Abstract
Reovirus infection leads to apoptosis in cultured cells and in vivo. Binding of viral attachment protein ς1 to both sialic acid and junction adhesion molecule is required for induction of apoptosis. However, it is not known whether viral engagement of receptors is sufficient to elicit this cellular response. To determine whether steps in reovirus replication subsequent to viral attachment are required for reovirus-induced apoptosis, we used inhibitors of viral disassembly and RNA synthesis, viral disassembly intermediates, temperature-sensitive (ts) reovirus mutants, and reovirus particles deficient in genomic double-stranded RNA (dsRNA). We found that reovirus-induced apoptosis is abolished in the presence of the viral disassembly inhibitors ammonium chloride and E64. Infectious subvirion particles (ISVPs), which are intermediates in reovirus disassembly that can be generated in vitro by protease treatment, are capable of inducing apoptosis in the presence or absence of these inhibitors. Treatment of cells with the viral RNA synthesis inhibitor ribavirin does not diminish the capacity of reovirus to induce apoptosis, and reovirus ts mutants arrested at defined steps in viral replication produce apoptosis with efficiency similar to that of wild-type virus. Furthermore, reovirus particles lacking dsRNA are capable of inducing apoptosis. Finally, we found that viral attachment and disassembly must occur within the same cellular compartment for reovirus to elicit an apoptotic response. These results demonstrate that disassembly of reovirus virions to form ISVPs, but not viral transcription or subsequent steps in viral replication, is required for reovirus to induce apoptosis.
Apoptotic cell death occurs in response to numerous developmental and environmental stimuli. Cells undergoing apoptosis display a variety of morphological and biochemical alterations that clearly distinguish this process from other forms of cell death (53). Induction of apoptosis is a common cellular response to viral infection (37, 43); however, little is known about mechanisms by which viruses elicit this response. Following infection by mammalian reoviruses, apoptotic cell death plays an important role in virus-induced cytopathic effect in cell culture (12, 41, 48) and virus-induced tissue injury in vivo (15, 36). Reoviruses are nonenveloped, icosahedral viruses with a genome consisting of 10 segments of double-stranded RNA (dsRNA). These viruses provide a useful experimental system for studies of viral pathogenesis and mechanisms of virus-induced cell death.
In studies to identify reovirus genes that segregate with strain-specific differences in the capacity to induce apoptosis, the S1 gene, which encodes viral attachment protein ς1 (31, 51), was identified as the primary genetic determinant of the magnitude of the apoptotic response (11, 41, 48). Subsequent studies revealed that the efficiency with which reovirus strains elicit apoptosis is determined by the capacity to bind different types of cell surface receptors (5, 11). Reovirus strains T3SA− and T3SA+ differ genetically by a single point mutation and phenotypically by the capacity to bind sialic acid (4). Sialic acid-binding strain T3SA+ is a significantly more efficient inducer of apoptosis than is non-sialic acid-binding strain T3SA− in both HeLa cells and L cells. Enzymatic removal of cell surface sialic acid with neuraminidase or competitive blockade of virus binding to cell surface sialic acid with sialyllactose abolishes the capacity of T3SA+ to induce apoptosis (11). Incubation of cells with antibody specific for junction adhesion molecule (JAM), a recently identified reovirus receptor (5), also blocks apoptosis induced by sialic acid-binding reovirus (5). These findings demonstrate that reovirus strains capable of binding both sialic acid and JAM induce maximal levels of apoptosis.
Although receptor binding is a critical determinant of the apoptotic response, there are two lines of evidence to suggest that aspects of the reovirus replication cycle subsequent to viral attachment potentiate signals that trigger apoptosis. First, reovirus infection leads to activation of the transcription factor NF-κB, which is required for reovirus-induced apoptosis (12). In HeLa cells, activation of NF-κB is first detectable approximately 4 h after infection and peaks 8 to 10 h after infection (12). However, activation of NF-κB as a direct result of receptor-ligand interactions typically occurs with more rapid kinetics (47). The delay in NF-κB activation raises the possibility that steps following viral attachment are required to activate NF-κB and elicit apoptosis. Second, in addition to the important role of the S1 gene in determining the magnitude of the apoptotic response, the M2 gene also contributes to the efficiency of apoptosis induction (41, 48). The M2 gene encodes μ1/μ1C, a viral outer capsid protein that plays an important role in reovirus entry into cells (25, 33, 35). Following viral attachment and receptor-mediated endocytosis, reovirus virions are proteolytically disassembled to form infectious subvirion particles (ISVPs), a process characterized by removal of outer capsid protein ς3, proteolytic cleavage of μ1/μ1C to form particle-associated fragments δ and φ, and conformational changes in ς1 (18, 44). ISVPs are capable of penetrating endosomal membranes, an event that is likely mediated by μ1/μ1C (25, 33, 35). The influence of the M2 gene on the efficiency of reovirus-induced apoptosis suggests that events during viral entry, subsequent to virus engagement of cellular receptors, are required for apoptosis.
To determine whether steps following reovirus binding to cellular receptors are required to elicit apoptosis, we utilized inhibitors of viral disassembly and RNA synthesis, viral disassembly intermediates, temperature-sensitive (ts) reovirus mutants with defined blocks in viral replication, and reovirus particles lacking genomic dsRNA. We found that steps in virion disassembly, but not steps in viral replication subsequent to membrane penetration, are required to induce apoptosis following reovirus infection. Furthermore, our findings indicate that receptor binding and disassembly must occur within the same cellular compartment to elicit an apoptotic response.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were maintained as previously described (12). Isolation and characterization of T3/C44-SA− (T3SA−) and T3/C44MA-SA+ (T3SA+) have been previously described (4). Reovirus ts mutants were grown from stocks originally obtained from Kevin Coombs (tsA201, tsB352, tsC447, tsD357, and tsG453) and Bernard Fields (tsE320). T3D is a laboratory stock. Purified reovirus virions (dsRNA-positive [dsRNA+]) and top-component particles (dsRNA−) (45) were generated with second- or third-passage L-cell lysate stocks of twice-plaque-purified reovirus (23). Viral particles were freon extracted from infected-cell lysates, layered onto 1.2- to 1.4-g/cm3 CsCl gradients, and centrifuged at 62,000 × g for 18 h. Virion (1.36-g/cm3) and top-component particle (1.29-g/cm3) bands (45) were collected and dialyzed in virion storage buffer (150 mM NaCl, 15 mM MgCl2, 10 mM Tris [pH 7.4]). Concentrations of reovirus dsRNA+ virions in purified preparations were determined from the equivalence 1 U of optical density at 260 nm = 2.1 × 1012 virions per ml (45). Concentrations of dsRNA− particles in purified preparations were determined from the equivalence 1 mg of viral protein per ml = 1.8 × 1013 particles per ml.
Generation of ISVPs.
