Abstract
Self-association of viral proteins is important for many of their functions, including enzymatic, transcriptional, and transformational activities. Epstein-Barr virus (EBV) nuclear antigen leader protein (EBNA-LP) contains various numbers of W1W2 repeats and a unique carboxyl-terminal Y1Y2 domain. It was reported that EBNA-LP associates with a variety of cellular proteins and plays a critical role in EBV-induced transformation. We report here that EBNA-LP self-associates in vivo and the domain responsible for the homotypic association is a multifunctional domain mediating nuclear localization, nuclear matrix association, and EBNA-2-dependent coactivator function of the protein. Our conclusions are based on the following observations. (i) EBNA-LP interacts with itself or its derivatives in the yeast two-hybrid system. (ii) A purified chimeric protein consisting of glutathione S-transferase fused to EBNA-LP specifically formed complexes with EBNA-LP transiently expressed in COS-7 cells. (iii) When Flag epitope-tagged EBNA-LP with either one or two W1W2 repeats and EBNA-LP containing four W1W2 repeats were coexpressed in COS-7 cells, the latter was specifically coimmunoprecipitated with the former. (iv) Mutational analyses of EBNA-LP with deletion mutants revealed that the region between codons 19 and 39 (relative to the first amino acid residue of the W2 domain) is essential for self-association of the protein. The mapped region almost completely overlaps with CR2 and CR3, regions conserved among a subset of primate γ-herpesviruses and critical for EBNA-2-dependent coactivator function. Amino acid substitutions in CR2 alone abolished the ability of the protein to self-interact. This laboratory previously reported that CR2 is also responsible for nuclear localization and nuclear matrix association (A. Yokoyama, Y. Kawaguchi, I. Kitabayashi, M. Ohki, and K. Hirai, Virology 279:401–413, 2001). (v) Sucrose gradient sedimentation showed that amino acid substitutions in CR2 reduced the ability of the protein to form protein complexes in B cells. These results suggest that self-association of EBNA-LP may be important for its various functions and interactions of the protein with multiple cellular proteins.
Epstein-Barr virus (EBV) is a ubiquitous human pathogen closely associated with infectious mononucleosis and a variety of neoplastic diseases, including Burkitt’s lymphoma, nasopharyngeal carcinoma, various other lymphomas, Hodgkin’s disease, and gastric carcinoma (24, 35). In vitro, EBV infection induces a very long-term proliferation of resting human B cells (24, 35). In the resulting lymphoblastoid cell lines, the EBV genome is maintained as an episomal form, and only a limited number of viral proteins (EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, EBNA leader protein [EBNA-LP], LMP1, LMP2A, and LMP2B) are expressed (24, 35). Among these latency-associated EBV proteins, EBNA-1, EBNA-2, EBNA-3A, EBNA-3C, EBNA-LP, and LMP1 are critical for EBV-induced B-cell immortalization, while EBNA-3B, LMP2A, and LMP2B are not (4, 10, 22, 27, 29, 43).
EBNA-LP, the subject of this report, is the first viral gene product to be expressed together with EBNA-2 during EBV-induced B-cell immortalization (1). EBNA-LP consists of a multirepeat domain (W1W2) and a unique carboxyl-terminal domain (Y1Y2) (Fig. 1A) and is therefore detected as a protein ladder in immunoblot analyses, possibly as the result of heterologous polypeptides with different numbers of W1W2 repeats (6). EBNA-LP is considered critical for EBV-induced B-cell immortalization, based on the observation that recombinant EBNA-LP mutants showed severely impaired transforming activity (2, 10, 27). Relevant background information that may account for the mechanisms by which EBNA-LP acts in EBV-induced B-cell immortalization is as follows.
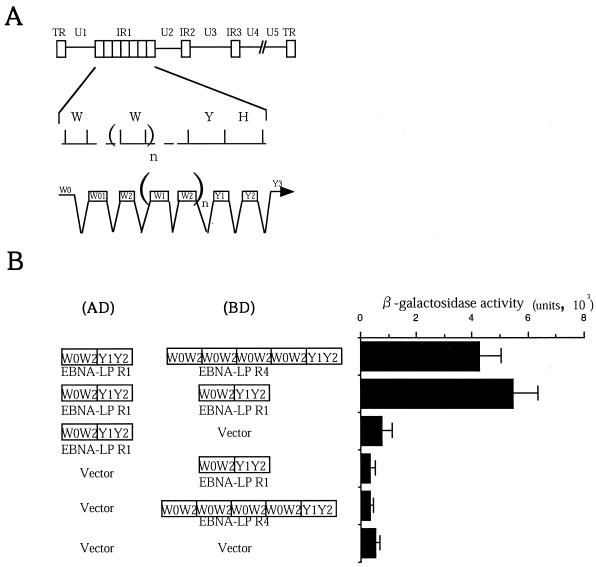
FIG. 1.
Homomeric interaction between molecules of EBNA-LP in the yeast two-hybrid system. (A) Schematic diagram of the sequence of the EBV genome and location of the EBNA-LP gene. The top line is a linear representation of the EBV genome. The unique sequences are designated U1 to U5. The terminal and internal repeats flanking the unique sequences are shown as open rectangles with their designations given above. The middle line shows an expanded section of the domain encoding the EBNA-LP gene. The exons of EBNA-LP open reading frames are derived from BamHI W and Y fragments. The bottom line shows the structures of the EBNA-LP transcript and coding regions. (B) EBNA-LP constructs used in the yeast two-hybrid system (left) and β-galactosidase activity detected for respective constructs in the yeast two-hybrid system (right). β-Galactosidase activity was determined by liquid assay procedures as described in Materials and Methods. The values are the means and standard deviations for three independent experiments.
