and 2 mM glutamine, endothelial cell basal medium-2 with the endothelial supplement EGM-2 MV (Clontics, San Diego, Calif.), and DMEM with 10% FCS and 4.5 g of glucose/liter, respectively, at 37°C in a 5% CO2 atmosphere.
Design and construction of longer-shafted Ad vectors.
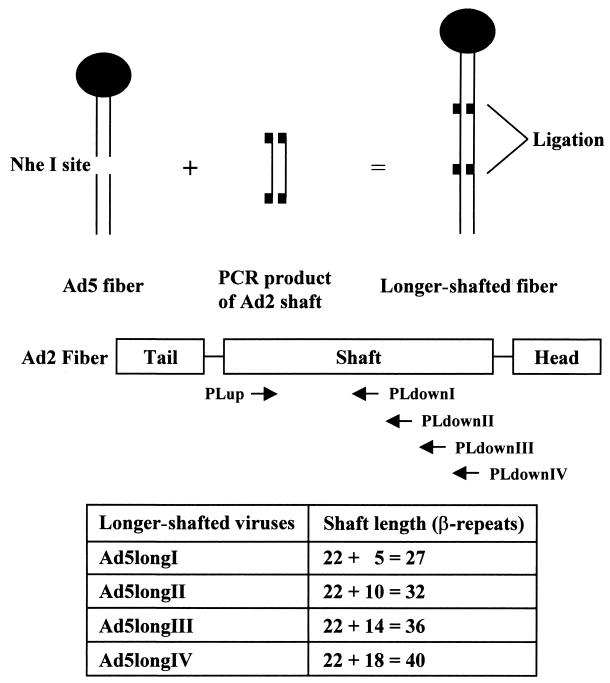
The construction of longer-shafted Ad vectors used in this study is shown in Fig. 1. According to a triple β-spiral model for Ad2 fiber shaft (32), we made Ad5 capsid-based longer-shafted Ad vectors by incorporating Ad2 shaft fragments of different lengths into the NheI site located in the eighth β-repeat of Ad5 shaft. In order to generate Ad2 shaft fragments (from the 4th to the 9th, the 14th, the 18th, and the 22nd β-repeat), PCR was performed. Template DNA was purified from Ad2 viral particles obtained from the American Type Culture Collection. The primer sequences used to amplify fragments of Ad2 fiber shaft were as follows: PLup (nucleotides [nt] 31312 to 31341; 5′-TCAAACCTAGGTTTGGACACCTCCGCACCA-3′), PLdownI (nt 31554 to 31525; 5′-TACACCTAGGGTGTCGCTGTCACTGCCAGA-3′), PLdownII (nt 31788 to 31759; 5′-AATTCCTAGGTTGTTTGATGAATCATAACC-3′), PLdownIII (nt 31986 to 31957; 5′-AACTCCTAGGTTTTTAGTATTGTTTGATGC-3′), and PLdownIV (nt 32202 to 32173; 5′-TATTCCTAGGGCCCCTGAGTTGTCAAAGCT-3′). Nucleotide numbers are according to the sequences obtained from GenBank (accession no. J01917). Each 5′ end of the primer was modified to create a restriction enzyme site for AvrII. These fragments were cloned into the NheI-digested pNEB.PK.FSP (Ad5 fiber shuttle vector [18]). The resultant plasmids were digested with PacI and KpnI and were used for homologous DNA recombination with SwaI-linearized pVK50 (17)-based backbone plasmids. The pVK50-based backbone plasmids contain cytomegalovirus promoter-driven green fluorescent protein (GFP) gene with or without cytomegalovirus promoter-driven firefly luciferase gene-expressing cassette in the E1 region of Ad5 genome. After homologous DNA recombination, these plasmids were linearized with PacI and transfected into 293 cells to generate Ad vectors. Ad vectors, which contain GFP, were designated by the suffix “G,” and Ad vectors, which contain GFP and firefly luciferase, were designated by the suffix “GL.” All Ad vectors were propagated on 293 cells and purified by double centrifugation in CsCl gradients by a standard protocol. Determination of virus particle (VP) titer was accomplished spectrophotometrically by the method of Maizel et al. by using a conversion factor of 1.1 × 1012 VPs per absorbance unit at 260 nm (21). To determine whether the genomes of longer-shafted Ad vectors indeed show the expected structure, PCR and DNA sequencing were performed. The primer sequences used to sequence the complete shaft region or to amplify the fiber gene were as follows: PSup (nt 31114 to 31143; 5′-ACTGTGCCTTTTCTTACTCCTCCCTTTGTA-3′), PSdown (nt 32277 to 32248; 5′-TCTACAGTTAGGAGATGGAGCTGGTGTGGT-3′), PFup (nt 31042 to 31071; 5′-ATGAAGCGCGCAAGACCGTCTGAAGATACC-3′), and PFdown (nt 32784 to 32755; 5′-TTCTTGGGCAATGTATGAAAAAGTGTAAGA-3′). Nucleotide numbers are according to the sequences obtained from GenBank (accession no. M73260). Ad vectors containing RGD (Arg-Gly-Asp) peptide in the HI loop of the fiber knob domain were made similarly. To generate these Ad vectors, pNEB.PK.FHIRGD was used instead of pNEB.PK.FSP (5).