Reovirus virion or top-component particles (2 × 1011) were digested in 100 μl of virion storage buffer containing 0.2 mg of chymotrypsin (Sigma-Aldrich, St. Louis, Mo.) per ml at 37°C for 90 min. Reactions were stopped by adding 2 μl of 100 mM phenylmethylsulfonyl fluoride and cooling to 4°C. Digested particles were electrophoresed in sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and stained with Coomassie blue to confirm removal of ς3 protein and digestion of μ1/μ1C to form δ (34).
Treatment with ammonium chloride (AC), E64, or ribavirin.
HeLa cells (5 × 104) grown in 24-well tissue culture plates (Costar, Cambridge, Mass.) were either treated with 10 ng of tumor necrosis factor alpha (TNF-α; Sigma-Aldrich) per ml or infected with reovirus virions or ISVPs at a multiplicity of infection (MOI) of 100 or 1,000 PFU per cell. After addition of TNF-α or viral adsorption at 4°C for 1 h, cells were incubated in the absence or presence of 10 mM AC (Fisher Scientific, Fair Lawn, N.J.), 200 μM E64 (Sigma-Aldrich), or 25 to 200 μM ribavirin (Sigma-Aldrich) at 37°C for various intervals. Cells were then processed for viral growth (50), apoptosis, immunoprecipitation, or electrophoretic mobility shift assays (EMSAs). For experiments using E64, cells were incubated with 200 μM E64 for 4 h prior to the addition of TNF-α or viral adsorption.
Quantitation of apoptosis by acridine orange staining.
Cells (5 × 104) grown in 24-well tissue culture plates were infected with reovirus virions or ISVPs at various MOIs. The percentage of apoptotic cells was determined using acridine orange staining as previously described (48). For each experiment, 200 to 300 cells were counted, and the percentage of cells exhibiting condensed chromatin was determined by epi-illumination fluorescence microscopy using a fluorescein filter set (Zeiss Photomicroscope III; Zeiss, Oberkochen, Germany).
Yields of reovirus ts mutants.
HeLa cells (5 × 104) grown in 24-well tissue culture plates were infected with reovirus ts mutants at an MOI of 10 PFU per cell and incubated at either 32 or 39°C. Viral titers were determined 0 and 48 h after infection by plaque assay using L-cell monolayers (50) incubated at 32°C. Viral yields were determined by dividing viral titer at 48 h by viral titer at 0 h. Efficiency of yield (EOY) was calculated by dividing viral yield at 39°C by viral yield at 32°C.
Immunoblotting for PARP cleavage.
Cells (5 × 106) grown in 75-cm2 tissue culture flasks (Costar) were infected with reovirus ts mutants at an MOI of 100 PFU per cell. After viral adsorption for 1 h, cells were incubated at 32 or 39°C for 24 h. Nuclear extracts were prepared as previously described (12). Extracts (50 μg of total protein) were electrophoresed in SDS-7% polyacrylamide gels (30) and transferred to nitrocellulose membranes. Immunoblotting was performed as previously described (42) using poly(ADP-ribose) polymerase (PARP)-specific primary antibody (Roche Molecular Biochemicals, Indianapolis, Ind.) followed by horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Amersham Pharmacia Biotech, Piscataway, N.J.), each diluted 1:2,000 in Tris-buffered saline containing 0.05% Tween 20 and 5% low-fat dry milk.
Immunoprecipitation of viral proteins from ribavirin-treated cells.
Cells (2 × 106) grown in 25-cm2 tissue culture flasks (Costar) were infected with reovirus at an MOI of 1,000 PFU per cell. After viral adsorption for 1 h, the inoculum was removed, and cell culture medium containing various concentrations of ribavirin and 100 μCi of [35S]methionine-cysteine (DuPont NEN Research Products, Boston, Mass.) per ml was added. Cells were incubated at 37°C for 24 h, and reovirus proteins were captured by immunoprecipitation using a rabbit polyclonal antiserum raised against reovirus strain type 1 Lang (T1L) as previously described (11). Immunoprecipitated viral protein was subjected to electrophoresis in SDS-10% polyacrylamide gels. Gels were dried under vacuum and exposed to Biomax MR film (Kodak, Rochester, N.Y.) or a phosphorimaging plate (Fuji, Edison, N.J.). Total immunoprecipitated viral protein was quantitated with a phosphorimager (Fuji BAS2000).
EMSAs.
Cells (5 × 106) grown in 75-cm2 tissue culture flasks were infected with reovirus at an MOI of 100 PFU per cell and either untreated or treated with 10 mM AC, 200 μM E64, or 200 μM ribavirin. After incubation at 37°C for 10 h, nuclear extracts were prepared as previously described (12). Nuclear extracts (10 μg of total protein) were assayed for NF-κB activation by EMSA using a 32P-labeled oligonucleotide consisting of the NF-κB consensus binding sequence (Santa Cruz Biotechnology, Santa Cruz, Calif.) as previously described (12).
HA assay.
Purified dsRNA+ and dsRNA− virions and ISVPs were aliquoted into 96-well U-bottomed microtiter plates (Costar) and serially diluted twofold in 50 μl of phosphate-buffered saline. Calf erythrocytes (Colorado Serum Co., Denver, Colo.) were washed twice in phosphate-buffered saline and resuspended at a concentration of 1% (vol/vol). Erythrocytes (50 μl) were added to wells containing viral particles and incubated at 4°C for 2.5 h. A partial or complete shield of erythrocytes on the well bottom was interpreted as a positive hemagglutination (HA) result; a smooth, round button of erythrocytes was interpreted as negative. The minimum number of viral particles sufficient to produce HA was designated to equal 1 HA unit.
Coinfection of T3SA− ISVPs and T3SA+ virions.
HeLa cells (5 × 104) grown in 24-well plates were mock infected, infected with T3SA− ISVPs or T3SA+ virions at an MOI of 100 PFU per cell, or infected with both T3SA− ISVPs and T3SA+ virions, each at an MOI of 100 PFU per cell. Coinfections were either simultaneous or offset by 4 h. Cells were incubated at 37°C for 48 h and processed for acridine orange staining.
RESULTS
Treatment of cells with inhibitors of reovirus disassembly blocks apoptosis induced by virions.