(i) EBNA-LP is known primarily as a transcriptional coactivator of EBNA-2. It has been reported that EBNA-LP and EBNA-2 cooperatively stimulate expression of cyclin D2 in resting B cells and the progression of these cells from G0 to G1 in the cell cycle (36). It has also been shown that EBNA-LP cooperates with EBNA-2 in up-regulating the expression of the essential viral transforming gene product LMP1 in B cells (12, 31). Recent studies have identified functional domains of EBNA-LP required for transcriptional cooperation with EBNA-2 (33, 51, 52). EBNA-LP has five conserved regions (CR1 to CR5) among related primate γ-herpesviruses (32). This laboratory has demonstrated that the conserved region called CR2 is responsible not only for nuclear localization but also for nuclear matrix association of the protein and that nuclear and/or nuclear matrix localization is critical to EBNA-LP’s ability to induce LMP1 in concert with EBNA-2 in B cells (52). Phosphorylation by cellular kinase(s) is also important for the function of EBNA-LP (34, 51). Mapped major sites of EBNA-LP phosphorylation by cellular kinase(s) have been mapped to serine 35 in the W2 repeat region, and it has been shown that replacement of the major phosphorylation sites in each W2 repeat with alanines abolished the cooperative induction of LMP1 with EBNA-2 in B cells, while replacement of the identified sites with glutamic acids restored the wild-type phenotype (51). The requirement of CR2 and serine 35 for the functions of EBNA-LP was also reported by Peng et al. (33).
(ii) EBNA-LP interacts with numerous cellular proteins. EBNA-LP is localized in both the nucleus and the cytoplasm in EBV-infected cells (19). So far it has been reported that EBNA-LP binds to p53 and pRb in vitro (39) and associates with the 70-kDa family of heat shock proteins (Hsp70s), HS1-associated protein X-1 (HAX-1), the DNA-dependent protein kinase catalytic subunit, HA95, α- and β-tubulin, and Hsp27 at the cellular level (11, 19, 25, 28). In lymphoblastoid cell lines, EBNA-LP is localized in nuclear structures called ND10 and translocated to the nucleolus together with Hsp70s under conditions of cellular stress, including heat shock and high cell density (40, 41). Although the functional consequence of the interactions remains largely unknown at present, these results suggest that EBNA-LP is not only a coactivator of EBNA-2 but also a multifunctional protein that interacts with and modulates various components of cellular machinery during EBV-induced transformation.
Self-association of viral proteins often plays significant roles in their functions, such as enzymatic, transcriptional, and transformational activities (16, 23, 26, 37, 47). For example, EBV EBNA-2 and ICP0 of herpes simplex virus, a regulatory protein for gene expression, both of which interact with multiple cellular proteins (5, 8, 9, 14, 15, 17, 20, 21, 44–46, 48–50), self-associate, and the domain of the homotypic-association is critical to their functions, possibly because it facilitates their interaction with multiple cellular targets simultaneously (3, 7, 13, 30, 47). As described above, EBNA-LP also interacts with multiple cellular proteins, raising the possibility that EBNA-LP self-associates and homodimerization of the protein is important for its function. In this report we show that EBNA-LP self-associates through CR2, which has been shown to be critical for nuclear localization, nuclear matrix association, and the coactivator function of the protein.
MATERIALS AND METHODS
Cells.
An African green monkey kidney epithelial cell line, COS-7, was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. An EBV-negative B-lymphoma cell line, BJAB, was grown in RPMI 1640 medium supplemented with 10% fetal calf serum.
Plasmids.
To construct pAS-EBNA-LPR1 and pAS-EBNA-LPR4, EcoRI and PstI fragments of pGBT9-EBNA-LPR1 and pGBT9-EBNA-LPR4 (19), respectively, were cloned into EcoRI and PstI sites of pAS2-1 (Clontech, Palo Alto, Calif.) in frame with the DNA-binding domain of GAL4. pACT-EBNA-LPR1 and pACT-EBNA-LPR4 were generated by cloning PstI-NcoI fragments of pAS-EBNA-LPR1 and pAS-EBNA-LPR4 (PstI sites were blunt ended), respectively, into NcoI and EcoRI sites of pACT2 (EcoRI sites were blunt ended) (Clontech) in frame with the activation domain of GAL4. Plasmids encoding the DNA-binding domain of GAL4 fused to various fragments of EBNA-LP (pAS-EBNA-LPΔY2, pAS-EBNA-LPΔY1Y2, pAS-EBNA-LPW0, pAS-EBNA-LPW2,pAS-EBNA-LPΔW0W2, pAS-EBNA-LPY2, pAS-EBNA-LPW2Δ1, pAS-EBNA-LPW2Δ2, pAS-EBNA-LPW2Δ3, pAS-EBNA-LPW2Δ4, pAS-EBNA-LPW2Δ5, and pAS-EBNA-LPRA) were obtained by PCR of pGBT9-EBNA-LPR1 with appropriate primer pairs. pACT2HAX-1.1, isolated in a yeast two-hybrid screen using EBNA-LPR4 as bait, contains a partial cDNA encoding HAX-1 as described previously (19). pGEX-EBNA-LPR1 was constructed by cloning the EcoRI-SalI fragment of pGBT9-EBNA-LPR1 into EcoRI and SalI sites of pGEX4T-1 (Amersham Pharmacia, Uppsala, Sweden) in-frame with glutathione S-transferase (GST). PCR fragments of pGBT9-EBNA-LPR2 and pGBT9-EBNA-LPR4 (19) were cloned into pBS-Flag-Stop to yield pBS-EBNA-LPR2(F) and pBS-EBNA-LPR4(F), respectively. Then the EcoRI-NotI fragments of pBS-EBNA-LPR2(F) and pBS-EBNA-LPR4(F) were cloned into EcoRI and NotI sites of pME18S (kindly provided by K. Maruyama), respectively. Construction of pME-EBNA-LPR4RA(F) was described previously (52).