FIG. 1.
Design and construction of longer-shafted Ad vectors. According to a triple β-spiral model for Ad2 fiber shaft, Ad5 capsid-based longer-shafted Ad vectors were made by incorporating Ad2 shaft fragments of different lengths into the NheI site of the Ad5 shaft. In order to generate Ad2 shaft fragments, PCR was performed with four pairs of primers (Ad5longI, PLup and PLdownI; Ad5longII, PLup and PLdownII; Ad5longIII, PLup and PLdownIII; Ad5longIV, PLup and PLdownIV). The resulting longer-shafted Ad vectors have a total of 27, 32, 36, and 40 β-repeats, respectively.
Western blot analysis.
To confirm that longer-shafted Ad vectors have different lengths of fiber proteins, Western blot analysis was performed. Purified Ad virions (Ad5, Ad5longIG, Ad5longIIG, Ad5longIIIG, or Ad5longIVG) were diluted in Laemmli Sample Buffer (Bio-Rad, Hercules, Calif.). Each sample was boiled and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoretic transfer to polyvinylidene difluoride membrane (Bio-Rad), the blot was blocked for 2 h at room temperature with blocking buffer (phosphate-buffered saline [PBS; pH 7.4] with 0.05% Tween 20 and 2% bovine serum albumin [BSA]). Mouse monoclonal antibody (MAb) 4D2 was diluted in binding buffer (PBS [pH 7.4] with 0.05% Tween 20 and 1% BSA) to 1:2,000 and incubated for 1 h at room temperature. The blot was washed four times with washing buffer (PBS [pH 7.4] with 0.05% Tween 20) and then incubated with a 1:2,000 dilution of goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Sigma, St. Louis, Mo.) for 1 h at room temperature. The blot was washed four times with washing buffer and incubated with alkaline phosphatase development buffer (Bio-Rad) as recommended by the manufacturer. A 0.5-μg standard of recombinant Ad5 fiber protein (5) was included for comparison to the proteins in the sample. Mouse MAb 4D2 directed against the tail domain of Ad5 fiber protein was generated at the University of Alabama at Birmingham Hybridoma Core Facility (4, 16).
ELISA.
Solid-phase binding enzyme-linked immunosorbent assay (ELISA) was performed by a method previously described (4, 5). Purified Ad virions (Ad5GL, Ad5longIGL, or Ad5longIIGL) were diluted in 50 mM carbonate buffer (pH 9.6) to a concentration of 1010 VPs/ml, and 100-μl aliquots were added to wells of a 96-well Nunc-Maxisorp ELISA plate. Plates were incubated overnight at 4°C and then blocked for 2 h at room temperature by the addition of 200 μl of blocking buffer (PBS [pH 7.4] with 0.05% Tween 20 and 2% BSA) to each well. Wells were then washed three times with washing buffer (PBS [pH 7.4] with 0.05% Tween 20). Purified recombinant soluble form of truncated CAR-His6-tagged protein (sCAR-His6 [4]) was diluted in binding buffer (PBS [pH 7.4] with 0.05% Tween 20 and 0.5% BSA) to concentrations ranging from 12 to 3,000 ng/ml. The 100-μl dilutions were added to the wells. After incubation at room temperature for 1 h, the wells were washed four times with washing buffer and then bound sCAR-His6 protein was detected by incubation for 1 h at room temperature with a 1:2,000 dilution of anti-sCAR-His6 MAb 2A3. The wells were washed again four times with washing buffer and then incubated with 1:1,000 dilution of goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Sigma). After incubation for 1 h, the wells were washed four times with washing buffer, and then the plate was developed with p-nitrophenyl phosphate (Sigma) as recommended by the manufacturer. Plates were read in a microtiter plate reader set at 405 nm. Recombinant sCAR-His6 protein was expressed in High Fives cells (Invitrogen, Carlsbad, Calif.) infected with recombinant baculovirus by a previously described method (4). Mouse MAb 2A3 to baculovirus-produced human sCAR-His6 protein was generated at the University of Alabama at Birmingham Hybridoma Core Facility (4).