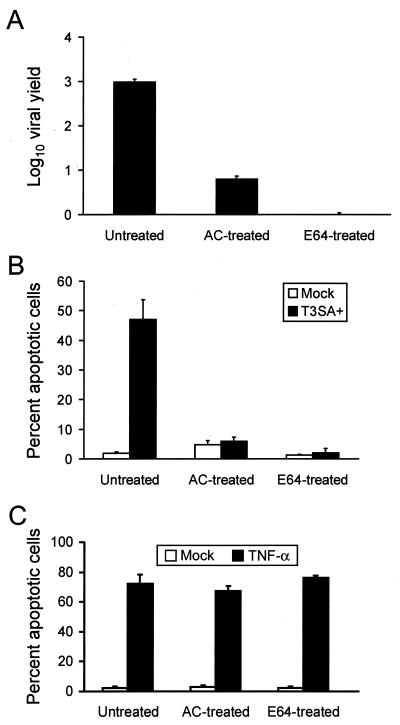
Conversion of reovirus virions to ISVPs in cellular endosomes requires acidic pH (8, 46) and the activity of cysteine-containing endocytic proteases (1, 2). To determine whether acid-dependent or protease-dependent steps in viral uncoating are required for reovirus-induced apoptosis, we assessed the capacity of reovirus to elicit apoptosis in the presence of AC, an inhibitor of endosomal acidification (46), or E64, an inhibitor of cysteine-containing proteases (3). E64 is internalized into cells by endocytosis, and its inhibitory activity is restricted to proteases in the endocytic pathway (3). HeLa cells were either mock infected or infected with T3SA+ at an MOI of 100 PFU per cell and incubated in the absence or presence of 10 mM AC or 200 μM E64, concentrations that block reovirus disassembly (data not shown) and growth in HeLa cells (Fig. 1A). Apoptosis was assessed by acridine orange staining 48 h after infection (Fig. 1B). T3SA+ induced 47% of cells to undergo apoptosis in the absence of inhibitors; however, apoptosis was reduced to the level of mock-infected cells in the presence of either AC or E64. Similar results were obtained with L cells treated with AC or E64 (data not shown). TNF-α induced equivalent levels of apoptosis in the absence or presence of AC or E64 (Fig. 1C), indicating that these inhibitors do not inhibit the activity of cellular proteins, including cysteine-containing proteases, required for apoptotic cell death. These results demonstrate that treatment of cells with either AC or E64 blocks reovirus-induced apoptosis, which suggests that virion disassembly is required for induction of apoptosis following reovirus infection.
FIG. 1.
(A) Growth of T3SA+ in cells treated with either AC or E64. HeLa cells (5 × 104) were infected with T3SA+ at an MOI of 100 PFU per cell and incubated in the absence or presence of 10 mM AC or 200 μM E64. Viral titers were determined 0 and 48 h after infection, and viral yields were calculated by dividing viral titer at 48 h by viral titer at 0 h. (B and C) Apoptosis induced by T3SA+ (B) or TNF-α (C) in cells treated with either AC or E64. HeLa cells (5 × 104) were mock infected, infected with T3SA+ at an MOI of 100 PFU per cell, or treated with 10 ng of TNF-α per ml and incubated in the absence or presence of 10 mM AC or 200 μM E64. After incubation for 48 h, cells were stained with acridine orange. The results are expressed as the mean viral yield (A) or mean percentage of cells undergoing apoptosis (B and C) for three independent experiments. Error bars indicate standard deviations of the means.
Reovirus ISVPs generated in vitro are capable of inducing apoptosis.
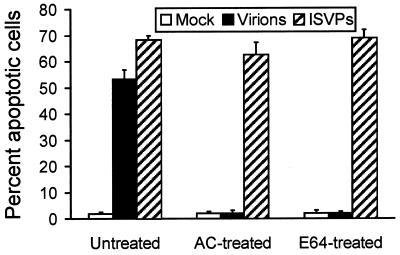
Experiments using AC and E64 indicate that conversion of reovirus virions to ISVPs is required for reovirus-induced apoptosis. It seemed possible that either ISVPs alone, or steps in reovirus replication subsequent to ISVP formation, potentiate signals elicited by receptor binding to induce apoptosis. Alternatively, events that occur during disassembly, such as release of ς3 from virions, could potentiate the apoptotic response. To distinguish between these possibilities, we tested the capacity of ISVPs generated in vitro by chymotrypsin treatment of virions to induce apoptosis. HeLa cells were either mock infected or infected with T3SA+ virions or ISVPs at an MOI of 100 PFU per cell, and apoptosis was assessed by acridine orange staining 48 h after infection (Fig. 2). T3SA+ virions induced 53% of cells to undergo apoptosis, while infection with T3SA+ ISVPs led to apoptosis of 68% of cells. Therefore, ISVPs are capable of inducing apoptosis when either generated within endosomes or adsorbed to the cell surface, suggesting that disassembly need not occur in cyto for apoptosis induction.
FIG. 2.
Apoptosis induced by T3SA+ ISVPs. HeLa cells (5 × 104) were either mock infected or infected with T3SA+ virions or ISVPs at an MOI of 100 PFU per cell and incubated in the absence or presence of 10 mM AC or 200 μM E64. After incubation for 48 h, cells were stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
To determine whether reovirus ISVPs are capable of bypassing the AC- and E64-mediated blocks to apoptosis induced by virions, we tested the capacity of ISVPs to induce apoptosis in the presence of these inhibitors. HeLa cells were infected with either T3SA+ virions or ISVPs at an MOI of 100 PFU per cell and either untreated or treated with 10 mM AC or 200 μM E64. Apoptosis was assessed 48 h after infection by acridine orange staining (Fig. 2). As anticipated, treatment with either AC or E64 blocked apoptosis induced by virions. However, apoptosis induced by ISVPs was not significantly altered in the presence of either inhibitor. Similar results were obtained with L cells treated with either AC or E64 (data not shown). These results provide additional evidence that the inhibition of apoptosis mediated by AC and E64 is due to the capacity of these inhibitors to block disassembly of reovirus virions to form ISVPs. Moreover, these findings indicate that reovirus-induced apoptosis is elicited by either the binding of ISVPs to receptors or the concerted action of receptor binding and steps in reovirus replication subsequent to ISVP formation.
Reovirus ts mutants with defined blocks in viral replication are capable of inducing apoptosis.
To test directly whether steps in reovirus replication following disassembly are required to induce apoptosis, we utilized a panel of reovirus ts mutants that were generated by chemical mutagenesis of reovirus prototype strain T3D (20, 26). These viral mutants have defects in synthesis of viral dsRNA, assembly of core and double-shelled particles, or cell entry of progeny virions (14, 20-22, 26, 27) (Table 1).
TABLE 1.
Characteristics of reovirus ts mutantsa
| Mutant | Gene | Protein | Replication block | EOY (39°C)b |
|---|---|---|---|---|
| tsA201 | M2 | μ1 | Entry of ts progeny | 0.028 |
| tsB352 | L2 | λ2 | Assembly of outer capsid | 0.000027 |
| tsC447 | S2 | ς2 | Assembly of core particles, dsRNA synthesis | 0.00012 |
| tsD357 | L1 | λ3 | dsRNA synthesis | 0.0001 |
| tsE320 | S3 | ςNS | dsRNA synthesis | 0.00014 |
| tsG453 | S4 | ς3 | Assembly of outer capsid | 0.00014 |
Reovirus ts mutants were generated by chemical mutagenesis of strain T3D (20, 26). Gene segment assignments and replication blocks of each ts mutant have been previously reported (14, 20, 21, 22, 26, 27).
The viral yield of each ts mutant was determined after 48 h of growth in HeLa cells incubated at either 32 or 39°C. EOY was calculated by dividing viral yield at 39°C by viral yield at 32°C.
