Abstract
This study examines the persistence and fitness of multidrug-resistant (MDR) viruses acquired during primary human immunodeficiency virus infection (PHI). In four individuals, MDR infections persisted over the entire study period, ranging from 36 weeks to 5 years, in the absence of antiretroviral therapy. In stark contrast, identified source partners in two cases showed expected outgrowth of wild-type (WT) virus within 12 weeks of treatment interruption. In the first PHI case, triple-class MDR resulted in low plasma viremia (1.6 to 3 log copies/ml) over time compared with mean values obtained for an untreated PHI group harboring WT infections (4.1 to 4.3 log copies/ml). Increasing viremia in PHI patient 1 at week 52 was associated with the de novo emergence of a protease inhibitor-resistant variant through a recombination event involving the original MDR virus. MDR infections in two other untreated PHI patients yielded viremia levels typical of the untreated WT group. A fourth patient's MDR infection yielded low viremia (<50 to 500 copies/ml) for 5 years despite his having phenotypic resistance to all antiretroviral drugs in his treatment regimen. In two of these PHI cases, a rebound to higher levels of plasma viremia only occurred when the M184V mutation in reverse transcriptase could no longer be detected and, in a third case, nondetection of M184V was associated with an inability to isolate virus. To further evaluate the fitness of MDR variants acquired in PHI, MDR and corresponding WT viruses were isolated from index and source partners, respectively. Although MDR viral infectivity (50% tissue culture infective dose) was comparable to that observed for WT viruses, MDR infections in each case demonstrated 2-fold and 13- to 23-fold reductions in p24 antigen and reverse transcriptase enzymatic activity, respectively. In dual-infection competition assays, MDR viruses consistently demonstrated a marked replicative disadvantage compared with WT virus. These results indicate that MDR viruses that are generated following PHI can establish persistent infections as dominant quasispecies despite their impaired replicative competence.
The transmission of dual- and triple-class multidrug resistance (MDR) to newly infected persons has been previously reported in several primary human immunodeficiency virus (HIV) infection (PHI) cohort studies (3, 17, 25, 34). The virological implications are of serious concern, since MDR can result in treatment failure and clinical progression (2, 5, 11, 15, 22, 25, 30, 32). Indeed, recent case reports indicate that PHI patients with MDR virus do not respond well to antiretroviral drugs (ARVs) (3, 14, 17, 25, 34).
Nevertheless, cumulative findings indicate that MDR viruses generally show diminished replicative capacity both in vivo and in vitro and should be outcompeted by drug-sensitive wild-type (WT) virus (8, 12, 13, 18-21, 27, 33; S. G. Deeks, T. Wrin, R. Hoh, J. Troiano, T. Liegler, M. Hayden, C. Petropoulos, N. Hellmann, J. Barbour, R. M. Grant, J. M. McCune, and M. Hellerstein, Abstr. 7th Conf. Retroviruses Opportunistic Infect., abstr. LB10, p. 236, 2000). In individuals developing MDR infections following prolonged ARV therapy, treatment interruption leads to the rapid reappearance of WT virus within 12 weeks (8, 10, 31; Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect.). On the basis of the results in chronic infection, it would be expected that MDR infections acquired during PHI should not persist over time in the absence of ARV therapy.
To better understand the virological implications of MDR in the PHI setting, this study evaluated the persistence and replicative fitness of MDR virus in four newly infected individuals and their identified source partners. The replicative capacity of MDR viruses in each instance was assessed in vitro and correlated with longitudinal virological progression. Our findings show that despite impaired replicative fitness, MDR variants transmitted in PHI persist over time in the absence of ARV therapy.
This study had two components. The first was an observational study of the progression of transmitted MDR infections in four patients from the Montreal PHI cohort (25). Three of the patients acquired their infections from identified source partners and remained untreated for the duration of study (9 months to 1 year). The fourth patient was monitored over 5 years while on ARV combination therapy. The plasma viremia (HIV RNA levels), CD4 cell count, and genotypic drug resistance profile were assessed at specified study visits (25).
The second aspect of this study was to isolate pure MDR and WT viruses from each case and to compare their replicative capacities and fitnesses. We attempted to isolate MDR virus from peripheral blood mononuclear cells (PBMCs) of PHI patients at all time points. In addition, MDR and WT viruses were isolated from identified source partners at the time of initial transmission and following planned treatment interruptions, respectively.
(The research performed by Marco Petrella was in partial fulfillment of the requirements for the Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montreal, Canada.)
MATERIALS AND METHODS
Genotypic and other analyses.
Blood samples were obtained at designated study visits with written informed consent. Following PHI, patients initiated or deferred ARV treatment based on decisions of the primary health care provider. Plasma HIV RNA was measured at each study visit by using the Amplicor assay (Roche Diagnostics, Mississauga, Canada) (25). CD4 cell numbers were determined by flow cytometric analysis (25).
Viral RNA was isolated from plasma samples by using the QIAamp viral extraction kit (Qiagen Inc., Chatsworth, Calif.). TRUGENE HIV type 1 (HIV-1) genotyping was used in conjunction with the Open Gene automated DNA sequencing system (Visible Genetics Inc., Toronto, Canada) to sequence the protease and reverse transcriptase (RT) regions of HIV-1 cDNA (25). In samples with low viremia (<500 RNA copies/ml of plasma), virus from plasma supernatants was concentrated by ultracentrifugation prior to RNA extraction and genotypic analysis (J. Lawrence, R. M. Lloyd, W. F. McCarthy, L. M. Hough, P. M. Feorino, and M. A. Thompson, presented at the Conference of the Infectious Diseases Society of America, Philadelphia, Pa., 18-21 November 1999). For some plasma samples, the line probe assay (LiPA) (kindly provided by Innogenetics Inc., Norcross, Ga.) was used as previously described to independently assess coexistence of WT and resistance codons in the RT gene at positions 41, 69, 70, 74, 184, 214, and 215 (26, 29). LiPA testing can detect variants present as 5% of quasispecies, compared with a 25% cutoff for sequencing analysis (26, 29). In all PHI cases, the presence of MDR or WT virus was confirmed using a recombinant virus phenotypic drug resistance assay (HIV PhenoSense; ViroLogic Inc., San Francisco, Calif.).