Yeast two-hybrid system.
The yeast two-hybrid system was used to detect dimerization of EBNA-LP or interaction of EBNA-LP with cellular proteins as described previously (21). Quantitative analyses were conducted by a liquid β-galactosidase assay as described in the Clontech yeast protocol handbook PT3024-1 with minor modifications. Briefly, yeast colonies were grown overnight at 30°C in synthetic defined medium lacking the appropriate amino acids. Two milliliters of each overnight culture was inoculated in 9 ml of yeast extract-peptone-dextrose and incubated at 30°C for 0.5 to 2 h until an optical density (600 nm) of 0.8 to 1 was reached. The culture was then centrifuged, resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4), frozen in liquid nitrogen, and supplemented with o-nitrophenyl-β-d-galactopyranoside (ONPG) and β-mercaptoethanol to 0.67 mg/ml and 0.2%, respectively. The reaction mixtures were incubated at 30°C for 1 to 8 h, and absorbance at 420 nm was measured to calculate β-galactosidase activity. One unit of β-galactosidase activity was defined as the amount which hydrolyzes 1 μmol of ONPG per min per cell. Yeast protein extracts were prepared as described in the Clontech yeast protocol handbook (PT3024-1) with minor modifications. Briefly, yeast colonies were grown overnight at 30°C in synthetic defined medium lacking the appropriate amino acids. Two milliliters of each overnight culture was inoculated in 9 ml of yeast extract-peptone-dextrose and incubated at 30°C until an optical density (600 nm) of 0.8 to 1 was reached. The culture was then centrifuged, and the pellet was washed once with ice-cold water and then frozen immediately in liquid nitrogen. The pellets were then resuspended in prewarmed complete cracking buffer (8 M urea, 5% sodium dodecyl sulfate, 40 mM Tris-HCl [pH 6.8], 0.1 mM EDTA, 0.4 mg of bromophenol blue per ml, 1% β-mercaptoethanol, 5 mM phenylmethylsulfonyl fluoride) containing acid-washed glass beads (Sigma), incubated at 70°C for 10 min, and vortexed vigorously for 1 min. Each sample was centrifuged, and the supernatant was removed into a new tube. The pellet was incubated at 100°C for 5 min, vortexed vigorously for 1 min, and centrifuged again. The second supernatant was combined with the first supernatant and subjected to immunoblotting.
Transfection.
COS-7 cells were transfected with plasmids by DEAE-dextran methods as described previously (19). At 48 to 72 h posttransfection, the cells were harvested and used for further experiments. For transfection of plasmids into BJAB cells, 1.5 × 107 cells in 0.4 ml of RPMI 1640 medium were electroporated with 15 μg of plasmid DNA at 0.2 kV and 960 μF using a Bio-Rad gene pulser. At 24 h after transfection, BJAB cells were harvested and used for further experiments.
Affinity precipitation with GST-EBNA-LP fusion protein, coimmunoprecipitation, and immunoblotting.
A GST fusion protein was expressed in Escherichia coli BL21 transformed with pGEX-EBNA-LPR1, purified on glutathione-Sepharose beads (Amersham Pharmacia), and quantified as described previously (21). Affinity precipitation with GST-EBNA-LP fusion protein, immunoprecipitation, and immunoblotting were performed as described previously (17, 18, 21).
Sucrose gradient.
BJAB cells were transfected with either pME-EBNA-LPR4RA(F) or pME-EBNA-LPR4(F) as described in Materials and Methods. At 24 h posttransfection, cells were harvested, washed twice with phosphate-buffered saline, and then lysed in NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, and 1 mM phenylmethylsulfonyl fluoride). The supernatants obtained after centrifugation of cell lysates were applied to a 10-to-30% sucrose gradient in 50 mM sodium phosphate buffer (pH 7.4) containing 0.1% NP-40 and 1 mM dithiothreitol. The gradient was run for 16 h at 35,000 rpm and 4°C in an SW41 rotor (Beckman Corp). The gradient was collected from the top as 35 0.3-ml fractions. Each fraction was subjected to electrophoresis on a 10% polyacrylamide gel containing sodium dodecyl sulfate, transferred to a nitrocellulose membrane, and reacted with a mouse monoclonal antibody to EBNA-LP (JF186) (kindly provided by G. Klein).
RESULTS
EBNA-LP self-associates in the yeast two-hybrid system.