Ad-mediated gene transfer assay.
Cells (HeLa, HUVEC, or RD) were seeded in 96-well plates in triplicate at 104/well the day prior to infection. After being washed with PBS, cells were infected with Ad5GL, Ad5longIGL, Ad5longIIGL, Ad5RGDGL, or Ad5RGDlongIIGL at 101, 102, and 103 VPs/cell in 2% FCS medium at 37°C for 3 h. The unbound viruses were washed away with PBS, and 10% FCS medium was added. After incubation at 37°C for 48 h, the infected cells were washed again with PBS and lysed by using a passive lysis buffer (Promega, Madison, Wis.). Luciferase activity in the cell lysates were determined by using a luciferase assay system kit (Promega) and a luminometer (Berthold, Gaithersburg, Md.) and normalized by protein concentration in the cell lysates by using a DC protein assay kit (Bio-Rad) with BSA as the standard. The blocking experiment by recombinant Ad5 knob protein (5) was performed similarly. All cells were preincubated for 10 min at room temperature in 2% FCS medium with or without recombinant Ad5 fiber knob protein at 100 μg/ml and then exposed for 30 min at room temperature to Ad5GL, Ad5longIGL, or Ad5longIIGL in 2% FCS medium at 103 or 104 VPs/cell. Cells were washed with PBS and unbound viruses were aspirated, and 10% FCS medium was added. After incubation at 37°C for 30 h, luciferase activity was measured.
RESULTS
Generation of longer-shafted Ad vectors.
Figure 1 shows the schematic design and construction of longer-shafted Ad vectors developed in this study. Theoretically, the longer-shafted Ad vectors—Ad5longI, Ad5longII, Ad5longIII, and Ad5longIV—express fibers with shaft lengths of 27, 32, 36, and 40 β-repeats, respectively.
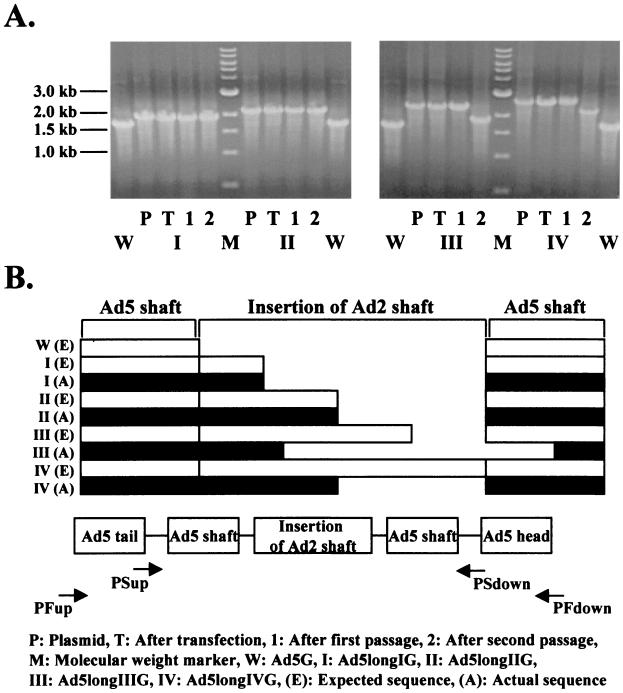
To confirm that the genomes of longer-shafted Ad vectors show the expected structure, we analyzed DNA isolated from viral particles. As shown in Fig. 2A, the PCR bands of the fiber region of both Ad5longIG and Ad5longIIG resolved the expected length at all individual steps of viral propagation (transfection, first passage, and second passage). In contrast, the PCR bands for both Ad5longIIIG and Ad5longIVG were shifted to smaller lengths after the second passage. Therefore, DNA isolated from CsCl-purified viral particles (second passage) was sequenced. The results of sequencing are shown as schematic DNA alignment in Fig. 2B. Interestingly, the causes of these shifts in size of PCR products were proven to be due to DNA deletions. Especially, in case of Ad5longIIIG, this deletion was based on homologous DNA recombination.
FIG. 2.
Generation of longer-shafted Ad vectors. To confirm that the genomes of longer-shafted Ad vectors show the expected structure, DNA isolated from viral particles was analyzed. (A) PCR was performed to detect the length of total fiber region by using a pair of primers (PFup and PFdown). (B) DNA sequencing of complete shaft region was performed by using a pair of primers (PSup and PSdown). The results of sequencing are shown as schematic DNA alignment. Ad5G was used as a control.
