Abstract
We recently reported the first crystal structure of a paramyxovirus hemagglutinin-neuraminidase (HN) from Newcastle disease virus. This multifunctional protein is responsible for binding to cellular sialyl-glycoconjugate receptors, promotion of fusion through interaction with the second viral surface fusion (F) glycoprotein, and processing progeny virions by removal of sialic acid from newly synthesized viral coat proteins. Our structural studies suggest that HN possesses a single sialic acid recognition site that can be switched between being a binding site and a catalytic site. Here we examine the effect of mutation of several conserved amino acids around the binding site on the hemagglutination, neuraminidase, and fusion functions of HN. Most mutations around the binding site result in loss of neuraminidase activity, whereas the effect on receptor binding is more variable. Residues E401, R416, and Y526 appear to be key for receptor binding. The increase in fusion promotion seen in some mutants that lack receptor binding activity presents a conundrum. We propose that in these cases HN may be switched into a fusion-promoting state through a series of conformational changes that propagate from the sialic acid binding site through to the HN dimer interface. These results further support the single-site model and suggest certain residues to be important for the triggering of fusion.
Viruses belonging to the family Paramyxoviridae are major causative agents for respiratory illnesses in humans, particularly in children. Members of the Paramyxovirinae subfamily include the human parainfluenza viruses (PIVs), mumps viruses, Newcastle disease virus (NDV), Sendai virus, and simian virus 5. Infection of host cells by paramyxoviruses is accomplished by the interaction of two surface glycoproteins, hemagglutinin-neuraminidase (HN) and the fusion (F) protein. HN possesses both the receptor recognition of sialic acid at the termini of host glycoconjugates and neuraminidase activity to hydrolyze sialic acid from progeny virion particles to prevent viral self-aggregation (14, 23, 24). In addition to these activities, HN has been shown to promote fusion through its interaction with the F protein, which involves residues from the stalk and the globular head region of HN (1, 2, 8, 26, 31, 34), thereby allowing the entry of viral RNA.
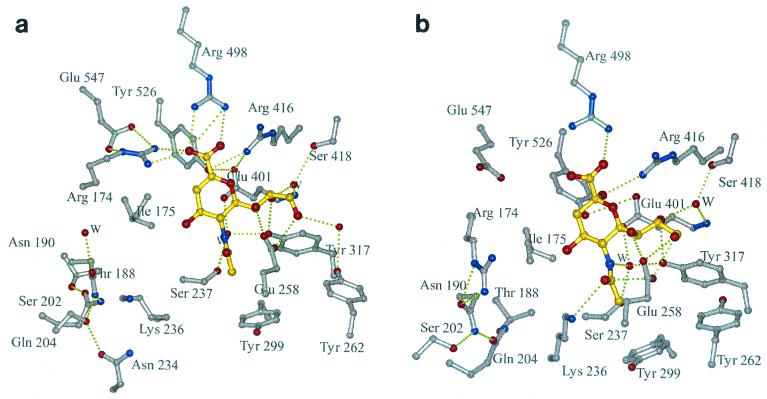
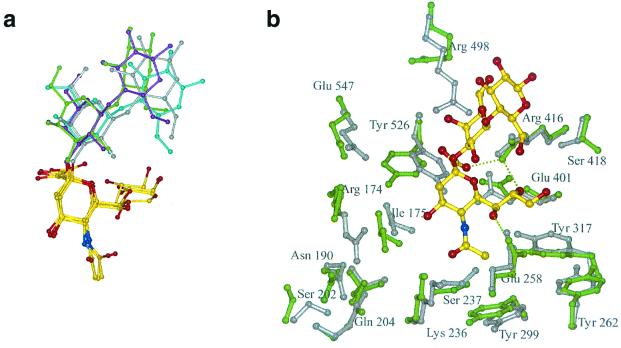
Recently, we determined the first crystal structure of the globular head region of the Newcastle disease virus HN (6). HN displays the six-bladed β-propeller fold typical of other sialidases/neuraminidases, whose structures are known (5, 7, 9, 35). Two crystal forms of the dimeric HN molecule were determined: a pH 6.5 hexagonal crystal form that would only grow in the presence of the inhibitor 2-deoxy-2,3-dehydro-n-acetyl-neuraminic acid (Neu5Ac2en) and a pH 4.6 orthorhombic crystal form grown in the absence of ligand but into which ligands could be soaked. The association of monomers in these two forms is very different, suggesting a flexible interface between the monomers that may have functional significance. The pH 6.5 inhibitor complex revealed an active site with many features in common with other sialidases/neuraminidases (Fig. 1a): a triarginyl cluster (R174, R416, and R498) that encircles the carboxyl moiety of the inhibitor, a glutamic acid (E547) that stabilizes R174, a tyrosine residue (Y526) whose hydroxyl group is positioned near the C1—C2 bond of the inhibitor and which interacts with E401, and a hydrophobic interaction between the methyl group of the inhibitor and the face of Y299. In contrast to other sialidases/neuraminidases, in NDV HN there are extensive interactions with all three hydroxyls of the inhibitor glycerol group, directly involving E258 and Y317, and through water-mediated interactions, involving Y262 and S418. A unique feature of the HN active site is the large acidic cavity around the inhibitor O4 that is lined with residues conserved across all HNs.