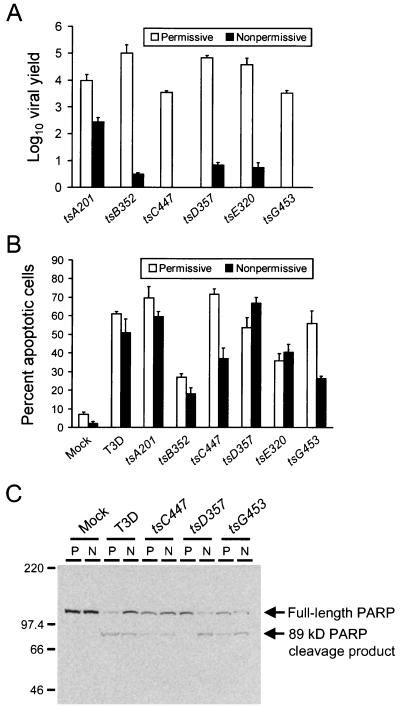
To establish feasibility for the use of reovirus ts mutants for apoptosis experiments using HeLa cells, we tested whether selected ts mutants display ts phenotypes in HeLa cells comparable to those previously reported for other cell types (13). HeLa cells were infected with ts mutants at an MOI of 10 PFU per cell and incubated at either 32°C (permissive temperature) or 39°C (nonpermissive temperature). Viral titers were determined 0 and 48 h after infection, and viral yields were calculated by dividing viral titer at 48 h by viral titer at 0 h (Fig. 3A). EOY values were calculated by dividing viral yield at 39°C by viral yield at 32°C (Table 1). We found that each ts mutant displayed significant defects in the production of viral progeny at the nonpermissive temperature. The ts phenotypes of both tsC447 and tsG453 were particularly severe, as demonstrated by viral yields of less than one progeny PFU per input PFU. Thus, reovirus ts mutants exhibit ts phenotypes after growth in HeLa cells at the nonpermissive temperature, suggesting that these mutants have defects in particle assembly, dsRNA synthesis, or entry of progeny virions as previously defined.
FIG. 3.
(A) Growth of reovirus ts mutants in HeLa cells. Cells (5 × 104) were infected with the reovirus ts mutants shown at an MOI of 100 PFU per cell and incubated at permissive or nonpermissive temperature. Viral titers were determined 0 and 48 h after infection, and viral yields were calculated by dividing viral titer at 48 h by viral titer at 0 h. (B) Apoptosis induced by reovirus ts mutants. HeLa cells (5 × 104) were either mock infected or infected with T3D or the reovirus ts mutants shown at an MOI of 100 PFU per cell and incubated at permissive or nonpermissive temperature. After incubation for 48 h, cells were stained with acridine orange. The results are expressed as the mean viral yield (A) or mean percentage of cells undergoing apoptosis (B) for three independent experiments. Error bars indicate standard deviations of the means. (C) PARP cleavage induced by reovirus ts mutants. HeLa cells (5 × 106) were either mock infected or infected with T3D, tsC447, tsD357, or tsG453 at an MOI of 100 PFU per cell and incubated at permissive (P) or nonpermissive (N) temperature. Nuclear extracts were prepared 24 h after infection, and 50 μg of total protein was resolved by SDS-PAGE, transferred to nitrocellulose, and blotted with PARP-specific antiserum. The positions of full-length PARP and the 89-kDa PARP cleavage product are indicated. Molecular mass markers are indicated in kilodaltons.
To determine whether steps subsequent to viral disassembly are required for reovirus-induced apoptosis, we assessed the capacity of ts mutant viruses to induce apoptosis at the nonpermissive temperature. HeLa cells were infected with T3D or one of six ts mutants at an MOI of 100 PFU per cell and incubated at either 32 or 39°C. Apoptosis was assessed by acridine orange staining 48 h after infection (Fig. 3B). T3D and each of the ts mutant viruses tested were capable of inducing apoptosis at both 32 and 39°C. Mutant virus tsD357 induced slightly higher levels of apoptosis at 39°C than at 32°C, whereas both tsC447 and tsG453 induced approximately 50% fewer cells to undergo apoptosis at the nonpermissive temperature than at the permissive temperature. Similar results were obtained when the capacity of ts mutants to induce apoptosis was assessed with L cells (data not shown).
We thought it possible that the diminution in apoptosis induced by tsC447 and tsG453 at the nonpermissive temperature might be due to the inability of these ts viruses to initiate secondary rounds of infection at 39°C. Therefore, to minimize these potential differences, we assessed apoptosis at an earlier time point by performing immunoblotting to detect the cleavage of caspase substrate PARP, which occurs within 12 h following reovirus infection (11). HeLa cells were either mock infected or infected with T3D, tsC447, tsD357, or tsG453 at an MOI of 100 PFU per cell and incubated at either the permissive or the nonpermissive temperature for 24 h. Nuclear extracts were prepared and used in immunoblots for detection of full-length PARP and the 89-kDa PARP cleavage product (Fig. 3C). T3D, tsC447, tsD357, and tsG453 were each capable of inducing cleavage of PARP at the nonpermissive temperature. In tsD357-infected cells, apoptosis appeared to be more efficient at 39°C, as demonstrated by the absence of the PARP cleavage product in cells incubated at 32°C, a finding consistent with results obtained by acridine orange staining assays of tsD357-infected HeLa cells (Fig. 3B) and L cells (data not shown). Therefore, ts mutant reoviruses are capable of eliciting both the morphological and biochemical hallmarks of apoptosis at the nonpermissive temperature.
Viral RNA synthesis is not required for reovirus-induced apoptosis.
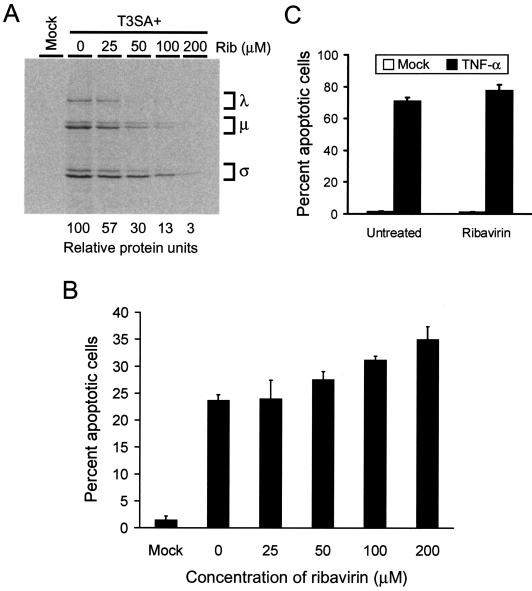
To determine whether viral RNA synthesis, including production of single-stranded RNA (ssRNA) during primary transcription, is required for reovirus-induced apoptosis, we tested the capacity of reovirus to induce apoptosis in the presence of ribavirin. Ribavirin is a guanosine analog that inhibits production of viral ssRNA and dsRNA at concentrations that do not significantly alter cellular RNA synthesis (40, 52). To determine the concentration of ribavirin required to inhibit reovirus RNA synthesis in HeLa cells, we tested the effect of various concentrations of ribavirin on levels of reovirus protein produced as a surrogate marker. HeLa cells were infected with T3SA+ at an MOI of 1,000 PFU per cell and incubated for 24 h in media containing 25 to 200 μM ribavirin and [35S]methionine-cysteine. Radiolabeled viral proteins in cell lysates were immunoprecipitated using a T1L-specific polyclonal serum, which recognizes all T3SA+ proteins with the exception of ς1. Immunoprecipitated viral proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography or quantitated by phosphorimager analysis (Fig. 4A). Reovirus protein synthesis was inhibited at each concentration of ribavirin tested, with levels of viral protein synthesis in cells treated with 200 μM ribavirin reduced to 3% of that in untreated cells. Growth of T3SA+ also was completely abolished in the presence of 200 μM ribavirin (data not shown). Cellular protein synthesis in reovirus-infected cells was not significantly altered by ribavirin concentrations of ≤200 μM (data not shown).