The degree of DNA sequence homology between viral isolates at different time points in PHI patients and source partners was ascertained. This included analysis of changes in resistance-conferring mutations, as well as silent mutations and natural polymorphisms.
Isolation of MDR and WT viral isolates.
We attempted to isolate virus from PHI patients and source partners at all study visits. PBMCs were isolated by Ficoll-Hypaque centrifugation and depleted of CD8 suppressor cells as previously described (4). Virus was amplified by coculture of PBMCs with phytohemagglutinin (PHA)- and interleukin-2 (IL-2)-prestimulated cord blood mononuclear cells (CBMCs) (24, 25). Cultures were grown for 4 weeks and replenished with fresh CBMCs every week. Levels of p24 antigen in the cocultures were monitored weekly (24, 25). After 4 weeks, p24-positive isolates were serially passaged in PHA- and IL-2-stimulated CBMCs until amplified viral stocks produced maximal levels of virus, as measured by p24 antigen capture and RT enzymatic assays (24, 25). Aliquots of amplified viral stocks were frozen at −70°C until required.
Pure MDR and WT isolates were obtained from PHI patients or identified source partners. Genotypic analysis of these isolates revealed no nucleotide sequence drift over time (>6 months of coculture). The infectivity of amplified viral stocks was determined in serially diluted samples of CBMCs, yielding 50% tissue culture infectivity doses (TCID50), measured both by p24 antigen capture assays and RT enzymatic assays (16, 24).
Phenotypic drug susceptibility testing.
The drug susceptibilities of viral isolates were measured by determining the extent to which select ARVs inhibited in vitro HIV replication (25). Briefly, CBMCs infected with pure MDR or WT isolates were grown in 96-well culture plates in both the absence and presence of serially diluted ARVs. After 7 days, RT enzymatic assays were used to determine the 50% drug inhibitory concentration (IC50), using Prism analytic software (GraphPad Inc.) (25). The observed IC50s determined for MDR or WT viral isolates were compared to known IC50s for WT drug-susceptible and drug-resistant laboratory control strains. The drugs used for phenotypic testing included zidovudine (ZDV), lamuvidine (3TC), abacavir (ABC), didanosine (ddI), delaviridine (DLV), nevirapine (NVP), efavirenz (EFV), saquinavir (SQV), indinavir (IND), nelfinavir (NFV), and amprenavir (APV) (22, 23). The GlaxoSmithKline, Boerhinger-Ingelheim, Agouron, Merck, DuPont, and Roche pharmaceutical companies, respectively, kindly provided 3TC, ABC, and APV; NVP; NFV; IND; EFV; and SQV.
Measurements of viral replicative capacity.
MDR and WT viral isolates were used to establish infections in CBMCs at equivalent multiplicities of infection (0.005 or 0.01) based on TCID50 infectivity values (16, 24). The replicative capacities of MDR and WT viral stocks were assessed on the basis of p24 antigen levels and RT enzymatic activities in culture supernatants at weekly rounds of infection. The kinetics of viral production in MDR or WT infections were similar, with p24 antigen and RT levels peaking at 6 to 7 days postinfection (unpublished data). Thereafter, new rounds of infection were initiated by the addition of fresh CBMCs at weekly serial passage (24).
The relative fitness of MDR and WT isolates was evaluated experimentally by dual-infection competition assays. Initial infections in CBMCs were established with MDR and WT viruses mixed at ratios ranging from 10:90 to 98:2, based on p24 antigen levels. In case studies 1 and 3, competitive fitness assays were performed with genotypically matched MDR and WT isolates. In cases 2 and 4, the replicative capacity of MDR virus was compared to that of WT virus obtained from case 1. Following infection, cells were washed three times with RPMI 1640 medium and grown in complete RPMI 1640 medium (24, 25). Cells were fed twice weekly and replenished weekly with fresh PHA- and IL-2-stimulated CBMCs to initiate new rounds of infections. Prior to weekly passage, RT enzymatic activity and p24 antigen levels in cell-free culture supernatants were monitored. In addition, proviral DNA was isolated from PBMCs and sequenced to determine the presence of WT, MDR, or mixed quasispecies.
RESULTS
Persistence of MDR infections acquired in PHI.
To date, 129 individuals have been recruited into the Montreal primary HIV cohort. Resistance mutations were observed in eight PHI cases (presently 6.2% of cases) (Table 1). These mutations conferred expected dual- or triple-class resistance to ARVs, as measured in recombinant vector phenotypic assays. The K101E mutation was also observed to result in low-level (6- to 10-fold) resistance to nonnucleoside RT inhibitors (NNRTIs). The present study monitored the persistence and clinical progression of MDR infections in four patients, three of whom had not initiated ARV therapy.
TABLE 1.
Genotypic profiles of patients acquiring MDR in PHI
| Patient | Codon(s) associated with resistance toa:
|
||
|---|---|---|---|
| NRTIs | NNRTIs | PIs | |
| 1 | 41L, 67N, 69N, 70R, 74V, 184V, 215F, 219Q | 100I, 103N | 10I, 36I, 54V, 63P, 71V, 73S, 82I, 90M |
| 2 | 41L, 184V, 215Y | 103N, 179E | 48V, 63P, 71V, 73S, 77I, 82A, 90M |
| 3 | None | 103N | 10I, 54V, 63P, 71V, 82T, 84V, 90M |
| 4 | 184V, 215Y | 103N | 63S, 71V |
| 6 | 41L, 67N, 210W, 215Y | None | 63P, 71V, 73S, 90M |
| 7 | 184V | 108I | 20R, 71V |
| 8 | 41L, 215Y | 101E | None |
| 9 | 41L, 215Y | 101E | None |
In all cases of resistance except that of patient 4 to PIs, dual- or triple-class MDR was confirmed by means of recombinant phenotypic testing of drug susceptibility.