FIG. 1.
The sialic acid binding site of NDV HN. Hydrogen bonding interactions are shown as dotted lines; water molecules are labeled W. (a) complex with Neu5Ac2en obtained at pH 6.3. (b) complex with sialic acid (in its β-anomeric form) obtained at pH 4.6.
These structural studies reveal no evidence for a second sialic acid binding site on HN and suggest that HN has a single site that is switchable between being a catalytic site and a binding site. Support for this comes from the low pH crystal form in which one of the critical arginines, R174, swings away from its “normal” position to take up residence in the cavity around O4 where it interacts with N190, one of the invariant residues lining the cavity (Fig. 1b). The movement of R174 and a concomitant rotation of I175 lead to the critical tyrosine, Y526, shifting out of its position below the C1—C2 bond of the inhibitor. As R174 moves into the cavity, invariant residue K236, part of the conserved hexapeptide NRKSCS, swings out, as if these two positively charged residues act in concert.
The structural comparisons of the two forms of HN therefore suggest that there is a single, switchable site for neuraminidase and sialic acid binding activities (6). A recent mutagenesis study focused on several conserved amino acids thought to comprise the neuraminidase site (11), which had been identified by homology modelling of HN based on the influenza virus neuraminidase (15). The study mutated 10 residues, of which R174, I175, D198, K236, R416, Y526 and E547 resulted in protein lacking both detectable neuraminidase and receptor recognition activity, measured by hemadsorption (HAd). The study also showed that addition of neuraminidase activity, either exogenously or by coexpression with another HN protein, partially rescues receptor recognition by the mutated HN proteins that lack both activities themselves.
In this study, we have extended our previous mutagenesis studies to test the single-site hypothesis, by making 24 mutants from 10 conserved sialic acid binding site residues in NDV HN (R174, I175, E258, Y299, Y317, E401, R416, R498, Y526, and E547) identified through our crystal structure studies. In addition to being tested for NA and HAd activities, the mutants were analyzed for their ability to promote fusion. The results are discussed in the light of the three-dimensional structure of NDV HN and suggest that particular amino acids are key to the several functions of HN.
MATERIALS AND METHODS
Cells, vectors, and recombinant plasmids.
HeLa and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). A cDNA fragment encoding the HN protein of NDV Kansas strain, with the mutation W123C, was inserted into the plasmid vector pTF1 (29) and also into the transient mammalian expression vector pCAGGS (20). Similarly, a cDNA fragment encoding the F protein from NDV Kansas strain was inserted into the expression vector pCAGGS. The mutation introduced at position 123 into HN was to allow the formation of disulfide-linked dimers of HN, which enhances the fusion promotion activity that is needed for the fusion assay.
Site-directed mutagenesis of NDV HN gene.
Mutation of NDV HN was achieved by oligonucleotide-directed mutagenesis using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Synthetic oligonucleotide primers were used to introduce single amino acid mutations in the HN gene amplified by PCR using the plasmid construct pTF1-NDV HN as the template. After sequencing to screen for the presence of the desired mutation, each mutated gene was isolated from the original construct and ligated to the transient mammalian expression vector pCAGGS (20). Plasmid DNA from ampicillin-resistant colonies of transformed DH5α cells were screened by restriction analysis for the presence of the HN gene. The entire mutated HN genes were fully sequenced to confirm the presence of only the desired base change(s).
Transient expression of mutated NDV HN gene.
Mammalian 293T cells were seeded at 8 × 105 cells/well in six-well plates and were grown overnight with culture medium, i.e., DMEM containing 10% FCS. Subconfluent monolayers of 293T cells were then transfected with 2 μg of cDNA (pCAGGS vectors containing NDV HN mutant genes) and 16 μg of Lipofectamine (Life Technologies) for 3 to 4 h at 37°C. Cells were washed with OptiMEM (Gibco BRL) and incubated for 18 h at 37°C with culture medium, before being assayed for neuraminidase and HAd activities.
Neuraminidase assay.
Neuraminidase activity was determined by the fluorimetric procedure of Potier et al. (21) with some modifications. Transfected 293T cells were washed with phosphate-buffered saline (PBS), pH 7.2, after 18 h of incubation at 37°C, before the addition of 0.1 M sodium acetate (pH 6.0) buffer containing 1 mM substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MU-Neu5Ac; Sigma Chemical Co., Poole, Dorset, United Kingdom). Plate contents were left to incubate for 1 h at 37°C. The reaction was stopped by the addition of 0.25 M glycine buffer, pH 11. The relative fluorescence of the methylumbelliferone released in the supernatants was determined fluorimetrically at 365 nm for excitation and 450 nm for emission.