FIG. 4.
(A) T3SA+ protein synthesis in cells treated with ribavirin. HeLa cells (2 × 106) were either mock infected or infected with T3SA+ at an MOI of 1,000 PFU per cell. Cells were incubated for 24 h in the presence of ribavirin (Rib) at the concentrations shown and [35S]methionine-cysteine. Viral proteins were immunoprecipitated from cell lysates, resolved by SDS-PAGE, dried, and exposed to film. Reovirus λ, μ, and ς proteins are indicated. Total viral protein was quantitated by phosphorimager analysis, and the relative amount of protein present is indicated (protein in untreated cells is arbitrarily set at 100). (B and C) Apoptosis induced by reovirus (B) or TNF-α (C) in cells treated with ribavirin. HeLa cells (5 × 104) were mock infected, infected with T3SA+ at an MOI of 1,000 PFU per cell, or treated with 10 ng of TNF-α per ml and incubated in the absence or presence of ribavirin at the concentrations shown. Mock-infected cells (B) and ribavirin-treated cells (C) were incubated in the presence of 200 μM ribavirin. After incubation for 24 h, cells were stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
To determine whether ribavirin treatment inhibits reovirus-induced apoptosis, HeLa cells were infected with T3SA+ at an MOI of 1,000 PFU per cell, incubated for 24 h in the absence or presence of various concentrations of ribavirin, and scored for apoptosis by acridine orange staining (Fig. 4B). This MOI was chosen to enable quantitation of apoptosis following a single round of infection. T3SA+ efficiently induced apoptosis in the absence of ribavirin and in the presence of each concentration of ribavirin tested. In control experiments, ribavirin treatment did not alter levels of apoptosis induced by treatment with TNF-α (Fig. 4C). These results suggest that neither synthesis of ssRNA during primary transcription nor steps in reovirus replication subsequent to primary transcription are required for apoptosis induction.
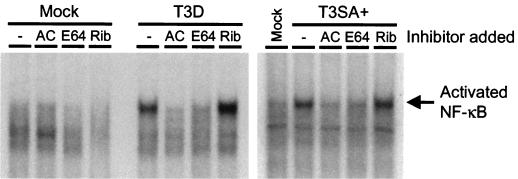
Reovirus-induced NF-κB activation is blocked by inhibition of viral disassembly but not by inhibition of viral RNA synthesis.
We have previously demonstrated that reovirus infection leads to activation of NF-κB and that this activation is required for reovirus-induced apoptosis (12). To determine whether inhibitors of either viral disassembly or RNA synthesis block activation of NF-κB, we used an NF-κB-specific oligonucleotide probe in EMSAs performed with nuclear extracts prepared from untreated cells and cells treated with AC, E64, or ribavirin. HeLa cells were either mock infected or infected with T3D, a relatively potent inducer of NF-κB activation (12), or T3SA+, a relatively weak inducer of NF-κB (11), at an MOI of 100 PFU per cell. Following adsorption, cells were incubated in the absence or presence of 10 mM AC, 200 μM E64, or 200 μM ribavirin for 10 h. Nuclear extracts were prepared and used in EMSAs (Fig. 5). Both T3D and T3SA+ induced NF-κB activation in untreated cells and in cells treated with ribavirin. However, NF-κB activation was blocked in cells treated with either AC or E64. TNF-α retains the capacity to activate NF-κB in the presence of each inhibitor (data not shown), indicating that the inhibition of NF-κB activation in reovirus-infected cells treated with AC or E64 is a direct consequence of the capacity of these inhibitors to block viral disassembly. These results suggest that NF-κB activation, a biochemical signal necessary for reovirus-induced apoptosis, is elicited following viral disassembly but prior to synthesis of viral ssRNA.
FIG. 5.
Reovirus-induced activation of NF-κB in the presence of AC, E64, and ribavirin. HeLa cells (5 × 106) were either mock infected or infected with T3D or T3SA+ at an MOI of 100 PFU per cell and incubated in the absence (−) or presence of 10 mM AC, 200 μM E64, or 200 μM ribavirin (Rib). After incubation for 10 h, nuclear extracts were prepared. Extracts (10 μg) were incubated with a 32P-labeled oligonucleotide comprised of the NF-κB consensus binding sequence. Incubation mixtures were resolved by acrylamide gel electrophoresis, dried, and exposed to film. The position of the activated NF-κB complex is indicated.
Reovirus particles lacking genomic dsRNA are capable of inducing apoptosis.
To conclusively demonstrate that postpenetration events during reovirus replication are dispensable for apoptosis induction, we tested the capacity of reovirus particles devoid of genomic dsRNA to induce apoptosis. Reovirus top-component (dsRNA−) particles contain viral proteins in stoichiometric amounts equivalent to virions, and yet they are deficient in dsRNA (17, 45). T3SA+ dsRNA− particles generated for apoptosis studies also exhibited these properties (data not shown). Since the capacity to bind sialic acid is critical for induction of apoptosis by reovirus (11), we first tested the capacity of T3SA+ dsRNA− virions and ISVPs to agglutinate bovine erythrocytes, a property dependent on sialic acid binding (10, 16) (Table 2). We found that HA titers of dsRNA− virions and ISVPs were within twofold of HA titers of dsRNA+ virions and ISVPs.
TABLE 2.
Characteristics of dsRNA+ and dsRNA− reovirus particles
| Particle type | Particle/PFU ratioa | HA titerb |
|---|---|---|
| dsRNA+ virions | 108 | 1.25 × 108 |
| dsRNA− virions | 18,182 | 2.5 × 108 |
| dsRNA+ ISVPs | 23 | 1.25 × 108 |
| dsRNA− ISVPs | 6,250 | 2.5 × 108 |
Particle/PFU ratios were determined by dividing the number of viral particles per milliliter by the number of PFU per milliliter.