Patient 1, a 30-year-old male, acquired a three-class MDR infection in February 1999 from a male partner infected since 1991. At PHI presentation, genotypic analysis revealed that viruses from both partners showed 99.2% sequence identity, with 18 mutations conferring resistance to ZDV, 3TC, and all commercialized NNRTIs and protease inhibitors (PIs) (Table 2). This MDR infection persisted in the newly infected partner for up to 36 weeks (Table 2). LiPA testing showed an absence of WT RT codons at these time points.
TABLE 2.
Persistence of MDR viral quasispecies over time in untreated PHI patients who acquired MDR from identified source partners
| Wk post-PHIa | Patient | Codon(s) associated with genotypic resistance tob:
|
||
|---|---|---|---|---|
| NRTIs | NNRTIs | PIs | ||
| 16* | PHI 1 | 41L, 67N, 69N, 70R, 74V, 184V, 215F, 219Q | 100I, 103N | 10I, 36I, 54V, 63P, 71V, 73S, 82I, 90M |
| 24 | PHI 1 | 41L, 67N, 69N, 74V, 70R, 184V, 215F, 219Q | 100I, 103N | 10I, 63P, 71V, 73S, 90M |
| 30 | PHI 1 | 41L, 67N, 69N, 74V, 70R, 184V, 215F, 219Q | 100I, 103N | 10I, 63P, 71V, 73S, 90M |
| 42 | PHI 1 | 70K/R, 184V | None | 10I, 63P, 71V, 73S, 90M |
| 44 | PHI 1 | 184V | None | 10I, 63P, 71V, 73S, 90M |
| 52 | PHI 1 | 184M/V | None | 10I, 63P, 71V, 73S, 90M |
| 16* | Source 1 | 41L, 67N, 69N, 70R, 74V, 184V, 215F, 219Q | 100I, 103N | 10I, 36I, 54V, 63P, 71V, 73S, 82I/V, 90M |
| Post-TI | Source 1 | None | None | None |
| 8* | PHI 2 | 41L, 184V, 215Y | 103N, 179E | 48V, 63P, 71V, 73S, 77I, 82A, 90M |
| 12 | PHI 2 | 41L, 184V, 215Y | 103N, 179E | 48G/V, 63P, 71V, 77I, 82A, 90M |
| 24, 28 | PHI 2 | 41L, 184V, 215Y | 103N, 179E | 63P, 71V, 77I, 82A, 90M |
| 32 | PHI 2 | 41L, 184V/M, 215Y | 103N, 179E | 63P, 71V, 77I, 82A, 90M |
| 36 | PHI 2 | 41L, 215Y | 103N, 179E | 63P, 71V, 77I, 82A, 90M |
| 47 | PHI 2 | 41L, 215C | 103N, 179E | 63P, 71V, 77I, 82A, 90M |
| 8* | Source 2 | 41L, 184V; 215Y | 103N; 179E | 48V, 63P, 71V, V77I, 73S, 82A, 90M |
| 14* | PHI 3 | None | 103N | 10I, 54V, 63P, 71V, 82T, 84V, 90M |
| 18-28 | PHI 3 | None | 103N | 10I, 54V, 63P, 71V, 82T, 84V, 90M |
| 32,36 | PHI 3 | None | 103N | 10I, 54V, 63P, 71V, 82T, 90M |
| 14* | Source 3 | None | 181C/Y, 103N | 10I, 46I, 54V, 63P, 71V, 82T, 84V, 90M |
| Post-TI | Source 3 | None | None | 63P |
| 10-234 | PHI 4 | 184V, 215Y | 103N | A71V, L63S |
| 234-250 | PHI 4 | 184V, 215Y | 103N, V108I | A71V, L63S |
| 8-20 | PHI 5 | 184V | None | V771 |
| 24 | PHI 5 | 184M/V | None | V771 |
| 26-36 | PHI 5 | None | None | V771 |
Genotypic analysis of plasma viral RNA was performed for PHI patients 1 to 5 at various times after the estimated date of initial infection (*), as well as for source partners at the time of initial PHI presentation (*) and following treatment interruption (TI).
In all cases of genotypic resistance except those of source partner 3 post-TI and of PHI patients 4 and 5 to PIs, viruses harboring phenotypic resistance to relevant NRTIs and NNRTIs or PIs were present.
Thereafter, genotypic analysis revealed a novel PI-resistant viral species in which all NNRTI and nucleoside RT inhibitor (NRTI) mutations except M184V were lost. It is presumed that a recombination event at week 42 gave rise to this virus, which demonstrated 96.9% sequence homology to the original MDR isolate. At week 52, partial reversion of codon 184 to WT led to M184M/V mixed quasispecies (Table 2). This dual MDR and PI-resistant infection persisted for over 1 year in newly infected patient 1. In contrast, rapid outgrowth of WT virus was observed in the source partner within 10 weeks following a planned treatment interruption (Table 1).
Patient 2, a 43-year-old male, acquired HIV-1 in April 1999 from a female partner who had been HIV positive since 1993. Both partners were infected with the same MDR variants (99.3% homology) with 12 mutations conferring phenotypic resistance to ZDV, 3TC, NNRTIs, and PIs (Tables 2 and 3). LiPA analysis confirmed the absence of WT codons at all NRTI codons in both partners. Apart from the reversion of M184V at week 36, this MDR infection persisted in PHI patient 2 for over 47 weeks in the absence of ARV therapy (Table 2).
TABLE 3.