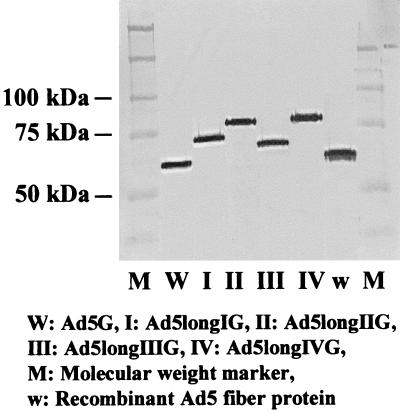
To verify that longer-shafted Ad vectors have the predicted different lengths of fiber proteins, viral particles were analyzed by Western blot analysis. Compared with control Ad5G and recombinant Ad5 fiber protein (5), the bands of the fibers of both Ad5longIG and Ad5longIIG appeared at upper positions relative to wild-type fibers. As expected from DNA sequencing, the size of the fiber of Ad5longIIIG was smaller than the fiber of Ad5longIG, while the fiber of Ad5longIVG appeared almost at the same position as the fiber of Ad5longIIG (Fig. 3). These analyses demonstrate that longer-shafted Ad vectors (up to 32 β-repeats) can be rescued and purified.
FIG. 3.
Generation of longer-shafted Ad vectors. To verify that longer-shafted Ad vectors have the predicted different lengths of fiber proteins, Western blot analysis was performed. Purified Ad virions were boiled and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then detected with anti-fiber MAb 4D2. Ad5G or recombinant Ad5 fiber protein was used as a control.
Knob-CAR interaction of longer-shafted Ad vectors.
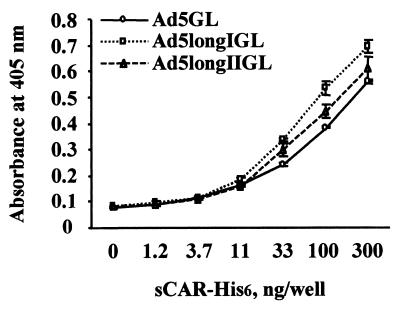
Interaction between Ad vectors and the soluble form of sCAR-His6 was examined by ELISA. It had already been established that sCAR-His6 protein efficiently binds to recombinant Ad5 fiber protein (4). Figure 4 shows that there was no significant difference in binding to sCAR-His6 between Ad5longIGL, Ad5longIIGL and control Ad5GL. This result suggests that shaft extension has no impact on knob-CAR interaction.
FIG. 4.
Knob-CAR interaction of longer-shafted Ad vectors. Purified Ad virions of Ad5GL (control), Ad5longIGL, and Ad5longIIGL immobilized in the wells of an ELISA plate were incubated with various concentrations of sCAR-His6 protein. Binding sCAR-His6 protein was then detected with anti-sCAR-His6 MAb 2A3.
Infectivity profiles of longer-shafted Ad vectors in cell lines expressing variable levels of CAR.
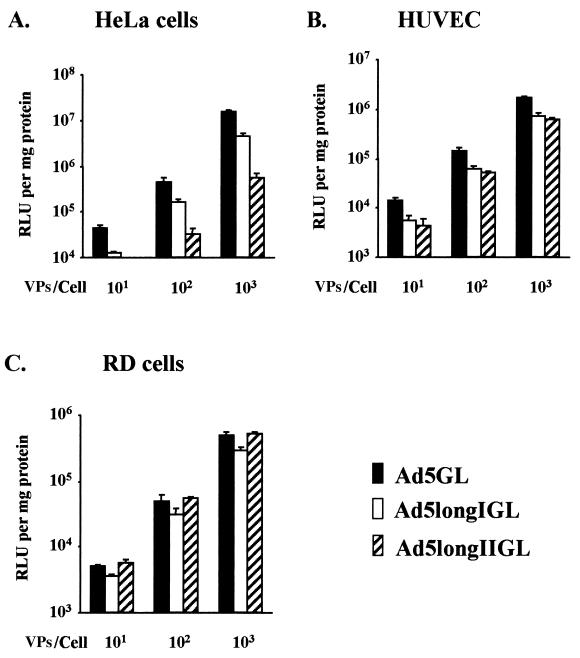
In order to examine the infectivity profiles of longer-shafted Ad vectors, gene transfer efficiencies of Ad5longIGL and Ad5longIIGL were tested. In this regard, we selected cell lines expressing high levels of αV integrins, but variable levels of CAR: HeLa cells (high CAR), HUVEC (moderate CAR), and RD cells (low CAR). The expression of CAR and αV integrins on these cell lines has well been established (5, 8, 30). Luciferase activity in the cell lysates was determined after infection as described in Materials and Methods. As shown in Fig. 5, longer-shafted Ad vectors displayed similar infectivity in HUVEC and RD cells, compared to control Ad5GL. Interestingly, they showed significantly lower infectivity in HeLa cells. These results suggest that the infectivity is independent of shaft extension in CAR-negative cell lines. However, infectivity is significantly inhibited by shaft extension in CAR-positive cell lines.