The objective of the first series of experiments was to determine whether EBNA-LP self-associates. To this end, the entire coding sequence of EBNA-LP containing either one (EBNA-LPR1) or four (EBNA-LPR4) W1W2 repeats was cloned next to the GAL4 activation and DNA-binding domains of the yeast two-hybrid vectors (Fig. 1B), and a yeast two-hybrid assay was performed. The plasmids pAS-EBNA-LPR4 and pACT2HAX-1.1, encoding HAX-1, which is a cellular protein known to physically interact strongly with EBNA-LP (19), were used in the assay as positive controls. As shown in Fig. 1B, EBNA-LPR1 interacts with itself or versions containing a different number of W1W2 repeats, while neither EBNA-LP interacted with the GAL4 activation or DNA binding domain alone in the yeast two-hybrid system. These results indicate that EBNA-LP self-interacts in yeast. The positive controls, EBNA-LP and HAX-1, yielded β-galactosidase activity as high as 9.3 × 10−2 U. Thus, the EBNA-LP self-interactions were relatively weak in yeast.
EBNA-LP self-aggregates in mammalian cells.
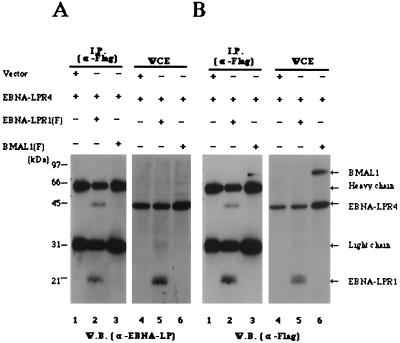
To determine whether EBNA-LP self-associates in mammalian cells, COS-7 cells transfected with expression vectors (Fig. 2 and 3) were immunoprecipitated with a mouse monoclonal antibody to Flag epitope (M2; Sigma). Immunoprecipitates were solubilized, separated on denaturing gels, transferred to nitrocellulose sheets, and reacted with mouse monoclonal antibody to EBNA-LP. The anti-Flag antibody coprecipitated EBNA-LPR4 with Flag epitope-tagged EBNA-LPR1 [EBNA-LPR1(F)] when EBNA-LPR4 and EBNA-LPR1(F) were coexpressed in COS-7 cells (Fig. 2A, lane 2). In contrast, when EBNA-LPR4 was expressed by itself (Fig. 2A, lane 1) or EBNA-LPR4 and unrelated Flag epitope-tagged protein (BMAL1) were coexpressed (lane 3), EBNA-LP was not coimmunoprecipitated with the antibody. The levels of expression of EBNA-LPR4 in whole-cell extracts of the transfected cells were equivalent (Fig. 2A, lanes 4 to 6), and the electrophoretic mobility of the protein was similar to that of the form coimmunoprecipitated with EBNA-LPR1(F). Figure 2B shows the results of immunoblotting of the same nitrocellulose membrane depicted in Fig. 2A but reprobed with the anti-Flag antibody. The results indicate that BMAL1 tagged with Flag epitope was appropriately expressed in COS-7 cells and immunoprecipitated with the antibody to the Flag epitope.
FIG. 2.
Interaction of EBNA-LPR1 with EBNA-LPR4 in mammalian cells. COS-7 cells transiently expressing the indicated proteins were immunoprecipitated with the mouse monoclonal antibody to the Flag epitope. The immunoprecipitates were subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, reacted with the mouse monoclonal antibody to EBNA-LP (A), and then reprobed with anti-Flag antibody (B). One-tenth of the COS-7 whole-cell extract (WCE) input in immunoprecipitation reactions for lanes 1, 2, and 3 was loaded in lanes 4, 5, and 6, respectively. Positions of molecular mass markers are on the left. W.B., Western blot.
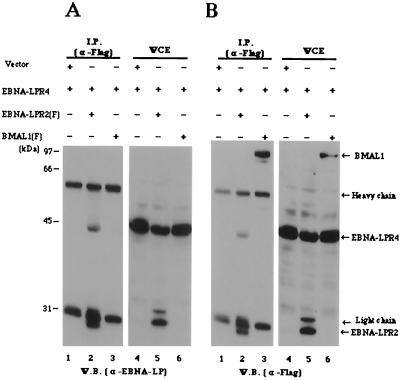
FIG. 3.
Interaction of EBNA-LPR2 with EBNA-LPR4 in mammalian cells. Experiments were done as described in the legend to Fig. 2. Positions of molecular mass markers are on the left. W.B., Western blot.
It has been reported that EBNA-LPR1 shows biological activity different from that of EBNA-LPs with multiple W1W2 repeats when it is transiently expressed in mammalian cells (19, 31, 33, 52). Unlike EBNA-LP with more than two W repeats, EBNA-LPR1 shows a distinct cellular localization and has no activity to function cooperatively with EBNA-2. Because the EBNA-LP in Fig. 2 was one with a single W1W2 repeat, it could be argued that self-association of EBNA-LP in mammalian cells is specific to EBNA-LPR1. To eliminate this possibility, we tested whether EBNA-LPs with multiple W1W2 repeats self-interact. The results (Fig. 3) were that the anti-Flag antibody specifically coimmunoprecipitated EBNA-LPR4 with EBNA-LPR2(F).
These observations indicate that EBNA-LP self-associates in mammalian cells and that self-association is independent of the number of W repeats. An earlier report from this laboratory showed that transiently expressed EBNA-LP is predominantly localized in the cytoplasm, while EBNA-LPs with multiple W1W2 repeats are mainly localized in the nucleus of COS-7 cells (19, 33). The results (Fig. 2 and 3) also suggest that self-association of EBNA-LP occurs in both the nucleus and cytoplasm of COS-7 cells.