Genotypes and phenotypes of viral stocks isolated from PHI patients and identified source partners
| Viral isolatesa | Viral replicative capacityb
|
IC50 (μM)c
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCID50 (log) | p24 (ng) | RT activity (cpm) | ZDV | 3TC | ABC | DLV | EFV | NVP | IDV | SQV | NFV | |
| Lab WT isolates (n = 12) | 6.17 ± 0.32 | 365 ± 47 | 225,700 ± 57,780 | 0.001 | 0.02 | 0.01 | 0.01 | 0.0001 | 0.02 | 0.002 | 0.002 | 0.002 |
| Lab MDR isolates (n = 8) | 5.38 ± 0.18 | 151 ± 22* | 46,880 ± 8,351* | |||||||||
| SP 1 MDR (wk 16 post-PHI) | 6.22 | 267 | 42,140 ± 6,240* | 0.010d | >100d | 0.08d | 10d | 0.832d | 1.28d | 0.451d | 0.422d | NDe |
| SP 1 WT (post-TI) | 6.60 | 481 | 548,400 ± 16,240 | 0.001 | 0.01 | 0.01 | 0.01 | 0.0001 | 0.003 | 0.003 | 0.002 | ND |
| PHI 2 MDR (wk 47 post-PHI) | 5.20 | 183 | 21,120 ± 6,667* | 0.002 | 0.02 | 0.01 | 0.62d | 0.151d | 1.82d | 0.017d | 0.015d | ND |
| PHI 3 MDR (wk 14 post-PHI) | 5.60 | 92 | 41,120 ± 7,597* | 0.002 | 0.02 | ND | 3.15d | 0.013d | >2d | 0.068d | ND | 0.069d |
| SP 3 MDR (wk 14 post-PHI) | 6.00 | 236 | 65,572 | 0.002 | 0.02 | ND | 4.42d | 0.010d | >2d | 1.152d | ND | 0.176d |
| SP 3 WT (post-TI) | 6.00 | 289 | 251,800 ± 9,035 | 0.001 | 0.01 | ND | 0.01 | 0.0001 | 0.03 | 0.002 | ND | 0.002 |
| PHI 4 MDR (wk 250 post-PHI) | 5.00 | 29 | 23,928 | 0.006d | >20d | ND | >20d | 0.320d | >2d | 0.015d | 0.002 | 0.036d |
WT or MDR virus from PHI patients or source partners (SP) were isolated at the indicated times following PHI or treatment interruption (TI), with genotypic resistance mutations reported in Table 2.
Results are means and standard errors of the means. *, P < 0.01 for comparison of MDR and WT isolates.
Phenotypic sensitivity of viral isolates to to ARVs in comparison to the mean value observed for laboratory WT strains.
Phenotypic resistance.
ND, not determined.
Patient 3, a 28-year-old male, acquired his dual-class MDR infection in February 2000 from a 30-year-old male partner who had been infected since 1997. Both partners had MDR viruses showing 98.4% sequence homology, with nine identical mutations conferring resistance to NNRTIs and PIs (Tables 2 and 3). This MDR infection persisted in PHI patient 3 for at least 36 weeks. This MDR infection continued in the source partner until a planned treatment interruption led to WT outgrowth within 12 weeks (Table 2).
Thus, in all three PHI patients with persistent MDR infections, the virus showed 98.4 to 99.3% sequence homology to the predominant MDR quasispecies expressed and transmitted by their respective source partners. In contrast, source partners showed expected rapid outgrowth of WT virus within 12 weeks of treatment interruption (8, 10, 31; Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect.).
Clinical progression of untreated MDR infections.
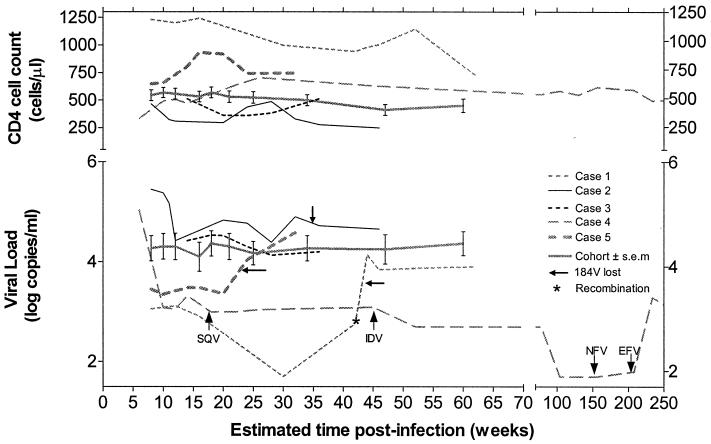
We evaluated changes in plasma HIV RNA levels and CD4 cell counts in patients acquiring MDR infections. The resulting viremia from the initial MDR infection in PHI patient 1 was several orders of magnitude lower than the viremia of the newly infected group harboring WT virus (Fig. 1). An increase in his viremia followed the emergence of the PI-resistant recombinant species at week 47 (Table 2; Fig. 1). Persistent MDR infections in PHI patients 2 and 3 yielded levels of viremia similar to those in the WT group (Fig. 1). PHI patient 5, who had the M184V and V77I mutations, was the only other untreated participant in our PHI cohort to possess unusually low viremia at presentation (2,244 copies/ml) that increased following the disappearance of M184V (Table 2; Fig. 1).
FIG. 1.
Natural history of primary MDR infections. Plasma viremia and CD4 cell counts were monitored in untreated patients 1, 2, and 3 in comparison with average values from an untreated PHI cohort harboring WT infections (n = 20; mean values ± standard errors of the means [s.e.m.] are shown). Viremia in PHI patient 4 (see Table 1) over time is also depicted, since he harbored phenotypic resistance to his treatment ARVs, including ZDV, 3TC, IDV, NFV, and EFV. Viremia in untreated PHI patient 5, in relation to his M184V infection, is also presented for comparative purposes.
Persistence and clinical progression of MDR infection in treated PHI patients.