FIG. 5.
Infectivity profiles of longer-shafted Ad vectors in cell lines expressing variable levels of CAR. Gene transfer efficiencies of Ad5GL (control), Ad5longIGL, and Ad5longIIGL were tested in cell lines expressing high levels of αV integrins but variable levels of CAR: HeLa cells (high CAR) (A), HUVEC (moderate CAR) (B), and RD cells (low CAR) (C). All cells were infected at 101, 102, and 103 VPs/cell. At 48 h postinfection, the cell lysates were assayed for luciferase activity, expressed as relative light units (RLU) per milligram of cellular protein. The results are the means ± the standard deviations (SD) of triplicate experiments.
Infectivity profiles of longer-shafted Ad vectors after blocking knob-CAR interaction.
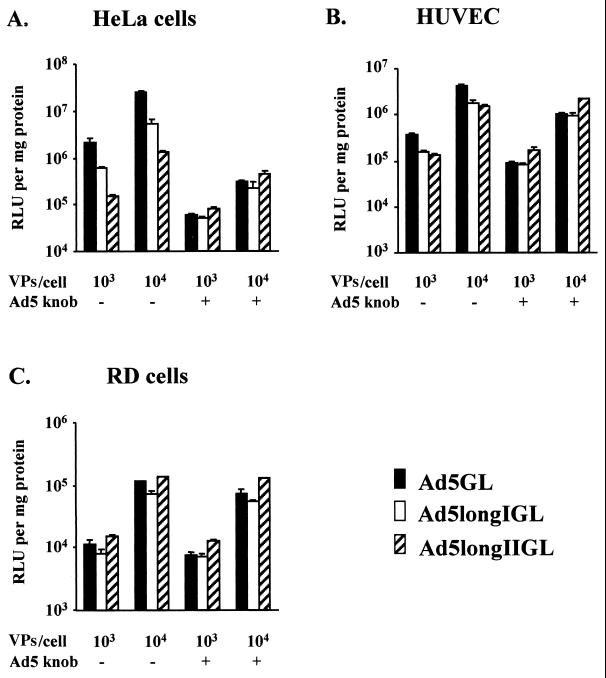
To monitor infectivity profiles of longer-shafted Ad vectors after blocking knob-CAR interaction, we examined gene transfer efficiencies in cells by using recombinant Ad5 knob protein (5). As shown in Fig. 6, we did not detect significant differences in infectivity between longer-shafted Ad vectors and the control Ad5GL after blocking knob-CAR interaction with Ad5 knob protein. This pattern is consistent with the results obtained for RD cells, which express low levels of CAR and therefore represent the same conditions as that of HeLa cells after blocking fiber-CAR interaction by Ad5 knob protein. These results suggest that shaft extension does not affect infectivity in the CAR-independent cell entry pathway.
FIG. 6.
Infectivity profiles of longer-shafted Ad vectors after blocking of knob-CAR interaction. HeLa cells (A), HUVEC (B), and RD cells (C) were preincubated with or without recombinant Ad5 fiber knob protein at 100 μg/ml for 10 min and then infected with Ad5GL (control), Ad5longIGL, and Ad5longIIGL at 103 or 104 VPs/cell. At 30 h postinfection, the cell lysates were assayed for luciferase activity, expressed as relative light units (RLU) per milligram of cellular protein. The results are the means ± the standard deviations of triplicate experiments.
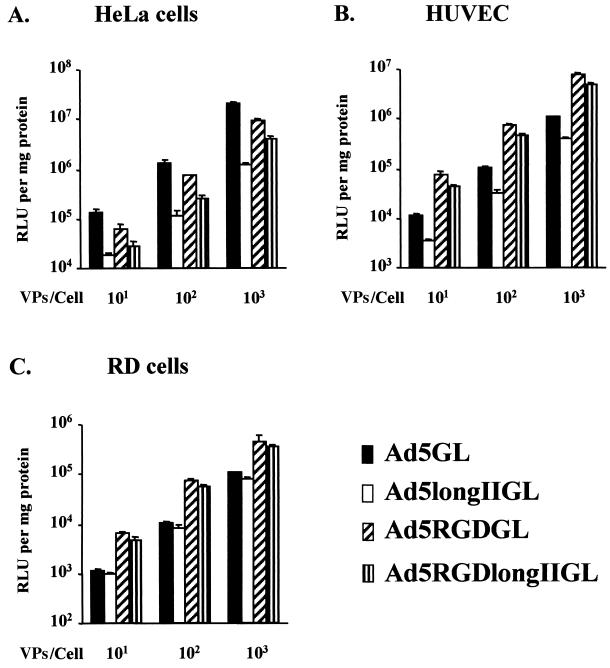
Infectivity profiles of longer-shafted RGD Ad vector.