Hemagglutinin binding.
Hemagglutination function was assayed as described by Morrison and McGinnes (18) with some modifications. Briefly, transfected 293T cells were washed with PBS buffer, pH 7.2, before addition of 2% (vol/vol) chicken erythrocyte suspension in PBS buffer and were left on ice for 30 min. After removal of unadsorbed erythrocytes by repeated gentle washing with PBS, bound cells were lysed using a cell lysis buffer containing 50 mM Tris, pH 7.4, 5 mM EDTA, 150 mM NaCl, and 0.5% NP-40. The absorbance of the supernatant was measured at 545 nm after centrifugation at 13,000 rpm in a Microcentaur benchtop centrifuge for 5 min.
Production of antibody-secreting hybridomas.
Hybridoma cell lines producing monoclonal antibodies (MAbs) against NDV HN were prepared as previously described (22). Culture fluids from fused cells were screened for antibody production by enzyme-linked immunosorbent assay (ELISA). Isotypes of the MAbs were determined by a Mouse Hybridoma Subtyping Kit (Roche).
Characterization of MAbs.
The MAbs in ascites fluids were titrated by standard ELISA using purified cleaved HN (cHN) (30) as the antigen. For measurement of hemagglutination inhibition and neuraminidase inhibition, standard procedures were employed (22). Four hemagglutinin units of NDV was incubated with antibody in the hemagglutination inhibition tests before the addition of chicken erythrocytes. For the neuraminidase inhibition test, purified virus (0.7 μg) was incubated with 1 μl of MAb for 30 min before the addition of N-acetylneuramin-lactose (Sigma). In the neutralization assay, 100 PFU of NDV was incubated with MAb for 30 min before infection to the MDBK cells. Infected cells were overlaid with medium containing 0.9% agar (Difco), and the plaques were counted 5 days postinfection.
Isolation of MAb escape mutants.
NDV was incubated with 100 μl of MAb for 1 h and was injected into a 10-day-old hen egg. After 3 days of incubation, virus in the allantoic fluid was harvested and subjected to the same procedure. The mutant NDV was then cloned by limited dilution. The purified escape mutants were used as antigens for ELISA against the various MAbs.
Cell surface expression of HN using ELISA.
To quantify the amount of HN expressed on cell surface of 293T cells, an ELISA was performed as described previously (1). Briefly, transfected 293T cells were washed with PBS after 18 h of incubation at 37°C and were reacted with a cocktail of anti-NDV HN MAbs (in PBS-0.1% [wt/vol] bovine serum albumin). The cells were left at room temperature for 30 min before cells were repeatedly washed with PBS. Horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Bio-Rad) was then added to the cells in the same buffer and was left again for 30 min at room temperature. The cells were washed with PBS before conjugate was reacted with substrate 2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid). The absorbance of the solution was measured at 405 nm.
Induction of cell fusion in HeLa cells by transfection with recombinant plasmids.
HeLa cells were seeded at 4 × 105 cells/well in six-well culture plates and were incubated at 37°C in DMEM containing 10% FCS overnight. One microgram of each of two recombinant pCAGGS plasmids, one containing the NDV HN gene and one containing the F gene, which encodes the fusion protein from NDV, was used to transfect subconfluent HeLa cells with 8 μl (16 μg) of Lipofectamine for 3 to 4 h. Cells were washed with OptiMEM before incubation with DMEM-FCS for 18 h at 37°C. Transfected HeLa cells were washed with PBS, fixed with a solution of 4% formalin, and stained briefly with 1% (wt/vol) crystal violet. Syncytium formation was observed using an inverted microscope, and fusion was determined by the number of syncytial cells formed compared to wild-type coexpression of NDV HN and F genes.
Molecular modelling and electrostatic calculations.
The trisaccharide sialyllactose (Neu5Ac-α2,3-Gal-β1,4-Glc) was modelled into the binding site by superimposing the sialic acid moiety of sialyllactose from influenza virus hemagglutinin (36) onto the sialic acid observed in the crystal structure of HN (6), Protein Databank code 1E8U. The complex was then subjected to a short, simulated annealing run (slowly cooling from 1,000 to 300 K in 25-K steps with a 0.0005-ps time step for the dynamics), with harmonic restraints being applied to the ligand and HN alpha-carbon coordinates throughout the run, using the CNS software package (3). Electrostatic calculations were performed using GRASP (19).