Similar to results observed for PHI patients 1 to 3, patient 4 harbored an MDR infection that persisted for almost 5 years (Table 2). This MDR infection was associated with consistently low viremia (1 to 2 log units lower than that in the WT cohort) for over 200 weeks despite having a virus showing phenotypic resistance to ZDV, 3TC, and the PIs included in his ongoing combination regimen (Fig. 1; Table 3). It is interesting that the K103N mutation was also retained for 5 years although this patient was NNRTI naive until week 210. At week 210, NFV was substituted for EFV due to gastrointestinal intolerance to the PI. This resulted in an increase in viremia concurrent with the de novo appearance of V108I, another NNRTI resistance-conferring mutation. Remarkably, apart from this mutation, the virus at week 240 showed complete RT and PI nucleotide sequence identity to the initial MDR virus acquired at PHI. Isolation of virus at week 240 confirmed that this virus was indeed resistant to all drugs in his regimen (Table 3).
The other PHI patients, i.e., patients 6 to 9, received various ARV combination therapy regimens (Table 1). To date, viremia in these individuals has remained below 50 copies/ml, precluding genotypic analysis. Therefore, in our experience, all patients acquiring MDR infections appeared to respond to ARV therapy.
Replicative capacity of MDR isolates.
To more completely understand the basis of persistence of MDR infections following PHI, the replicative capacities and fitnesses of MDR viral isolates were investigated. We attempted to isolate MDR virus from PBMCs of PHI patients at all time points. Where possible, we isolated homologous WT isolates from source partners.
In PHI patient 1, consistently low viremia (50 to 500 copies/ml) precluded the isolation of an MDR virus. However, a homologous MDR viral isolate was obtained from PBMCs of the source partner at the time of initial infection. This MDR viral isolate demonstrated >99% nucleotide sequence homology to the MDR virus present in PHI patient 1 for 36 weeks.
This MDR isolate with 18 mutations conferring high-level resistance to ARVs showed no genotypic drift for more than 6 months (Table 3). In addition, a matched WT isolate was generated from PBMCs of the source partner following an extended treatment interruption. The WT viral isolate showed sequence homology in all nucleotides apart from the 18 resistance-conferring mutations.
There was no difference in the infectivities (TCID50) of these MDR and WT viruses (Table 3). When these two viruses were used to separately infect CBMCs at an equivalent multiplicity of infection of 0.01, levels of viral p24 were reduced by 2-fold, whereas RT enzymatic activity was 13-fold less for the MDR isolate (Table 3). This marked reduction in RT activity was consistently observed at subsequent rounds of infection despite similarity in overall levels of viral production, reflected in levels of p24 antigen at serial passage (Fig. 2).
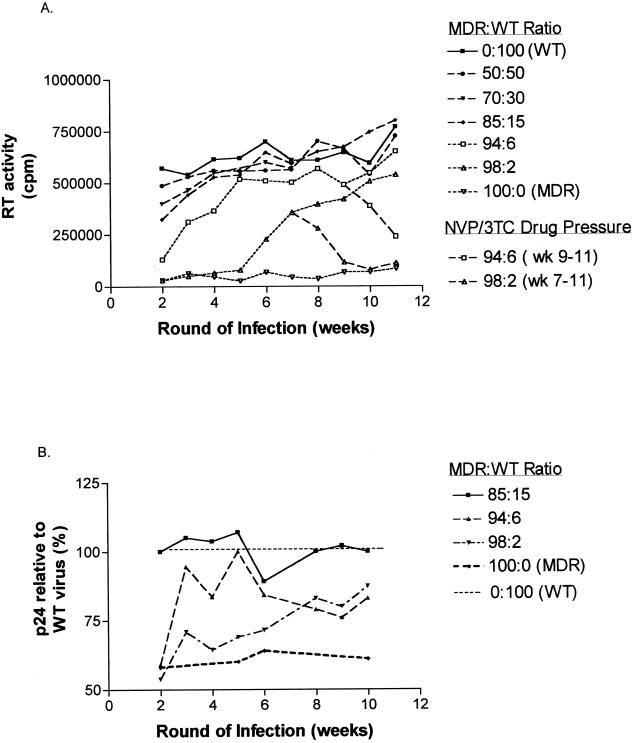
FIG. 2.
Effects of mixed MDR and WT populations on HIV-1 replicative capacity. PHA-stimulated CBMCs were coinfected with various ratios of homologous MDR and WT viral species from case 1. New rounds of infections were initiated by the weekly addition of fresh CBMCs. Proviral DNA was sequenced to determine the presence of MDR or WT virus (Table 4). Viral replicative capacity was ascertained by weekly determination of p24 antigen and RT activity in cell-free supernatants prior to addition of fresh CBMCs. (A) RT activity over time in different coinfections. In the infections with MDR/WT ratios of 94:6 and 98:2, NVP and 3TC drug pressure was reintroduced after WT virus was observed to predominate. (B) The levels of p24 antigen, expressed as percentages of those with the pure WT infection, are compared with those in pure MDR infections. In the case of infections in which MDR species represented less than 85% of the inocula, levels of p24 antigen paralleled those determined for the WT infection, as shown for the experiment with an MDR/WT ratio of 85:15.
In case study 2, MDR virus could not be expanded until the loss of M184V at week 36 (Table 1). The inability to culture virus prior to this time could not be attributed to low viremia, since plasma viral RNA was high at all study time points (Fig. 1). The MDR viral isolate isolated from PBMCs at weeks 36 and 47 showed sequence identity to plasma viral RNA in PHI patient 2, with nine mutations conferring triple-class resistance (Tables 2 and 3). Infection with this MDR isolate resulted in 10- to 26-fold reductions in RT activities relative to those of WT control viruses (Table 3). Similarly, MDR viruses expanded from both partners in PHI case 3 showed sequence identity, with eight mutations conferring phenotypic resistance to NNRTIs and PIs (Table 3). Compared to WT virus infection, MDR infections resulted in sixfold reductions in RT activity (Table 3).