Mapping of the domains of EBNA-LP responsible for self-association.
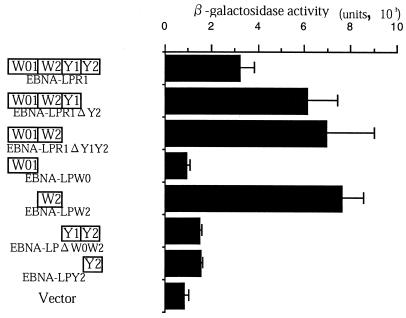
To map the domain of EBNA-LP responsible for the homodimerization of EBNA-LP, various EBNA-LPR1 deletion clones fused to the GAL4 DNA binding domain were constructed (Fig. 4) and tested for interaction with EBNA-LPR4 fused to GAL4 activation domain in the yeast two-hybrid system. As shown in Fig. 4, deletion of W2 abolished the interaction with EBNA-LPR4, while the W2 domain alone was able to interact with EBNA-LPR4, indicating that this domain is involved in homodimerization of EBNA-LP.
FIG. 4.
Mapping of the domain of EBNA-LP for self-association. The EBNA-LP deletion mutants (left) and β-galactosidase activity for each construct in the presence of EBNA-LPR4 in the yeast two-hybrid system (right) are shown. β-Galactosidase activity was determined by liquid assay procedures as described in the legend to Fig. 1.
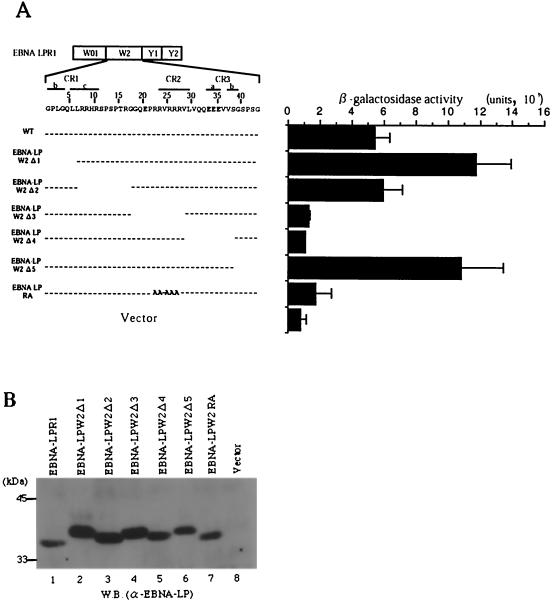
To further map the site responsible for homodimerization, a series of internal deletion mutants of the W2 domain fused to the GAL4 DNA binding domain were constructed (Fig. 5A) and tested for interaction with EBNA-LPR1 fused to the GAL4 activation domain in the yeast two-hybrid system. The site of the W2 domain of EBNA-LP required for self-association mapped to the amino acid region between Gly-19 and Ser-39 (relative to the first amino acid residue of the W2 domain) (Fig. 5A), which overlaps with the conserved regions, named CR2 and CR3, among a subset of primate γ-herpesviruses (32). A mutant with amino acid substitutions of alanines for clustered arginines in CR2 that we previously generated to map a domain responsible for nuclear matrix association and nuclear localization (52) was also tested for interaction with EBNA-LPR1. The experiments with this mutant (EBNA-LPRA) showed that the amino acid substitutions resulted in abolishment of the interaction with EBNA-LPR1 (Fig. 5A). Figure 5B shows the expression levels of the EBNA-LP mutants in Fig. 5A; the wild-type and mutant proteins were expressed in yeast to approximately similar levels, eliminating the possibility that the negative data with EBNA-LPΔ3, -Δ4, and -RA mutants in Fig. 5A were the consequence of insufficient expression of each mutant. These results indicated that the amino acid region between Gly-19 and Ser-39 contained the major determinant for self-association of EBNA-LP and the amino acid substitutions in CR2 alone abolished the homotypic interaction of the protein.
FIG. 5.
Fine mapping of the region responsible for self-interaction of EBNA-LP. (A, left) Line 1 is a schematic representation of the exon structures of EBNA-LPR1. Line 2 shows the predicted amino acid sequence of the EBNA-LP W2 domain. Conserved regions defined by Peng et al. (32) are indicated as lines with their designation given above. Internal deletion and amino acid substitution mutants of the EBNA-LPW2 domain are shown below the amino acid sequence of W2. (A, right) β-Galactosidase activity detected for each construct in the yeast two-hybrid system, determined by liquid assay procedures as described in the legend to Fig. 1. (B) Immunoblot of electrophoretically separated lysates from yeasts transfected with expression plasmids for wild-type and mutant EBNA-LPR1 proteins shown in panel A. Yeast protein lysates were prepared as described in Materials and Methods and subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the anti-EBNA-LP monoclonal antibody. Positions of molecular mass markers are on the left. W.B., Western blot.
Mutations in EBNA-LP CR2 resulted in impaired complex formation mediated by EBNA-LP in B cells.
Self-association of viral proteins often mediates the formation of a multiprotein complex by forming oligomers and by interacting with various viral and cellular proteins (13). Therefore, we tested whether self-association of EBNA-LP plays a role in the formation of protein complexes mediated by the protein. Since EBNA-LP species expressed in EBV-infected cells contain multiple W1W2 repeat domains, we used pME-EBNA-LPR4RA (52), which expresses a mutant containing four W1W2 repeat domains in which the clustered arginines of CR2 in each W2 domain were replaced by alanines.