On the basis of these findings, we hypothesized that the decreased gene transfer associated with longer-shafted Ad vectors could be circumvented by routing the virus via non-CAR pathways. In this regard, we have shown that incorporation of the integrin-binding peptide RGD4C at the fiber knob HI loop confers such CAR-independent gene delivery capacity to Ad vectors (5). We thus derived a variant of our longer-shafted Ad vectors (i.e., Ad5RGDlongIIGL), which also included this genetic modification. In these studies, it can be appreciated that the incorporated peptide allows augmented gene transfer in the context of the CAR-deficient cell lines HUVEC and RD cells (Fig. 7B and C). Of note, whereas the longer shaft appears to impair gene delivery in high-CAR HeLa cells, the HI loop modification restores infectivity to levels comparable to the corresponding normal shaft variant (Ad5RGDGL, Fig. 7A). Such a differential between the wild type and the longer-shafted variant of the HI loop-modified Ad was not noted in low-CAR contexts (Fig. 7B and C). It thus appears that the longer shaft impairment of gene transfer to high-CAR cellular targets can be circumvented when the virus possesses the capacity to accomplish CAR-independent delivery.
FIG. 7.
Infectivity profiles of longer-shafted RGD Ad vector. Gene transfer efficiencies of Ad5GL, Ad5longIIGL, Ad5RGDGL, and Ad5RGDlongIIGL were tested in cell lines expressing high levels of αV integrins but variable levels of CAR: HeLa cells (A), HUVEC (B), and RD cells (C). All cells were infected at 101, 102, and 103 VPs/cell. At 48 h postinfection, the cell lysates were assayed for luciferase activity expressed as relative light units (RLU) per milligram of cellular protein. The results are the means ± the standard deviations of triplicate experiments.
DISCUSSION
To retain the three-dimensional structure of the shaft of newly designed longer-shafted Ad vectors, we used a triple β-spiral model (32) to identify start and end positions of β-repeats in the shaft of Ad2 and in the shaft of Ad5. In addition, we avoided not only the head-shaft boundary regions but also well-defined hinge positions in the shaft, because similar putative hinge regions have been described in the very long bovine Ad fiber shaft (27) and in certain avian Ad fibers (14). Therefore, we expect our modifications to maintain high stability. We selected Ad2 shaft fragments to be incorporated into the Ad5 shaft because of the homology between these two Ads in amino acid sequences. The nucleic acid sequences, however, are distinct, thus avoiding homologous DNA recombination between insertion and native shaft. We constructed Ad genomes encoding four variations in shaft length (5, 10, 14, and 18 β-repeats longer than the wild-type shaft length). This range of β-repeats seemed to be critical for Ad tropism based on differences in infectivity of human Ad subgroups, which range from 6 to 23 β-repeats (24, 26).
In the process of propagation of longer-shafted Ad vectors, we observed a slower growth of viruses relative to the extension of the shaft length. Although same amount of viral DNA was successfully transfected into 293 cells, longer-shafted Ad vectors grew more slowly and with smaller plaques, with increasing extension of the shaft length. In all individual steps of viral propagation, differences in viral growth were observed. Interestingly, these differences in growth were less obvious at the second passage of Ad5longIIIG and Ad5longIVG (data not shown). This decrease in relative growth differences might be explained by a loss of DNA insertions. Indeed, the PCR products of the fiber region of both Ad5longIIIG and Ad5longIVG were shifted to a smaller length after the second passage. Sequencing analysis proved that the causes of these shifts in size of PCR products were due to DNA deletions. We postulate that the limitation of artificial shaft extension is supported by the fact that the length of the shaft has been limited from 6 β-repeats to 23 β-repeats in the process of evolution (2).