Infections with MDR virus from case study 4 also yielded reduced levels of RT activity (Table 3). Thus, in all four PHI cases, the infectivity of MDR viruses was not significantly impaired, whereas levels of RT activity were markedly compromised (Table 3).
Dual-infection and competition assays with paired MDR and WT isolates.
Several laboratories have performed dual-infection experiments to compare the relative fitnesses of virus isolates (13, 18, 23). We have conducted similar studies with matched MDR and WT viral isolates in cases 1 and 3. For cases 2 and 4, dual infections were performed with WT virus isolated from case 1.
Dual infections were performed at MDR/WT ratios extending from 10:90 to 98:2. Single infections with MDR and WT viral isolates were performed in parallel for comparative purposes. Viruses were harvested weekly, and proviral DNA was sequenced prior to new rounds of infection. Levels of RT activity and p24 antigen in clarified culture supernatants were assayed at each serial passage.
Table 4 and Fig. 2 depict representative results obtained in case study 1, where MDR virus comprised 10 to 98% of the MDR-WT quasispecies mixture. Genotypic analysis of proviral DNA harvested at week 1 revealed the predicted initial mixture of WT and MDR viruses. This finding indicated that MDR and WT viruses were equally competent to establish infection. In all cases where MDR virus was present at less than 50% of the MDR-WT mixture, WT viral outgrowth occurred within 2 weeks. In experiments where MDR virus comprised greater than 90% of the mixture, MDR infections were preferentially established and subsequently persisted for six to nine rounds of infection (Table 4). Nevertheless, WT viral outgrowth eventually occurred, indicating a replicative advantage of the minority WT variant present at earlier rounds of infection.
TABLE 4.
Genotypic analysis of viral species at various weeks following dual infection with MDR and WT viruses
| Initial MDR/ WT ratioa | Wk of coculture | Mutations that confer resistance to:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NRTIs
|
NNRTIs
|
PIs
|
|||||||||||||||||||
| 41 | 67 | 69 | 70 | 74 | 184 | 215 | 219 | 100 | 103 | 10 | 36 | 54 | 63 | 71 | 73 | 82 | 90 | ||||
| 0:100 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 10:90 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 20:80 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 30:70 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 40:60 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 50:50 (WT) | 2-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 70:30 (MDR-WT) | 2-3 | M/L | D/N | T/N | K/R | L/V | M/V | T/F | K/Q | L/I | K/N | L/I | M/I | I/V | L/P | A/V | G/S | V/I | L/M | ||
| 70:30 (WT) | 4-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 85:15 (MDR-WT) | 2-3 | M/L | D/N | T/N | K/R | L/V | M/V | T/F | K/Q | L/I | K/N | L/I | M/I | I/V | L/P | A/V | G/S | V/I | L/M | ||
| 85:15 (WT) | 4-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 94:6 (MDR) | 2 | L | N | N | R | V | V | F | Q | I | N | I | I | V | P | V | S | I | M | ||
| 94:6 (MDR-WT) | 3-5 | M/L | D/N | T/N | K/R | L/V | M/V | T/F | K/Q | L/I | K/N | L/I | M/I | I/V | L/P | A/V | G/S | V/I | L/M | ||
| 94:6 (WT) | 6-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 98:2 (MDR) | 2-5 | L | N | N | R | V | V | F | Q | I | N | I | I | V | P | V | S | I | M | ||
| 98:2 (MDR-WT) | 6-8 | M/L | D/N | T/N | K/R | L/V | M/V | T/F | K/Q | L/I | K/N | L/I | M/I | I/V | L/P | A/V | G/S | V/I | L/M | ||
| 98:2 (WT) | 9-10 | M | D | T | K | L | M | T | K | L | K | L | M | I | L | A | G | V | L | ||
| 100:1 (MDR) | 2-10 | L | N | N | R | V | V | F | Q | I | N | I | I | V | P | V | S | I | M | ||
PHA-stimulated CBMCs were coinfected with MDR and WT variants at the indicated ratios. By adding fresh uninfected CBMCs weekly, ongoing infections were continued. CBMC proviral DNA was genotyped weekly for WT, MDR, or MDR-WT quasispecies mixtures as indicated.
RT activities and p24 antigen were monitored at each round of infection concomitant with genotypic analysis for the presence of MDR and/or WT quasispecies. Single infection with MDR virus yielded substantially reduced levels of RT activity at each round of infection relative to those with WT virus (Fig. 2A). In contrast, levels of viral production, measured by p24 antigen, were not severely compromised in MDR infections, since WT virus showed only a 1.4-fold replicative advantage (Fig. 2B). Therefore, the observed 13-fold reductions in RT activity could not be explained by reduced or delayed viral production.
When the proportion of MDR virus in the mixture was less than 85%, p24 antigen and RT levels were observed to reflect those produced by WT infections (Fig. 2). This observation was consistent with the rapid outgrowth of WT virus within two to four rounds of infection. When MDR virus represented greater than 94% of the infecting quasispecies, levels of RT activity at initial rounds of infection reflected the substantially reduced levels of RT activity observed for pure MDR infections (Fig. 2A). Nevertheless, by week 7, RT activity sharply increased, reflecting outgrowth of the WT minority quasispecies (Fig. 2A; Table 4).
We also examined the effect of introducing selective drug pressure in those dual infections where MDR was initially established (i.e., MDR/WT ratios of 94:6 and 98:2) but where WT outgrowth occurred following seven and nine rounds of infection, respectively. In the presence of 0.1 μM NVP and 3TC, we observed rapid reexpression of all previous MDR mutations (Table 4). As previously observed, reappearance of the initial MDR variant was associated with a reduction in RT activity (Fig. 2A).