RESULTS
To characterize the active site mutations of NDV HN proteins, the mutated cDNAs were expressed in 293T cells by using the transient expression vector pCAGGS. After 18 h of posttransfection, the monolayers were assayed for neuraminidase activity and cell binding by HAd, and these results were compared to those for wild-type HN. Cell surface expression of active site mutants was quantified by ELISA by using a cocktail of MAbs (N1, N3, N6, and N7) which are conformationally specific for NDV HN (Table 1). Various levels of expression were obtained for the 24 mutants (Table 2), with five having <10% of wild-type expression levels. In this study, mutants showing less than 10% cell surface expression were not characterized further. In addition, fusion promotion abilities of the active site mutants were determined using HeLa cells instead of 293T cells, because 293T cells produce a high background level of fusion.
TABLE 1.
Characterization of MAb against NDV HN
| MAb | Subtype | ELISA titera | HI titerb | NI%c | Neutralization titerd | Reactivity of escape mutantse
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N3 | N4 | N6 | N7 | ||||||
| N1 | IgG2a | 6,000,000 | 5,120 | 25 | 16,000 | − | + | + | + | − |
| N3 | IgG2b | 6,000,000 | 640 | 96 | 6,000 | + | − | + | + | + |
| N6 | IgG2a | 660,000 | 240 | 41 | 4,200 | + | + | + | − | + |
| N7 | IgG1 | 2,000,000 | 5,120 | 36 | 12,000 | + | + | − | + | − |
Titers are expressed as the reciprocal of the highest dilution of antibody that showed higher OD value than negative control.
Titers are expressed as the reciprocal of the highest dilution of antibody that completely blocked four hemagglutination units in a standard hemagglutination inhibition (HI) test.
Percentage inhibition of viral neuraminidase activity by indicated antibody. NI, neuraminidase inhibition.
Titers are expressed as the reciprocal of the dilution of antibody that reduced 50% of plaque formation.
−, ELISA titer less than 10% of wild-type virus.
TABLE 2.
Comparison of biological activities of active site mutants
| Mutant | Cell surface expressiona | NA activityb | HAd activityb |
|---|---|---|---|
| R174A | 40.6 ± 0.9 | 0.5 ± 0.2 | 7.9 ± 0.3 |
| R174D | 2.0 ± 0.4 | NDd | ND |
| R174E | 4.1 ± 0.2 | ND | ND |
| R174K | 38.5 ± 1.2 | 0.8 ± 0.3 | 10.2 ± 1.2 |
| R174Q | 9.6 ± 3.3 | ND | ND |
| R416A | 15.1 ± 4.2 | 0.1 ± 0.01 | <1 |
| R416D | 49.0 ± 4.7 | 2.3 ± 0.9 | 3.3 ± 1.5 |
| R416E | 89.0 ± 7.2 | 0.4 ± 0.2 | 13.3 ± 4.2 |
| R416K | 19.2 ± 5.7 | 0.6 ± 0.1 | 3.3 ± 1.2 |
| R416Q | 8.4 ± 1.6 | ND | ND |
| R498A | 144 ± 15.1 | 1.6 ± 0.1 | 58.9 ± 7.4 |
| R498K | 4.2 ± 1.9 | ND | ND |
| R498Q | 48.3 ± 12.3 | 0.4 ± 0.2 | 22.3 ± 8.5 |
| I175E | 124.7 ± 4.4 | 13.4 ± 0.4 | 40.1 ± 0.1 |
| I175L | 90.8 ± 2.8 | 85.9 ± 2.5 | 39.9 ± 1.8 |
| I175T | 102.8 ± 6.8 | 65.5 ± 1.7 | 59.8 ± 0.7 |
| E258D | 135 ± 15.3 | 3.7 ± 0.2 | 46.8 ± 3.7 |
| E258S | 127.2 ± 2.6 | 5.5 ± 0.5 | 45.9 ± 4.5 |
| Y299A | 89.8 ± 3.9 | 8.5 ± 1.3 | 72.5 ± 0.6 |
| Y317F | 119.3 ± 3.3 | 27.6 ± 2.3 | 65.1 ± 3.7 |
| E401D | 29.2 ± 3.8 | —c | <1 |
| E547D | 42.5 ± 0.3 | 0.1 ± 0.02 | 1.5 ± 0.5 |
| Y526F | 101.7 ± 3.7 | 0.1 ± 0.04 | 3.1 ± 1.3 |
| Y526H | 118.7 ± 5.6 | 0.7 ± 0.2 | 6.6 ± 0.6 |
Cell surface expression values of active site mutants of HN were related to those of wild-type HN.
Neuraminidase (NA) and HAd activity values were expressed as a percentage of activity of wild-type NDV HN. All values represented were the average of three independent experiments.
—, either no activity or binding.
ND, not determined due to expression levels of HN being less than 10% to obtain significant assay data.
Triarginyl cluster (R174, R416, and R498).