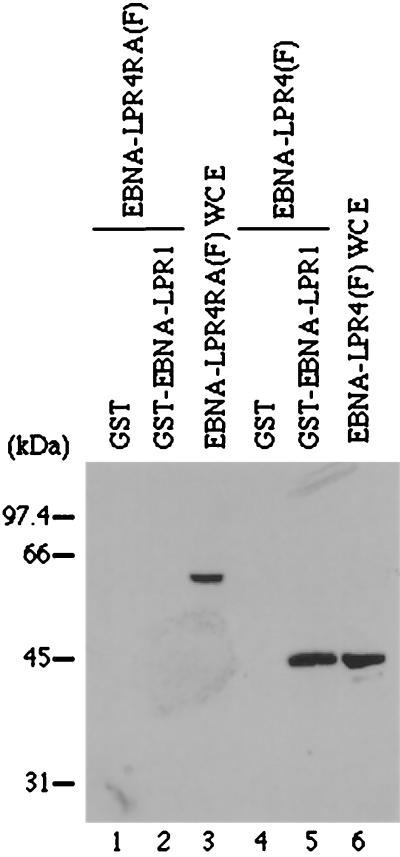
First, we examined whether the EBNA-LPR4RA mutant lacks the ability to self-interact by GST pull-down experiments. The GST-EBNA-LPR1 fusion protein was expressed and purified in E. coli, and an equal amount of GST-EBNA-LPR1 or GST bound to glutathione-Sepharose beads (Amersham Pharmacia) was reacted with an extract from COS-7 cells transfected with pME-EBNA-LPR4(F) or pME-EBNA-LPR4RA(F) (data not shown). After extensive rinsing, the protein complex captured on the beads was solubilized, subjected to electrophoresis in a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the anti-Flag antibody. GST-EBNA-LPR1 pulled down Flag epitope-tagged EBNA-LPR4 (Fig. 6, lane 5), whereas GST alone did not (lane 4). The electrophoretic mobility of the EBNA-LP that complexed with GST-EBNA-LPR1 (Fig. 6, lane 5) was similar to that found in whole-cell extracts of the transfected cells (lane 6). In contrast, neither GST-EBNA-LPR1 nor GST pulled down EBNA-LPR4RA(F) (Fig. 6, lanes 1 and 2). These results indicated that EBNA-LPR4RA is deficient in the ability to self-associate and that CR2 is critical for self-association of EBNA-LP.
FIG. 6.
Immunoblot of transfected-cell proteins bound to GST or chimeric GST-EBNA-LPR1, electrophoretically separated in a denaturing gel, and reacted with a mouse monoclonal antibody to the Flag epitope. Lysates of COS-7 cells transfected with pME-EBNA-LPR4(F) or pME-EBNA-LPR4RA(F) were reacted with GST or GST-EBNA-LPR1 chimeric protein immobilized on glutathione-Sepharose beads. The beads were pelleted, rinsed extensively, subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the anti-Flag epitope antibody. Lanes 1 and 2, whole-cell extracts (WCE) from COS-7 cells transfected with pME-EBNA-LPR4RA (F) bound to GST and GST-EBNA-LPR1, respectively; lane 3, 10% of WCE from COS-7 cells transfected with pME-EBNA-LPR4RA(F) used in the binding reaction; lanes 4 and 5, WCE from COS-7 cells transfected with pME-EBNA-LP R4(F) bound to GST and GST-EBNA-LPR1, respectively; lane 6, 10% of WCE from COS-7 cells transfected with pME-EBNA-LPR4(F) used in the binding reaction. Positions of molecular mass markers are on the left.
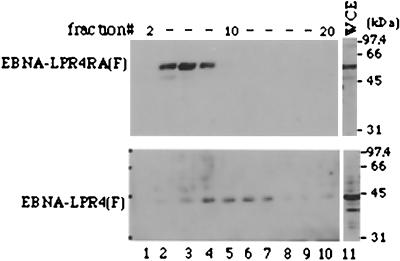
In the second series of experiments, BJAB cells were transfected with the expression vector for EBNA-LPR4(F) or EBNA-LPR4RA(F), harvested 1 day after transfection, solubilized, and applied to a 10-to-30% sucrose gradient. After the gradient had been run for 16 h, fractions were collected, subjected to electrophoresis in denaturing gels, transferred to nitrocellulose sheets, and reacted with the anti-EBNA-LP monoclonal antibody. As shown in Fig. 7, in the gradients, wild-type EBNA-LPR4(F) was detected between fractions 8 and 14, while EBNA-LPR4RA(F) showed a lower molecular mass and was detected between fractions 4 and 8. These results suggest that the self-association of EBNA-LP is important for formation of protein complex mediated by the protein in B cells.
FIG. 7.
Immunoblots of the sucrose gradient sedimentations of wild-type EBNA-LPR4(F) and EBNA-LPR4RA(F). BJAB cells were transfected with the expression vector for EBNA-LPR4(F) or EBNA-LPR4RA(F), harvested 1 day after transfection, solubilized, and applied to a 10-to-30% sucrose gradient. The gradient was run for 16 h, and then samples were collected from the top as 35 0.3-ml fractions, subjected to electrophoresis in denaturing gels, transferred to nitrocellulose sheets, and reacted with anti-EBNA-LP monoclonal antibody. Lane 11, 10% of whole-cell extract (WCE) used in the sucrose gradient sedimentations. Positions of molecular mass markers are on the right.