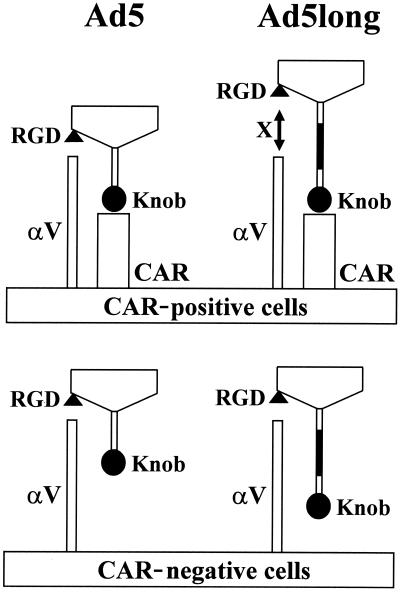
We showed that shaft extension had no impact on knob-CAR interaction (Fig. 4). Nevertheless, longer-shafted Ad vectors showed significantly lower infectivity in CAR-positive cell lines (Fig. 5). The reduced infectivity of longer-shafted Ad vector could be a consequence of the inhibition of penton base-αV integrin interaction after knob-CAR interaction or, alternatively, could be due to deficient biological steps later in the viral entry process. In this regard, we showed that shaft extension does not affect infectivity in CAR-independent cell entry pathway (Fig. 5C and 6). Therefore, consequences of post-cell entry are not strongly expected. We propose a model that extension of the shaft length disturbs penton base-αV integrin interaction in a context whereby longer-shafted Ad vectors must exclusively use CAR for attachment (Fig. 8). This implies that the high-affinity interaction between knob and CAR paradoxically prevents effective infection when the shaft length is longer than wild-type Ad. This model is consistent with the result that addition of Ad5 knob protein did not efficiently inhibit infectivity of longer-shafted Ad vectors in CAR-positive HeLa cells (Fig. 6A). This model is also consistent with the observation that longer-shafted Ad vectors grow more slowly and with smaller plaques in 293 cells (data not shown), which also express high-CAR and high-αV integrins (5).
FIG. 8.
Model for Ad infectivity based on fiber shaft extension. Artificial extension of Ad fiber shaft disturbs penton base-αV integrin interaction in a context whereby longer-shafted Ad vectors must exclusively use CAR for attachment. On the other hand, shaft extension does not affect infectivity in CAR-independent cell entry pathway.
Although Ad5 capsid-based short-shafted Ad vectors (29) and fiberless Ad (19) also showed lower infectivity compared to wild-type Ad, lower infectivity appeared in all cell types expressing various levels of CAR. In contrast, longer-shafted Ad vectors keep similar infectivity in cells expressing low amounts of CAR. We think that extension of the shaft length will lead to new strategies of changing Ad tropism without modification of knob-CAR interaction. Moreover, Ad has a particular tropism for the liver and induces hepatic toxicity in gene therapy applications. The low infectivity of longer-shafted Ad vectors observed in HeLa cells could provide a strategy of liver untargeting, since both liver and HeLa cells express high levels of CAR (7, 8, 30). In this regard, extension of the shaft length might offer a new approach to improve gene therapy applications in the near future.
Acknowledgments
We thank Victor Krasnykh for providing plasmids pNEB.PK.FSP and pVK50 and David Dion and Alexander Landar for expert assistance in obtaining reagents. We also thank Joel N. Glasgow for reviewing the manuscript and the CFAR DNA Sequencing Core of the University of Alabama at Birmingham for sequencing.
This study was supported by National Institutes of Health grants R01 CA86881, R01 CA74242, and R01 CA83821 and contract no. 1 CO 97110.