The highly conserved active site arginine triad, R174, R416, and R498, synonymous to the arginine triad of all known neuraminidases (33), appears very sensitive to mutation. These residues have been shown to interact with the carboxyl group of the inhibitor Neu5Ac2en (Fig. 1a). The change of charge at position 174 to a glutamate, aspartate, or glutamine appeared to be the most disruptive, resulting in very low levels of protein expressed at the cell surface (Table 2). These observations suggest these mutants were probably misfolded and therefore could not be efficiently transported to the cell surface. When R174 replaced lysine or alanine, these mutants were expressed at significant levels, around 40% of wild type, and both showed loss of neuraminidase activity but retained HAd activity, albeit at reduced levels (Fig. 2). Interestingly, all of the R174 mutants abolished fusion promotion: a typical observation for R174A is shown in Fig. 3. Mutation of R416 resulted in the loss of both neuraminidase and HAd activities, while mutation of R498 resulted in loss of neuraminidase activity and retention of significant HAd for the mutants R498Q and R498A.
FIG. 2.
Biological activities of active site mutants of NDV HN. Bar chart represents normalized percentage values of neuraminidase activity and HAd (HA) activity relative to percentage amount of HN expressed at the cell surface. Values shown are the mean result of three independent experiments.
FIG. 3.
Membrane fusion induced by coexpressed HN and F. HeLa T4 cells were transfected with NDV F cDNA and wild-type (WT) or mutant NDV HN cDNA and were incubated for 48 h at 37°C. Cells were stained with crystal violet.
Residues E547, Y526, E401, and I175.
From the crystal structure of NDV HN complexed with the inhibitor Neu5Ac2en (Fig. 1a), residue E547 appears to play an important role in stabilizing R174 and is an interaction seen in all other bacterial and influenza virus sialidase/neuraminidase structures (33). Mutation to a shorter aspartic acid effectively abolished both activities and significantly reduced fusion promotion. Other residues which sit close to the carboxyl end of the inhibitor, Y526 and E401, are also shown to play a major role in HN function. The hydroxyl group of Y526 is shown to interact with E401, and it has been predicted that these two residues stabilize a sialosyl cation transition state intermediate in other sialidases (4). Mutants E401D, Y526F, and Y526H led to almost complete elimination of both neuraminidase and HAd functions. Despite this lack of neuraminidase and HAd functionality, the Y526 mutants displayed enhanced levels of fusogenic activity of HeLa cells (Table 3).
TABLE 3.
Fusion promotion ability of active site NDV HN mutantsa
| Mutant or other classification | Fusionc |
|---|---|
| Wild type | 100 |
| R174A | —b |
| R174K | 6 ± 0.6 |
| R416A | 35 ± 2.5 |
| R416D | 89 ± 4.3 |
| R416E | 97 ± 5.2 |
| R416K | 35 ± 3.5 |
| R498A | 92 ± 4.1 |
| R498Q | 81 ± 2.8 |
| I175E | 151 ± 11.6 |
| I175L | 102 ± 9.3 |
| I175T | 86 ± 5.1 |
| E258D | 82 ± 3.1 |
| E258S | 79 ± 5.2 |
| Y299A | 78 ± 1.2 |
| Y317F | 83 ± 3.7 |
| E401D | 107 ± 6.2 |
| E547D | 13 ± 3.9 |
| Y526F | 129 ± 9.3 |
| Y526H | 149 ± 15.4 |
Cell fusion was calculated from the ratio of the total number of nuclei found in syncytia to the total number of nuclei counted in three or four randomly chosen fields under a light microscope. The values indicated represent the average percentage found in three independent experiments of fusion to wild-type NDV HN when it is coexpressed with the gene encoding NDV F protein in HeLa cells.
—, no syncytia observed.
Residue I175 appears to play a role in cushioning the position of Y526, as the two crystal forms reveal a rotation of the side chain concomitant with the movement of Y526. Mutants were designed that investigated the effect of branched and nonbranched side chains at this position. Mutants of I175 exhibited a slightly different pattern from those of all other mutants. Mutation to a leucine maintained wild-type neuraminidase activity, but HAd was halved (Table 2 and Fig. 2). Conversely, mutation to a glutamic acid gave levels of neuraminidase and HAd of less than 10 and 30% of wild-type, respectively, but displayed greatly increased fusogenic activity (Table 3 and Fig. 3). Interestingly, replacement by threonine appeared not to have a major effect on neuraminidase or HAd function, albeit levels were over a third lower than those for wild-type HN. Despite these differences, none of the I175 mutations appeared to have significantly affected their expression levels.
Residues E258, Y299, and Y317.