DISCUSSION
Most viral regulatory proteins examined to date express their functions by associating with themselves or with cellular or viral proteins. Here we demonstrated that EBNA-LP, an important regulator in EBV-induced B-cell immortalization, self-associates. The salient features of this study are as follows.
(i) In the yeast two-hybrid system, EBNA-LP interacts with itself or with a derivative having a different number of W1W2 repeat domains (Fig. 1). The self-association of EBNA-LP was also demonstrated by using the GST-EBNA-LP fusion protein in GST pull-down experiments (Fig. 6), which supported the evidence of physical self-interaction of EBNA-LP. Consistent with the binding data obtained with yeast and in vitro, transiently expressed EBNA-LP containing four W1W2 repeats was coimmunoprecipitated with chimeric protein having one or two W repeat domains when both proteins were transiently coexpressed in COS-7 cells (Fig. 2 and 3). These results indicate that the self-association of EBNA-LP occurs at the cellular level in mammalian cells.
(ii) Mutational analyses using deletion and substitution mutants in the yeast two-hybrid system revealed that CR2 is required for self-association of EBNA-LP. This was confirmed by additional in vitro biochemical assays in which the GST-EBNA-LP fusion protein did not pull down a EBNA-LPR4RA mutant with arginine-to-alanine substitutions in CR2 in each W repeat domain, while it did pull down the wild-type EBNA-LPR4 (Fig. 6). This laboratory previously reported that CR2 is important for nuclear matrix association and nuclear localization of EBNA-LP and that the mutant EBNA-LPR4RA is inactive in its transcriptional coactivator function with EBNA-2 (52). From these observations, we concluded that CR2 of EBNA-LP is a multifunctional domain mediating self-association, nuclear localization, nuclear matrix association, and coactivator function with EBNA-2.
(iii) Mutations in EBNA-LP CR2 and CR3, which are critical for self-association of the protein, abolished its cooperative transcriptional activation with EBNA-2 (33, 52). At present it is unknown whether there is any connection between EBNA-LP homodimerization and the EBNA-2-dependent coactivator function of the protein, because we were not able to completely distinguish the domains of EBNA-LP mediating self-association, nuclear matrix association, and nuclear localization of the protein. However, this is understandable in that many transcription factors express their transcriptional activity followed by homodimerization (13, 16, 26, 30, 42). For instance, the human T-cell lymphoma virus type 1 Tax transcriptional regulator, which like EBNA-LP has no intrinsic DNA-binding activity, interacts with a host transcription factor, CREB (cyclic AMP response element-binding protein), and the homodimerization of Tax protein is required for it to associate with CREB (38, 42). It has been shown that the Tax-CREB complex enhances human T-cell lymphoma virus type 1 transactivation.
(iv) Another possible role of the EBNA-LP self-association is to facilitate the formation of a multiple-protein complex mediated by EBNA-LP. Recently, Harada et al. reported that EBNA-2, which functions through interaction with multiple host cellular proteins, self-associates and that the self-association is likely to be critical for the ability of the protein to interact with multiple cellular proteins simultaneously (13, 47, 49). Supporting this hypothesis, EBNA-LP has also been shown to interact with a variety of cellular proteins (11, 19, 25, 28, 39) and the mutant EBNA-LP deficient in the ability to self-associate was demonstrated here to form complexes with a lower molecular mass that those formed by wild-type EBNA-LP in B cells (Fig. 7). Furthermore, it is conceivable that EBNA-LP forms high-order oligomers because in infected cells, EBNA-LP contains multiple W1W2 repeat domains, which we identified in this study as the region responsible for self-association of EBNA-LP. EBNA-LP oligomerization may enable the protein to form higher-order protein complexes.
In conclusion, homotypic associates form between molecules of EBNA-LP and CR2, which is responsible for the self-association and is a multifunctional domain mediating several aspects of EBNA-LP function. A series of recent reports including this study indicate that key functional domains of EBNA-LP center on the W repeat domain (11, 19, 31, 33, 51, 52). The EBNA-LP mutant viruses examined to date lack the ability to express the Y1Y2 domain, and studies of such viruses showed a much reduced but not a complete loss of transforming activity, leading to the consensus that EBNA-LP is not essential for the EBV-induced transformation process. It is, therefore, of interest and importance to determine whether EBNA-LP is truly a nonessential transforming protein by using a mutant virus that is unable to express the W1W2 domain of EBNA-LP.
Acknowledgments
We thank E. Kieff for the EBNA-LP cDNA, G. Klein for the mouse monoclonal antibody to EBNA-LP, and K. Maruyama for pME18S.