HA titer is defined as the minimum number of viral particles required to agglutinate bovine erythrocytes.
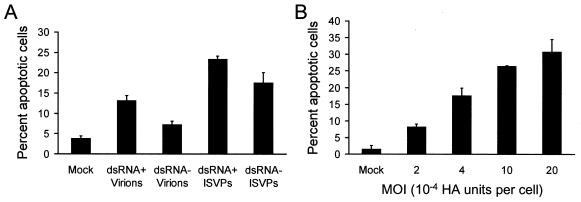
To quantitatively compare the capacities of dsRNA+ and dsRNA− particles to induce apoptosis, HeLa cells were adsorbed with dsRNA+ or dsRNA− virions or ISVPs at an MOI of 5 × 10−4 HA units per cell, and apoptosis was assessed by acridine orange staining 24 h after infection (Fig. 6A). This MOI was equivalent to 62,500 dsRNA+ particles per cell (approximately 900 PFU of virions per cell) or 125,000 dsRNA− particles per cell (approximately 7 PFU of virions per cell). This MOI was chosen to allow for quantitation of apoptosis following a single round of infection. Both dsRNA+ and dsRNA− virions induced low levels of apoptosis, approximately 13 and 7%, respectively. However, ISVPs of dsRNA+ and dsRNA− particles induced higher levels of apoptosis, approximately 23 and 18%, respectively. To determine whether apoptosis elicited by dsRNA− particles is dose dependent, HeLa cells were adsorbed with various MOIs of dsRNA− ISVPs, and apoptosis was assessed by acridine orange staining 24 h after infection (Fig. 6B). We found that levels of apoptosis increased with increasing MOI, suggesting that the magnitude of the apoptotic response is determined by viral dose. These results demonstrate that reovirus particles lacking genomic dsRNA are capable of inducing apoptosis, which provides further support for the idea that steps in viral replication subsequent to penetration of cellular membranes are dispensable for eliciting apoptosis by reovirus.
FIG. 6.
(A) Apoptosis induced by T3SA+ particles lacking genomic dsRNA. HeLa cells (5 × 104) were either mock infected or infected with dsRNA+ or dsRNA− virions or ISVPs of T3SA+ at an MOI of 5 × 10−4 HA units per cell. After incubation for 24 h, cells were stained with acridine orange. (B) Dose-dependent apoptosis induced by T3SA+ dsRNA− particles. HeLa cells (5 × 104) were either mock infected or infected with T3SA+ dsRNA− ISVPs at the MOIs shown. After incubation for 24 h, cells were stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
Receptor engagement and virion disassembly must occur within the same cellular compartment for reovirus to induce apoptosis.
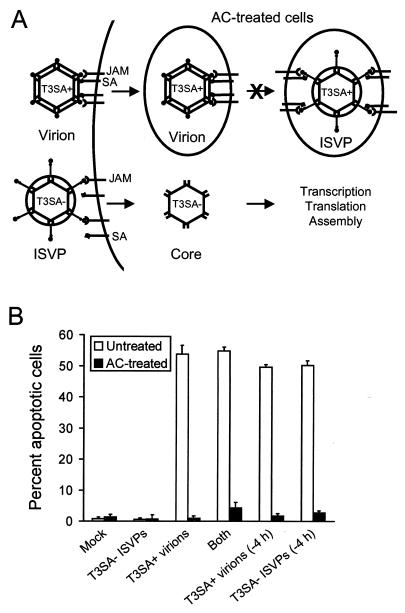
Our findings presented thus far indicate that both receptor binding and conversion of virions to ISVPs are absolute requirements for the induction of apoptosis by reovirus. It is possible that these events function as discrete signals or that engagement of cellular receptors by ISVPs might provide the sole signal required to induce apoptosis. To distinguish between these possibilities, we tested the capacity of T3SA− ISVPs to complement the block to apoptosis observed after infection of AC-treated cells with T3SA+ virions (Fig. 7A). T3SA− and T3SA+ differ by a single amino acid that determines the capacity to bind sialic acid (4). ISVPs of T3SA− do not induce apoptosis but complete all viral replication steps subsequent to virion disassembly in the presence or absence of AC (data not shown). We reasoned that if receptor binding and viral disassembly provide two separate signals, then T3SA+ virions, which are capable of binding both sialic acid and JAM (4, 5), should provide one signal, and T3SA− ISVPs, which are capable of bypassing the AC-mediated block to viral entry, should provide the other. Alternatively, if receptor engagement and viral disassembly must occur within the same cellular compartment, then T3SA− would be incapable of inducing apoptosis of AC-treated cells infected with T3SA+ virions. HeLa cells were either mock infected or infected with T3SA− ISVPs alone, T3SA+ virions alone, or both T3SA− ISVPs and T3SA+ virions at an MOI of 100 PFU per cell. In cells infected with both T3SA− ISVPs and T3SA+ virions, inoculations were performed either simultaneously or offset by 4 h to avoid competition for cellular receptors. Cells were incubated in the absence or presence of 10 mM AC, and apoptosis was assessed by acridine orange staining 48 h after infection (Fig. 7B). In untreated cells, equivalent levels of apoptosis were induced by infection with T3SA+ virions alone and by coinfection with T3SA+ virions and T3SA− ISVPs. However, apoptosis was completely blocked in AC-treated cells infected with T3SA+ virions, T3SA− ISVPs, and the combination of T3SA+ virions and T3SA− ISVPs. These results indicate that viral receptor binding and postbinding disassembly steps are not dissociable events in apoptosis induction. Instead, our data suggest that virus binding and disassembly must occur in the same cellular compartment to induce apoptosis.
FIG. 7.
(A) Model for coinfection of T3SA− ISVPs and T3SA+ virions. In AC-treated cells, T3SA+ virions bind JAM and sialic acid (SA) and enter cells by receptor-mediated endocytosis but do not disassemble or undergo subsequent steps in the viral replication cycle. In contrast, T3SA− ISVPs bind JAM, penetrate cells at the cell surface or from within endocytic vesicles, and complete all subsequent steps in viral replication. (B) Apoptosis induced by coinfection of T3SA− ISVPs and T3SA+ virions in AC-treated cells. HeLa cells (5 × 104) were either mock infected or infected with T3SA− ISVPs, T3SA+ virions, or both T3SA− ISVPs and T3SA+ virions. Coinfections were performed simultaneously or offset by 4 h. Cells were incubated in the absence or presence of 10 mM AC for 48 h and stained with acridine orange. The results are expressed as the mean percentage of cells undergoing apoptosis for three independent experiments. Error bars indicate standard deviations of the means.