In the Neu5Ac2en complex of HN, residue E258 hydrogen bonds to O7 and O9 of the glycerol group of the inhibitor and Y317 hydrogen bonds to O8. Y317 also interacts with N5 of the inhibitor acetamido group via a water molecule and thus suggests the importance of this residue in substrate recognition. As expected, mutation of E258 to either an aspartic acid or serine significantly affected neuraminidase activity (less than 6% of the wild-type level), with HAd levels about 35% of the wild-type level. Expression levels here were greater than for wild-type HN. Replacement of Y317 by phenylalanine demonstrated neuraminidase and HAd activities of 20 and 50% of wild-type levels, respectively, with expression levels comparable to wild-type levels. Residue Y299, which makes a hydrophobic interaction with the acetamido methyl group of Neu5Ac2en, also displayed severely reduced neuraminidase activity when this residue replaced alanine, with only a marginal effect on HAd activity. None of the mutations of these three residues affected fusion promotion (Table 3 and Fig. 3).
Model of receptor binding.
Sialyllactose is a typical receptor for NDV HN, but so far we have been unable to observe crystallographically the binding of this receptor to HN. This trisaccharide has been observed in the crystal structures of several other sialic acid binding proteins, including influenza virus hemagglutinin (36), leukoagglutinin (10), sialoadhesin (16), and murine polyomavirus (28). Superposition of the ligands from these four structures shows that the galactose sits at a preferred orientation relative to the sialic acid sugar (Fig. 4a). The simulated annealing run on NDV HN in the presence of sialyllactose, taken from the influenza virus hemagglutinin, revealed a movement of R498 to avoid steric clashes with the galactose (Fig. 4b). The α-anomer of sialic acid appears to be accommodated within the binding site, with the sialic acid carboxyl group hydrogen bonding to R416, which also hydrogen bonds to O7 of the ligand. The side chains of Y526, E547, R174, and I175 all move away from the ligand relative to their positions in the uncomplexed NDV HN structure (grey structure, Fig. 4b).
FIG. 4.
Sialyllactose modelling. (a) Four sialyllactose ligands taken from protein-ligand complexes have been superimposed on their sialic acid moieties, shown in atom coloring: yellow carbons, blue nitrogens, and red oxygens. The galactose and glucose residues are colored grey for the sugars from their complex with the influenza virus hemagglutinin, cyan for leukoagglutinin, magenta for sialoadhesin, and green for murine polyomavirus. (b) Sialyllactose modelled into the binding site of NDV HN. The side chains of the unliganded crystallographic structure are shown in grey, and the modelled structure is shown in green.
DISCUSSION
The purpose of this study was to determine the importance of certain conserved residues to the three activities of HN: sialic acid binding activity measured by HAd, promotion of fusion measured by syncytium formation, and finally neuraminidase activity. We have examined the effect of mutating binding site residues that are largely conserved across all paramyxoviruses, whose positions were previously defined in crystal structure complexes of NDV HN with sialic acid and the inhibitor Neu5Ac2en (6). Of the 10 residues targeted, 24 mutants were constructed, and only five of these failed to express sufficiently for detection by the panel of MAbs used in this study. Most of the mutant proteins led to a severe reduction in NA function and reduced HAd, with some mutants exhibiting loss of both functions (R174, R416, E401, E547, and Y526). These findings strongly support the notion of a single site with dual function as previously discussed (6).
The results of previous site-directed mutagenesis studies on conserved residues thought to form the active site for HN appear to be consistent, in most cases, with the data from the active site mutants that have been characterized in this study (12, 13, 17, 27). The most recent study made mutants R174L, R416L, R498L, E547Q, Y526L, I175E, and K236R, which complement those in this study (11). Of these, only R498L gave any measurable neuraminidase and HAd activities, 4 and 36% of wild-type levels, respectively. Our results on the I175E mutant differ in that we obtained significant neuraminidase and HAd activities. This could be the result of the different NDV strain used and/or the different cell types used for expression in the two studies. Mutants N500D and V502A, which were made in this recent study, had no effect on NA or HAd activities, which from our structural studies is not surprising: N500 is a surface-exposed residue remote from both the active site and dimer interface, while V502 is an alanine in the Kansas strain of NDV HN. Another mutational study carried out on conserved residues of NDV HN described the effect of mutation of active site residue E401 on receptor binding and catalysis of substrate (25). Their results suggest greater than wild-type binding and 30% neuraminidase activities; however, they only obtained 10% cell surface expression. In our study negligible levels of both hemagglutinin and NA activities were observed when HN was expressed at 29% of the wild-type level.
Catalysis requires precise positioning of amino acid side chains in the active site, and assuming that the neuraminidase mechanism of HN is similar to other neuraminidases, this requires tethering of sialic acid in order to induce strain in the sugar ring conformation prior to hydrolysis. The major anchor points are the arginine triad (R174, R416, and R498) interacting with the sialic acid carboxyl group, the residues interacting with the glycerol moiety (E258 and Y317), and the residues interacting with the methyl group of the acetamido moiety (Y299). Sialic acid binding as part of cell attachment is, like much of carbohydrate recognition, of low affinity (millimolar Ki) (32) and will not involve as many precise interactions as catalysis. The results of the mutations described in this report, together with those of previous studies, support such a view for the binding site of HN with its proposed dual functionality.