Similarly, dual infections involving MDR and WT viruses from PHI patient 3 and his source partner revealed a replicative advantage of WT virus. MDR infections could be established only when the virus was present at greater than 90% of the infecting quasispecies. In this instance, outgrowth of the minority WT variant occurred within 5 weeks (data not shown). As previously demonstrated, the presence of MDR infections was associated with characteristic reductions in RT activity. In addition, MDR virus from PHI patients 2 and 4 showed a marked replicative disadvantage, with WT outgrowth within 3 weeks when MDR virus represented greater than 90% of the quasispecies (data not shown).
Taken together, these in vitro results suggest that WT viral outgrowth should have occurred following PHI if it was present at the initial transmission.
DISCUSSION
We have evaluated the persistence and clinical progression of MDR infections acquired in PHI both in vivo and in vitro. In the four cases studied, MDR viruses persisted in patients for 36 weeks to 5 years in the absence of selective ARV drug pressure. These data complement and contrast with previous studies that showed a rapid disappearance of MDR variants within 12 weeks following treatment interruption in individuals with long-established infections (1, 6-9, 31; Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect.).
In the PHI setting, MDR virus may represent the principal quasispecies transmitted at the time of infection. Three of the patients whom we studied acquired MDR populations that were genotypically identical, in regard to both resistance-associated mutations and polymorphisms, to those of identified source partners who had themselves developed MDR viruses while on ARVs. LiPA testing and direct sequencing of proviral DNA extracted from the PBMCs of PHI patients did not reveal archived WT virus in their viral reservoirs. Finally, in our dual-infection fitness assays, WT viruses outcompeted MDR variants when present at levels of as low as 2%. These observations further support the presumed absence of WT virus in the infecting quasispecies in PHI transmission.
These results are also consistent with data on the vertical transmission of HIV-1 resistance-associated mutations when the latter were present as a major species (7, 10, 21). When maternal viruses consisted of mixed WT and ZDV-resistant quasispecies, a WT infection was more likely to be established in infants (7). In cases in which maternal viruses showed homogeneity in regard to resistance-associated mutations, the infected infants were shown to carry drug-resistant viruses (7). Thus, circulating viral pools in source partners can influence the nature of the viral population that succeeds in establishing a new infection.
The absence of archived WT virus in our untreated PHI patients helps to explain the persistence of MDR viruses following PHI, in spite of their replicative disadvantage. Higher numbers of resistance-associated mutations within the viral genome in PHI makes the occurrence of back mutations at each relevant locus less likely. In contrast, it is not unexpected that viruses harboring a single mutation, such as M184V in case 5, reverted at a relatively early time, as also observed in cases 1 and 2.
It is interesting to note that the presence of the M184V mutation in reverse transcriptase, associated with high-level resistance to 3TC, seems to have been associated with the persistence of low viral load. In two of our PHI cases, rebounds to a high level of plasma viremia occurred only at times when the M184V mutation in RT could no longer be detected. A third PHI patient maintained low plasma viremia over 5 years, and his virus also contained the M184V mutation throughout this time. In an additional individual, high viral loads were present at times after primary infection in spite of the M184V mutation, but virus could only be isolated from this individual in coculture experiments after loss of the M184V mutation. These data are consistent with previous findings on loss of fitness conferred by the M184V mutation in reverse transcriptase, alongside multiple other pleiotropic effects, including diminished processivity, diminished rates of nucleotide excision, and diminished rates of initiation of reverse transcription (13, 33).
Other studies suggest that despite reduced ARV susceptibility, MDR infections may be of some immunological and virological benefit due to the impaired replicative capacity of MDR variants (1, 6, 8-10, 28, 31; Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect.). The observed low plasma viremia associated with MDR infection in cases 1 and 4 is consistent with this notion. Moreover, in all cases, RT assays and competitive fitness assays showed MDR viruses to have severely compromised fitness. This finding is similar to those of other studies that used recombinant vector assays and reported a fivefold reduction of MDR viral fitness (8; Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect.).
The levels of viremia observed here for cases 2 and 3 were discordant with the impaired fitnesses of the respective MDR variants. It should also be noted that the infectivity of MDR viruses (reflected by TCID50 and p24 antigen determinations) was not severely compromised. This can explain, in part, the persistence of the MDR infections that we have observed in this PHI study.
The absence of genotypic changes in these viruses over time further supports the concept of expansion of predominant MDR quasispecies during primary infection. Remarkably, a recombination event was observed to have occurred in case 1. Equally interesting was the observation in PHI patient 4 that PI resistance-conferring mutations did not accumulate over a 5-year time period despite the presence of ARV drug pressure. This may be due, in part, to the multiple pleiotypic effects of the M184V mutation, as well as the impaired replicative fitness of MDR mutated viruses (33).
In conclusion, these findings underscore that MDR viruses can be expected to yield a wide array of phenotypes, including those associated with significantly impaired viral replication. Further studies of these viruses in PHI are needed to further understand the long-term virological and clinical implications of these infections.
Acknowledgments
This research was supported by the National Institutes of Health (NIAID RO1 AI43271-01), the Canadian Institutes for Health Research (CIHR) (T-14738), and the Fonds de la Recherche en Santé du Québec, for which J.-P. Routy is a Research Scholar.
We thank Mario Legault, Jean-Pierre Fortin, Rémi Cheynier, and Jean-François Poulin for technical assistance and dedication to this project.