REFERENCES
- 1.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chroboczek, J., R. W. H. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199:163–200. [DOI] [PubMed] [Google Scholar]
- 3.Crompton, J., C. I. Toogood, N. Wallis, and R. T. Hay. 1994. Expression of a foreign epitope on the surface of the adenovirus hexon. J. Gen. Virol. 75:133–139. [DOI] [PubMed] [Google Scholar]
- 4.Dmitriev, I., E. Kashentseva, B. E. Rogers, V. Krasnykh, and D. T. Curiel. 2000. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J. Virol. 74:6875–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706–9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas, J. T., B. E. Rogers, M. E. Rosenfeld, S. I. Michael, M. Feng, and D. T. Curiel. 1996. Targeted gene delivery by tropism-modified adenoviral vectors. Nat. Biotechnol. 14:1574–1578. [DOI] [PubMed] [Google Scholar]
- 7.Fechner, H., A. Haack, H. Wang, X. Wang, K. Eizema, M. Pauschinger, R. G. Schoemaker, R. van Veghel, A. B. Houtsmuller, H.-P. Schultheiss, J. M. J. Lamers, and W. Poller. 1999. Expression of coxsackie adenovirus receptor and alphav-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 6:1520–1535. [DOI] [PubMed] [Google Scholar]
- 8.Fechner, H., X. Wang, H. Wang, A. Jansen, M. Pauschinger, H. Scherübl, J. M. Bergelson, H.-P. Schultheiss, and W. Poller. 2000. Trans-complementation of vector replication versus Coxsackie-adenovirus-receptor overexpression to improve transgene expression in poorly permissive cancer cells. Gene Ther. 7:1954–1968. [DOI] [PubMed] [Google Scholar]
- 9.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman, C. K., B. E. Rogers, J. T. Douglas, B. A. W. Ying, G. P. Siegal, A. Baird, J. A. Campain, and D. T. Curiel. 1997. Targeted gene delivery to Kaposi’s sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 57:1447–1451. [PubMed] [Google Scholar]
- 11.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477–486. [DOI] [PubMed] [Google Scholar]
- 12.Green, N. M., N. G. Wrigley, W. C. Russell, S. R. Martin, and A. D. McLachlan. 1983. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 2:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry, L. J., D. Xia, M. E. Wilke, J. Deisenhofer, and R. D. Gerard. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J. Virol. 68:5239–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess, M., A. Cuzange, R. W. Ruigrok, J. Chroboczek, and B. Jacrot. 1995. The avian adenovirus penton: two fibres and one base. J. Mol. Biol. 252:379–385. [DOI] [PubMed] [Google Scholar]
- 15.Hidaka, C., E. Milano, P. L. Leopold, J. M. Bergelson, N. R. Hackett, R. W. Finberg, T. J. Wickham, I. Kovesdi, P. Roelvink, and R. G. Crystal. 1999. CAR-dependent and CAR-independent pathways of adenovirus vector-mediated gene transfer and expression in human fibroblasts. J. Clin. Investig. 103:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karayan, L., B. Gay, J. Gerfaux, and P. A. Boulanger. 1994. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology 202:782–795. [DOI] [PubMed] [Google Scholar]
- 17.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 72:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasnykh, V., G. V. Mikheeva, J. T. Douglas, and D. T. Curiel. 1996. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J. Virol. 70:6839–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legrand, V., D. Spehner, Y. Schlesinger, N. Settelen, A. Pavirani, and M. Mehtali. 1999. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J. Virol. 73:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis, N., P. Fender, A. Barge, P. Kitts, and J. Chroboczek. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J. Virol. 68:4104–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maizel, J. V. J., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115–125. [DOI] [PubMed] [Google Scholar]
- 22.Mathias, P., T. Wickham, M. Moore, and G. Nemerow. 1994. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 68:6811–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 72:6014–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roelvink, P. W., I. Kovesdi, and T. J. Wickham. 1996. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J. Virol. 70:7614–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roelvink, P. W., G. M. Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568–1571. [DOI] [PubMed] [Google Scholar]
- 26.Roelvink, P. W., A. Lizonova, J. G. M. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruigrok, R. W. H., A. Barge, S. K. Mittal, and B. Jacrot. 1994. The fibre of bovine adenovirus type 3 is very long but bent. J. Gen. Virol. 75:2069–2073. [DOI] [PubMed] [Google Scholar]
- 28.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573–2604. [DOI] [PubMed] [Google Scholar]
- 29.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274–10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stouten, P. F. W., C. Sander, R. W. H. Ruigrok, and S. Cusack. 1992. New triple-helical model for the shaft of the adenovirus fibre. J. Mol. Biol. 226:1073–1084. [DOI] [PubMed] [Google Scholar]
- 32.van Raaij, M. J., A. Mitraki, G. Lavigne, and S. Cusack. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935–938. [DOI] [PubMed] [Google Scholar]
- 33.Von Seggern, D. J., C. Y. Chiu, S. K. Fleck, P. L. Stewart, and G. R. Nemerow. 1999. A helper-independent adenovirus vector with E1, E3, and fiber deleted: structure and infectivity of fiberless particles. J. Virol. 73:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham, T. J., M. E. Carrion, and I. Kovesdi. 1995. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 2:750–756. [PubMed] [Google Scholar]
- 35.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570–1573. [DOI] [PubMed] [Google Scholar]
- 36.Wickham, T. J., D. M. Segal, P. W. Roelvink, M. E. Carrion, A. Lizonova, G. M. Lee, and I. Kovesdi. 1996. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J. Virol. 70:6831–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickham, T. J., E. Tzeng, L. L. Shears, P. W. Roelvink, Y. Li, G. M. Lee, D. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221–8229. [DOI] [PMC free article] [PubMed] [Google Scholar]