DISCUSSION
During the course of viral replication, there are numerous obligate interactions between virus and cell capable of triggering apoptosis, including virus-receptor engagement, fusion with or penetration of cellular membranes, and disruption of host cell transcriptional and translational machinery (19). For some viruses, such as human immunodeficiency virus type 1 and avian leukosis virus (ALV), binding to cellular receptors is sufficient to induce apoptosis. Human immunodeficiency virus type 1 attachment protein gp120, when either cross-linked by anti-gp120 antibodies or presented on cellular membranes, is capable of inducing apoptosis of both infected and uninfected CD4+ T cells by binding to CD4 and CXCR4 (6, 38). The receptor for ALV subgroups B and D, cytopathic ALV receptor 1 (CAR1), has homology to TNF receptor proteins, including a region with similarity to the death domains of TNFR1 and Fas (7). Cells expressing CAR1 undergo apoptosis following engagement of an Env-immunoglobulin fusion protein, suggesting that CAR1 mediates the apoptotic response following infection by ALV (7).
For other viruses, such as Sindbis virus, apoptosis is triggered by events that occur during cell entry subsequent to attachment (28, 29). Inhibitors of endosomal acidification, which block Sindbis virus fusion with endosomal membranes, prevent virus-induced apoptosis (29). In addition, interaction of Sindbis virus glycoproteins E1 and E2 with endosomal membranes results in activation of sphingomyelinases and the subsequent accumulation of ceramide, which has been hypothesized to mediate the apoptotic response following Sindbis virus infection (28). These findings pinpoint the initiation of Sindbis virus-induced apoptosis to fusion of the viral envelope with endosomal membranes. Interestingly, like apoptosis induced by reovirus, Sindbis virus-induced apoptosis requires activation of NF-κB (32).
We have previously demonstrated that the efficiency with which reovirus induces apoptosis is determined by receptor utilization. Specifically, binding of reovirus attachment protein ς1 to both cell surface sialic acid and JAM is required to achieve maximal levels of apoptosis following reovirus infection (5, 11). However, results reported here indicate that receptor engagement is not sufficient to initiate this cellular response. We found that treatment of cells with either AC or E64, both of which inhibit disassembly of reovirus virions, abolishes apoptosis induced by reovirus infection. ISVPs generated in vitro by chymotrypsin treatment are capable of inducing apoptosis in the presence or absence of each of these inhibitors. We also found that NF-κB activation, which is required for reovirus-induced apoptosis, is blocked in cells treated with either AC or E64. These data demonstrate that virion disassembly is an absolute requirement to elicit both NF-κB activation and apoptosis following reovirus infection.
In contrast to the requirement for viral disassembly, three lines of evidence suggest that viral RNA synthesis is dispensable for reovirus-induced apoptosis. First, each of six reovirus ts mutants, which have distinct blocks in the viral replication cycle, is capable of causing the morphological and biochemical features of apoptosis. Second, the capacity of reovirus to activate NF-κB and induce apoptosis is not diminished in cells treated with the viral RNA synthesis inhibitor ribavirin. Third, reovirus particles deficient in genomic dsRNA are capable of inducing apoptosis. Although these experiments do not exclude a contribution of postdisassembly events to enhancing the magnitude of reovirus-induced apoptosis, these findings strongly suggest that viral RNA synthesis and subsequent steps in reovirus replication are not required to elicit this process.
We thought it possible that reovirus attachment and disassembly might provide two distinct signals, both of which are required for apoptosis induction. On the other hand, engagement of reovirus ISVPs with cellular receptors, either at the cell membrane or within endocytic vesicles, might act as the sole signal to initiate the apoptotic response. To distinguish between these possibilities, we tested the capacity of T3SA− ISVPs to complement the AC-mediated block to apoptosis in cells infected with T3SA+ virions. We found that coinfection with T3SA− ISVPs and T3SA+ virions failed to rescue the block to apoptosis in AC-treated cells. This finding supports the idea that individual viral particles must coordinately engage cellular receptors and disassemble to trigger apoptosis. Alternatively, if receptor binding and virion disassembly induce two discrete signals required for apoptosis, both signals must occur temporally and in close proximity to elicit an apoptotic response.
Our results indicate that ISVPs capable of binding both sialic acid and JAM can induce apoptosis by binding these receptors at the cell surface or in endocytic vesicles. We hypothesize that conformational changes in ς1 that occur during viral disassembly (18, 23) enhance the affinity of ς1 for sialic acid, JAM, or both receptors, leading to receptor aggregation and stimulation of intracellular signaling. Proteolytic processing of μ1/μ1C during virion disassembly also may influence virus-receptor interactions, which would provide a mechanistic explanation for the contribution of the M2 gene to the efficiency of apoptosis induction (41, 48).
Although our findings demonstrate that the binding of reovirus ISVPs to cellular receptors is required for induction of apoptosis, it is possible that this interaction is not sufficient to elicit an apoptotic response. Instead, the interaction of ISVPs with cellular membranes might trigger signaling events that synergize with signals produced by ISVP-receptor engagement. Membrane perturbations mediated by μ1/μ1C (25, 33, 35) also might account for the influence of the M2 gene in apoptosis magnitude. In support of this idea, incubation of reovirus strain T3D with a μ1/μ1C-specific monoclonal antibody reduces levels of reovirus-induced apoptosis in L cells (48). Membrane disruption mediated by μ1/μ1C may result in activation of lipid second messengers that function to promote apoptosis (24, 39), as is observed with Sindbis virus (28, 29).
Regardless of the precise biochemical mechanism used by reovirus virions to induce apoptosis, it is clear that reovirus receptors do not initiate the signaling events that elicit apoptosis from the cell surface, but rather from endocytic vesicles. Since it is possible that ISVPs enter cells by endocytosis or direct penetration of cell membranes, these particles may be capable of activating cellular signaling pathways at either the cell surface or within endosomes. Studies of heterotrimeric guanine nucleotide-binding protein-coupled receptors and receptor tyrosine kinases (e.g., the epidermal growth factor receptor) (49) demonstrate that endocytosis plays a key role in determining the capacity of these receptors to initiate intracellular signaling as well as the potency of the signal induced (9). Endocytosis can influence receptor-initiated signaling by regulating receptor trafficking or properly localizing activated receptors near downstream signaling components (9). Similar effects also may be operant in the signaling events induced by the binding of reovirus to its receptors.
Findings reported here demonstrate that viral disassembly is an essential determinant of the outcome of reovirus infection and provide the first evidence that events subsequent to viral attachment are critical in mediating an apoptotic response. This work now establishes a foundation to define the signaling events that occur following reovirus engagement of sialic acid and JAM that culminate in apoptosis.
Acknowledgments
We thank Jim Chappell and Craig Forrest for critical review of the manuscript and Nikkia Tyler for expert technical assistance.