The electrostatic potential surfaces calculated for the inhibitor and “substrate” crystal structures of NDV HN, calculated in the absence of ligand (Fig. 5a and b), are quite different, both in charge distribution and magnitude of the charge. In the inhibitor complex (Fig. 5a), there is an intense charge distribution with strong positive potential arising from the arginine triad. In the sialic acid complex (Fig. 5b), this is modulated such that the most positively charged area is associated with R416. The mutation results reflect this observation: mutation of R416 results in a severe loss of HAd. Mutation of R174 also severely reduces HAd, whereas mutation of R498 can leave some significant HAd activity. Mutation of any of the three arginines, however, eliminates NA activity. We therefore propose that R416 is a key binding residue for recognition of sialic acid receptors involved in cell attachment. The crystallographic studies revealed the thermodynamically more stable β-anomer of sialic acid bound, as the sialyllactose used in crystal soaking experiments had undergone acid hydrolysis at the low pH of the crystals, resulting in the binding of free sialic acid in the binding site. The fact that the β-anomer was observed suggested a flexible binding site in HN (6). Modelling of sialyllactose into the binding site (Fig. 4b) suggests that R416 hydrogen bonds to the ligand's carboxyl group, further supporting the key role of R416 in receptor recognition. R174 can also be considered to be, indirectly, key for sialic acid binding. While we propose from the structural studies that R174 is not involved in sialic acid recognition during receptor binding, its location (Fig. 1b) may be important for the transfer of a conformational change to the dimer interface. R498 in the sialyllactose model complex moves to avoid steric clashes with the galactose, supporting the finding that mutation of R498 does not destroy receptor binding.
FIG. 5.
Surface electrostatic potential around the binding site. Blue indicates positive potential; red indicates negative potential, calculated in both cases for the complete HN dimer but omitting the ligand, using GRASP. (a) Complex with Neu5Ac2en. (b) Complex with sialic acid (in its β-anomeric form).
Of the mutants that interact with the glycerol group of the ligand, mutations of E258 have a profound effect on NA activity but still retain significant HAd activity. This suggests that that E258 in particular is critical for ligand recognition for catalysis but is less critical for HAd. Mutation of Y317 to a phenylalanine removes its ability to hydrogen bond to O9 and a water molecule that interacts with N5 of the ligand. The level of reduction in neuraminidase and HAd activities suggests that the hydroxyl of this conserved residue is not critical but that the aromatic ring may be required, in particular to stabilize conserved residue E401. Mutation of Y299, which interacts with the acetamido methyl group of the ligand, suggests that Y299 is very important for neuraminidase activity but not for HAd. Taken together, E258, Y299, and Y317 appear important for recognition of ligand for catalysis, which requires precise positioning of the ligand prior to hydrolysis, whereas they are important but not key for sialic acid recognition.
The side chains of residues I175, Y526, and E401 lie in a line below the sugar ring of the ligand. Y526 and E401 are hydrogen bonded to one another in both states of the binding site (Fig. 1) and are likeliest to be involved in the stabilization of the transition state during the catalytic cycle. Mutation of either of these residues eliminates both NA and HAd functions. Elimination of NA activity is not surprising, but elimination of sialic acid binding suggests a role for the hydroxyl of Y526 and the carboxyl of E401 in the recognition of the carboxyl group of α-anomer of sialic acid. We proposed that I175 plays a role in modulating the position of Y526 (6). The I175L and I175T mutants were chosen, as the equivalent residue is a leucine in Sendai virus and PIV type 1 (PIV1) and a threonine in PIV3. These mutants in NDV HN maintain neuraminidase activity, whereas I175E significantly reduces neuraminidase activity. Significant, although reduced, HAd activity is maintained in all three I175 mutants. This suggests that the rotation around the Cα—Cβ bond in I175 may be an elaboration in simian virus 5, PIV2, mumps virus, and NDV, all of which conserve this isoleucine.