REFERENCES
- 1.Batisse, D., M. Karmochkine, A. Mohamed, C. Piketty, M. D. Kazatchkine, and L. Belec. 1998. Persistence of HIV-1 variants harbouring the zidovudine mutation at pol codon 215 in patients who respond to triple combination therapy. AIDS 12:824-825. [PubMed] [Google Scholar]
- 2.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, and T. C. Merigan. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 3.Boden, D., A. Hurley, L. Zhang, Y. Cao, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, B. G., C. Gryllis, M. Gornitsky, and M. A. Wainberg. 1993. Changes in natural immunity during the course of HIV-1 infection. Clin. Exp. Immunol. 93:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevenbergh, P., J. Durant, P. Halfon, P. del Giudice, V. Mondain, N. Montagne, J. M. Schapiro, C. A. Boucher, and P. Dellamonica. 2000. Persisting long-term benefit of genotype-guided treatment for HIV-infected patients failing HAART. The Viradapt Study: week 48 follow-up. Antiviral Ther. 5:65-70. [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variability, pathogenesis and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Colgrove, R. C., J. Pitt, P. H. Chung, S. L. Welles, and A. J. Japour. 1998. Selective vertical transmission of HIV-1 antiretroviral resistance mutations. AIDS 12:2281-2288. [DOI] [PubMed] [Google Scholar]
- 8.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, J. Troiano, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. New Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 10.Dickover, R. E., E. M. Garratty, S. Plaeger, and Y. J. Bryson. 2001. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durant, J. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 12.Frost, S. D. W., M. Nijhuis, R. Schuurman, C. A. B. Boucher, and A. J. Leigh-Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrigan, P. R., S. Bloor, and B. A. Larder. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 70:5930-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht, G. M., R. M. Grant, C. J. Petropoulos, B. Dillon, M. A. Chesney, H. Tien, N. S. Hellman, N. I. Bandrapalli, L. Digilio, B. Branson, J., and O. Kahn. 1998. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N. Engl. J. Med. 339:307-311. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch, M. S., B. Conway, R. T. D'Aquila, V. A. Johnson, F. Brun-Vézinet, C. Clotet, L. M. Demeter, S. M. Hammer, D. M. Jacobson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, S. Vella, and D. D. Richman. 1998. Antiretroviral drug resistance testing in adults with HIV infection. Implications for clinical management. JAMA 279:1984-1991. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, V. A., and R. E. Byington. 1994. Quantitative assays for virus infectivity, p. 71-76 In ACTG virology manual for HIV laboratories. National Institute for Allergy and Infectious Diseases, Bethesda, Md.
- 17.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick. J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 18.Marée, A. F. M., W. Keulen, C. A. B. Boucher, and R. J. de Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. d'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 21.Niehues, T., H. Walter, G. Homeff, V. Wahn, and B. Schmidt. 1999. Selective vertical transmission of HIV: lamuvidine-resistant maternal clone undetectable by conventional resistance testing. AIDS 13:2482-2484. [DOI] [PubMed] [Google Scholar]
- 22.Nijhuis, M., R. Schurrman, D. de Jong, J. Erikson, E. Gustchina, J. Albert, P. Schipper, S. Gulnick, and C. A. B. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 23.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomon, H., A. Belmonte, K. Nguyen, Z. Gu, M. Gelfand, and M. A. Wainberg. 1994. Comparison of cord blood and peripheral blood mononuclear cells as targets for viral isolation and drug sensitivity studies involving human immunodeficiency virus type 1. J. Clin. Microbiol. 32:2000-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salomon, H., M. A. Wainberg, B. G. Brenner, Y. Quan, D. Rouleau, P. Coté, R. Leblanc, E. Lefebvre, B. Spira, C. Tsoukas, R.-P. Sekaly, B. Conway, D. Mayers, J.-P. Routy, and Investigators of the Quebec Primary Infection Study. 2000. Prevalence of HIV-1 viruses resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or intravenous drug use. AIDS 14:F17-F23. [DOI] [PubMed] [Google Scholar]
- 26.Servais, J., C. Lambert, E. Fontaine, J.-M. Plesséria, I. Robert, V. Arendt, T. Staub, F. Schneider, R. Hemmer, G. Burtonboy, and J.-C. Schmit. 2001. Comparison of DNA sequencing and a line probe assay for detection of a human immunodeficiency virus type 1 drug resistance mutations in patients failing highly active antiretroviral therapy. J. Clin. Microbiol. 39:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of a human immunodeficiency virus type 1 didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, M. S., K. L Koerber, and J. S. Pagano. 1994. Long term persistence of zidovudine resistance mutations of plasma isolates of human immunodeficiency virus type 1 of dideoxyinosine treated patients removed from zidovudine therapy. J. Infect. Dis. 169:184-188. [DOI] [PubMed] [Google Scholar]
- 29.Stuyver, L., A. Wyseur, A. Rombout, J. Iouwagie, T. Scarcez, C. Verhofstede, D. Rimland, R. F. Schinazi, and R. Rossau. 1997. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob. Agents Chemother. 41:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Vaerenbergh, K., K. Van Laethem, E. Van Wijngaerden, J. C. Schmit, F. Schneider, L. Ruiz, B. Clotet, C. Verhofstede, F. Van Wanzeele, G. Muyldermans, P. Simons, L. Stuyver, P. Hermans, C. Evans, E. De Clercq, J. Desmyter, and A. M. Vandamme. 2000. Baseline HIV type 1 genotypic resistance to a newly added nucleoside analog is predictive of virologic failure of the new therapy. AIDS Res. Hum. Retroviruses 16:529-537. [DOI] [PubMed] [Google Scholar]
- 31.Verhofstede, C., F. V. Wanzeele, B. Van Der Gucht, N. De Cabooter, and J. Plum. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541-2546. [DOI] [PubMed] [Google Scholar]
- 32.Wainberg, M. A., and G. Friedland. 1998. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 279:1977-1983. [DOI] [PubMed] [Google Scholar]
- 33.Wainberg, M. A., M. Hsu, Z. Gu, G. Borkow, and M. A. Parniak. 1996. Effectiveness of 3TC in HIV clinical trials may be due in part to the M184V substitution in 3TC-resistant HIV-1 reverse transcriptase. AIDS 10:S3-S10. [DOI] [PubMed] [Google Scholar]
- 34.Yerly, S., L. Kaiser, E. Race, J.-P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]