This work was supported by Public Health Service award AI50080 from the National Institute of Allergy and Infectious Diseases and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 for the Vanderbilt Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
REFERENCES
- 1.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, G. S., D. H. Ebert, C. J. Chung, A. H. Erickson, and T. S. Dermody. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73:9532-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, A. J., A. A. Kembhavi, M. A. Brown, H. Kirschke, C. G. Knight, M. Tamai, and K. Hanada. 1982. l-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 201:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multi-step adhesion-strengthening. J. Biol. Chem. 276:2200-2211. [DOI] [PubMed] [Google Scholar]
- 5.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 6.Berndt, C., B. Mopps, S. Angermuller, P. Gierschik, and P. H. Krammer. 1998. CXCR4 and CD4 mediate a rapid CD95-independent cell death in CD4+ T cells. Proc. Natl. Acad. Sci. USA 95:12556-12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 8.Canning, W. M., and B. N. Fields. 1983. Ammonium chloride prevents lytic growth of reovirus and helps to establish persistent infection in mouse L cells. Science 219:987-988. [DOI] [PubMed] [Google Scholar]
- 9.Ceresa, B. P., and S. L. Schmid. 2000. Regulation of signal transduction by endocytosis. Curr. Opin. Cell Biol. 12:204-210. [DOI] [PubMed] [Google Scholar]
- 10.Chappell, J. D., J. Duong, B. W. Wright, and T. S. Dermody. 2000. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 74:8472-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly, J. L., E. S. Barton, and T. S. Dermody. 2001. Reovirus binding to cell-surface sialic acid potentiates virus-induced apoptosis. J. Virol. 75:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly, J. L., S. E. Rodgers, P. Clarke, D. W. Ballard, L. D. Kerr, K. L. Tyler, and T. S. Dermody. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J. Virol. 74:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs, K. M. 1998. Temperature-sensitive mutants of reovirus. Curr. Top. Microbiol. Immunol. 233:69-107. [DOI] [PubMed] [Google Scholar]
- 14.Cross, R. K., and B. N. Fields. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. J. Virol. 50:799-809. [DOI] [PubMed] [Google Scholar]
- 15.DeBiasi, R., C. Edelstein, B. Sherry, and K. Tyler. 2001. Calpain inhibition protects against virus-induced apoptotic myocardial injury. J. Virol. 75:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dermody, T. S., M. L. Nibert, R. Bassel-Duby, and B. N. Fields. 1990. A ς1 region important for hemagglutination by serotype 3 reovirus strains. J. Virol. 64:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dryden, K. A., D. L. Farsetta, G. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 245:33-46. [DOI] [PubMed] [Google Scholar]
- 18.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 20.Fields, B. N., and W. K. Joklik. 1969. Isolation and preliminary genetic and biochemical characterization of temperature-sensitive mutants of reovirus. Virology 37:335-342. [DOI] [PubMed] [Google Scholar]
- 21.Fields, B. N., R. Laskov, and M. D. Scharff. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of peptides. Virology 50:209-215. [DOI] [PubMed] [Google Scholar]
- 22.Fields, B. N., C. S. Raine, and S. G. Baum. 1971. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology 43:569-578. [DOI] [PubMed] [Google Scholar]
- 23.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannun, Y. A., and C. Luberto. 2000. Ceramide in the eukaryotic stress response. Trends Cell Biol. 10:73-80. [DOI] [PubMed] [Google Scholar]
- 25.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikegami, N., and P. J. Gomatos. 1968. Temperature-sensitive conditional-lethal mutants of reovirus 3. I. Isolation and characterization. Virology 36:447-458. [DOI] [PubMed] [Google Scholar]
- 27.Ito, Y., and W. K. Joklik. 1972. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology 50:189-201. [DOI] [PubMed] [Google Scholar]
- 28.Jan, J. T., S. Chatterjee, and D. E. Griffin. 2000. Sindbis virus entry into cells triggers apoptosis by activating sphingomyelinase, leading to the release of ceramide. J. Virol. 74:6425-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jan, J. T., and D. E. Griffin. 1999. Induction of apoptosis by Sindbis virus occurs at cell entry and does not require viral replication. J. Virol. 73:10296-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lee, P. W., E. C. Hayes, and W. K. Joklik. 1981. Protein sigma 1 is the reovirus cell attachment protein. Virology 108:156-163. [DOI] [PubMed] [Google Scholar]
- 32.Lin, K. I., J. A. DiDonato, A. Hoffmann, J. M. Hardwick, and R. R. Ratan. 1998. Suppression of steady-state, but not stimulus-induced NF-κB activity inhibits alphavirus-induced apoptosis. J. Cell Biol. 141:1479-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nibert, M. L., J. D. Chappell, and T. S. Dermody. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved ς1 protein. J. Virol. 69:5057-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberhaus, S. M., R. L. Smith, G. H. Clayton, T. S. Dermody, and K. L. Tyler. 1997. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol. 71:2100-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Brien, V. 1998. Viruses and apoptosis. J. Gen. Virol. 79:1833-1845. [DOI] [PubMed] [Google Scholar]
- 38.Ohnimus, H., M. Heinkelein, and C. Jassoy. 1997. Apoptotic cell death upon contact of CD4+ T lymphocytes with HIV glycoprotein-expressing cells is mediated by caspases but bypasses CD95 (Fas/Apo-1) and TNF receptor 1. J. Immunol. 159:5246-5252. [PubMed] [Google Scholar]
- 39.Okazaki, T., T. Kondo, T. Kitano, and M. Tashima. 1998. Diversity and complexity of ceramide signalling in apoptosis. Cell. Signal. 10:685-692. [DOI] [PubMed] [Google Scholar]
- 40.Rankin, U. T., Jr., S. B. Eppes, J. B. Antczak, and W. K. Joklik. 1989. Studies on the mechanism of the antiviral activity of ribavirin against reovirus. Virology 168:147-158. [DOI] [PubMed] [Google Scholar]
- 41.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a ς1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen, Y., and T. E. Shenk. 1995. Viruses and apoptosis. Curr. Opin. Genet. Dev. 5:105-111. [DOI] [PubMed] [Google Scholar]
- 44.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanism of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 45.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top components and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 46.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Traenckner, E. B., H. L. Pahl, T. Henkel, K. N. Schmidt, S. Wilk, and P. A. Baeuerle. 1995. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 14:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyler, K. L., M. K. T. Squier, S. E. Rodgers, B. E. Schneider, S. M. Oberhaus, T. A. Grdina, J. J. Cohen, and T. S. Dermody. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by viral attachment protein ς1. J. Virol. 69:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira, A., C. Lamaze, and S. Schmid. 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274:2086-2089. [DOI] [PubMed] [Google Scholar]
- 50.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner, H. L., R. F. Ramig, T. A. Mustoe, and B. N. Fields. 1978. Identification of the gene coding for the hemagglutinin of reovirus. Virology 86:581-584. [DOI] [PubMed] [Google Scholar]
- 52.Wikowski, J. T., R. K. Robins, and R. W. Sidwell. 1972. The design, synthesis and broad-spectrum antiviral activity of 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide and related nucleosides. J. Med. Chem. 15:1150-1154. [DOI] [PubMed] [Google Scholar]
- 53.Wyllie, A. H. 1997. Apoptosis: an overview. Br. Med. Bull. 53:451-465. [DOI] [PubMed] [Google Scholar]