This study was supported in part by Grants for Scientific Research (Y.K. and Y.Y.) and Grants for Scientific Research in Priority Areas (Y.K. and Y.Y.) from the Ministry of Education, Science, Sports and Culture of Japan. Y.K. was supported by a grant from the Uehara Memorial Foundation.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595–608. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. J., G. J. Inman, B. D. Parker, D. T. Rowe, and P. J. Farrell. 1992. Cell growth effects of Epstein-Barr virus leader protein. J. Gen. Virol. 73:1547–1551. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J., C. Panagiotidis, and S. Silverstein. 1992. Multimerization of ICP0, a herpes simplex virus immediate-early protein. J. Virol. 66:5598–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific proteases is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkle, J., M. Rowe, B. Kallin, I. Ernberg, A. Rosen, J. Dillner, and G. Klein. 1987. Monoclonal and polyclonal antibodies against Epstein-Barr virus nuclear antigen 5 (EBNA-5) detect multiple protein species in Burkitt’s lymphoma and lymphoblastoid cell lines. J. Virol. 61:3870–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasser, F. A., P. Haiss, S. Gottel, and N. Muller-Lantzsch. 1991. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J. Virol. 65:3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman, S. R., E. Johansen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Muller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Bar virus nuclear protein EBNA2 and EBNA3C. J. Biol. Chem. 274:19136–19144. [DOI] [PubMed] [Google Scholar]
- 10.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393–397. [DOI] [PubMed] [Google Scholar]
- 11.Han, I., S. Harada, D. Weaver, Y. Xue, W. Lane, S. Orstavik, B. Skalhegg, and E. Kieff. 2001. EBNA-LP associates with cellular protein including DNA-PK and HA95. J. Virol. 75:2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada, S., R. Yalamanchili, and E. Kieff. 2001. Epstein-Barr virus nuclear protein 2 has at least two N-terminal domains that mediate self-association. J. Virol. 75:2482–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92–95. [DOI] [PubMed] [Google Scholar]
- 15.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, N. 1990. Transcriptional regulation by dimerization: two sides to an incestuous relationship. Cell 61:9–11. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1998. Eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J. Virol. 72:1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi, Y., K. Nakajima, M. Igarashi, T. Morita, M. Tanaka, M. Suzuki, A. Yokoyama, G. Matsuda, K. Kato, M. Kanamori, and K. Hirai. 2000. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J. Virol. 74:10104–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi, Y., M. Tanaka, A. Yokoyama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1α regulatory protein ICP0 functionally interacts with cellular transcriptional factor BMAL1. Proc. Natl. Acad. Sci. USA 96:1877–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khu, Y.-L., E. Koh, S. P. Lim, Y. H. Tan, S. Brenner, S. G. Lim, W. J. Hong, and P.-Y. Goh. 2001. Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol. 75:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff, E. 1996. Epstein-Barr virus and its replication, p.2343–2396. In B. N. Fields, D. M. Knipe, P. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Kitay, M. K., and D. T. Rowe. 1996. Protein-protein interactions between Epstein-Barr virus nuclear antigen-LP and cellular gene products: binding of 70-kilodalton heat shock proteins. Virology 220:91–99. [DOI] [PubMed] [Google Scholar]
- 26.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764. [DOI] [PubMed] [Google Scholar]
- 27.Mannick, J. B., J. I. Cohen, M. Birkenbach, A. Marchini, and E. Kieff. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannick, J. B., X. Tong, A. Hemnes, and E. Kieff. 1995. The Epstein-Barr virus nuclear antigen leader protein associates with hsp72/hsc73. J. Virol. 69:8169–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini, A., J. I. Cohen, F. Wang, and E. Kieff. 1992. A selectable marker allows investigation of a nontransforming Epstein-Barr virus mutant. J. Virol. 66:3214–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meredith, M., A. Orr, M. Elliott, and R. Everett. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209:174–187. [DOI] [PubMed] [Google Scholar]
- 31.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, R., A. V. Gordadze, E. M. F. Panana, F. Wang, J. Zong, G. S. Hayward, J. Tan, and P. D. Ling. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved region in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563–574. [DOI] [PubMed] [Google Scholar]
- 35.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p.2397–2446. In B. N. Fields, D. M. Knipe, P. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 36.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smelkova, N. V., and J. A. Boroweic. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, T., J.-I. Fujisawa, M. Toita, and M. Yoshida. 1993. The trans-activator Tax of human T-cell leukemia virus type 1 (HTLV) interacts with cAMP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair enhancer of HTLV-1. Proc. Natl. Acad. Sci. USA 90:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szekely, L., G. Selovanova, K. P. Magnusson, G. Klein, and K. G. Wiman. 1993. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc. Natl. Acad. Sci. USA 90:5455–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szekely, L., K. Pokrovskaja, W.-Q. Jiang, H. D. The, N. Ringertz, and G. Klein. 1996. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J. Virol. 70:2562–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szekely, L., W.-Q. Jiang, K. Pokrovskaja, K. G. Wiman, G. Klein, and N. Ringertz. 1995. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA5 and hsp70 proteins after exposure to heat shock or cell density congestion. J. Gen. Virol. 76:2423–2432. [DOI] [PubMed] [Google Scholar]
- 42.Tie, F., N. Adya, W. C. Greene, and C.-Z. Giam. 1996. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J. Virol. 70:8368–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong, X., R. Drapkin, R. Yalamanchili, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15:4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsui, S., and W. H. Schubach. 1994. Epstein-Barr virus nuclear protein 2A forms oligomers in vitro and in vivo through a region required for B-cell transformation. J. Virol. 68:4287–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634–641. [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama, A., M. Tanaka, G. Matsuda, K. Kato, M. Kanamori, H. Kawasaki, H. Hirano, I. Kitabayashi, M. Ohki, K. Hirai, and Y. Kawaguchi. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): the function of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 75:5119–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokoyama, A., Y. Kawaguchi, I. Kitabayashi, M. Ohki, and K. Hirai. 2001. The conserved domain CR2 of Epstein-Barr virus nuclear antigen leader protein is responsible not only for nuclear matrix association but also for nuclear localization. Virology 279:401–413. [DOI] [PubMed] [Google Scholar]