I175, Y526, and E401 are particularly interesting for their effect on fusion activity. If binding of sialic acid to HN is the prelude to fusion promotion, how can it be that Y526F/H and E401D have enhanced fusion promotion activity yet show almost no HAd activity? Why does I175E show the highest level of fusion promotion? One possibility is suggested by a recent study where the addition of exogenous neuraminidase restored HAd activity to mutants of NDV HN that lacked such activity (11). The 293T cells used in our assay of HAd would carry sialic acid receptors, which, if not removed by neuraminidase-active HN expressed on the surface, would be available for binding to HN molecules, thereby blocking their ability to bind to red blood cells. Another possibility is that these mutations invoke structural changes in HN that mimic the structural changes that occur upon sialic acid binding to HN. Comparison of the two structural forms of HN shows that most differences are confined to a small number of residues that form a connection between the carboxyl end of the ligand and the HN dimer interface. It is possible that upon sialic acid binding, a series of linked conformational changes occur that lead to a change in the dimer association, which may trigger the fusion protein into its fusogenic state. This could either be by revealing a part of the HN dimer interface, which is then free to interact with the F protein, or by inducing a change in the stalk regions of the dimer, which in turn interacts with the F protein to trigger fusion. Residues in both the head and stalk regions of HN have been implicated in fusion promotion (1, 2, 8, 26, 31, 34). In this model, the position of Y526 appears key. Removal of the ability of Y526 to hydrogen bond to E401, implicit in the Y526H/F and E401D mutants, might lead to the movement of the histidine or phenylalanine to the left as seen in Fig. 1b, thereby propagating the conformational change to the dimer interface leading to fusion promotion. The molecular modelling results support this hypothesis, as shown in Fig. 4b, where the binding of sialyllactose appears to promote the movement of Y526, I175, E547, and R174 in the direction of the dimer interface, towards the left in the picture.
In summary, the mutation studies on NDV HN support the single-site model with sialic acid binding and hydrolysis occurring at the same site. Most of the residues conserved across the paramyxovirus family that form direct interactions with sialic acid are hypersensitive to mutation with respect to neuraminidase function. These same residues show various degrees of sensitivity with respect to sialic acid binding but confirm the notion of a single site on HN with dual functionality. The two states of HN observed in the crystal studies appear to represent the catalytic state and the receptor binding state. In the latter state, we suggest that binding of sialic acid disrupts the hydrogen bonding between Y526 and E401, leading to the movement of Y526, which in turn moves E547, I175, and R174 and so propagates a conformational change to the dimer interface which somehow triggers the fusion protein into its fusogenic state. These studies provide the basis for further structural and mutagenesis studies to probe the mechanism of action of this multifunctional protein, whose binding site, unlike that of the influenza virus neuraminidase, appears to have remarkable flexibility to achieve its many functions.
Acknowledgments
This work was supported by grants from the United Kingdom Medical Research Council and United Kingdom Biotechnology and Biological Sciences Research Council and by U.S. NIAID grants AI-11949 and AI-38956.
REFERENCES
- 1.Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204:506-514. [DOI] [PubMed] [Google Scholar]
- 2.Bousse, T., T. Takimoto, and A. Portner. 1995. A single amino acid change enhances the fusion promotion activity of human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Virology 209:654-657. [DOI] [PubMed] [Google Scholar]
- 3.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Chong, A. K. J., M. S. Pegg, N. R. Taylor, and M. Vonitzstein. 1992. Evidence for a sialosyl cation transition-state complex in the reaction of sialidase from influenza virus. Eur. J. Biochem. 207:335-343. [DOI] [PubMed] [Google Scholar]
- 5.Crennell, S., E. Garman, G. Laver, E. Vimr, and G. Taylor. 1994. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 2:535-544. [DOI] [PubMed] [Google Scholar]
- 6.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 7.Crennell, S. J., E. F. Garman, W. G. Laver, E. R. Vimr, and G. L. Taylor. 1993. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. USA 90:9852-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, R. T., Z. Y. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 9.Gaskell, A., S. Crennell, and G. Taylor. 1995. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure 3:1197-1205. [DOI] [PubMed] [Google Scholar]
- 10.Imberty, A., C. Gautier, J. Lescar, S. Perez, L. Wyns, and R. Loris. 2000. An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides. J. Biol. Chem. 275:17541-17548. [DOI] [PubMed] [Google Scholar]
- 11.Iorio, R. M., G. M. Field, J. M. Sauvron, A. M. Mirza, R. Deng, P. J. Mahon, and J. P. Langedijk. 2001. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 75:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio, R. M., and R. L. Glickman. 1992. Fusion mutants of Newcastle disease virus selected with monoclonal antibodies to the hemagglutinin-neuraminidase. J. Virol. 66:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio, R. M., R. J. Syddall, R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Identification of amino acid residues important to the neuraminidase activity of the Hn glycoprotein of Newcastle disease virus. Virology 173:196-204. [DOI] [PubMed] [Google Scholar]
- 14.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 577-604. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 15.Langedijk, J. P. M., F. J. Daus, and J. T. Van Oirschot. 1997. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J. Virol. 71:6155-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May, A. P., R. C. Robinson, M. Vinson, P. R. Crocker, and E. Y. Jones. 1998. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol. Cell 1:719-728. [DOI] [PubMed] [Google Scholar]
- 17.Mirza, A. M., R. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison, T. G., and L. W. McGinnes. 1989. Avian cells expressing the Newcastle disease virus hemagglutinin-neuraminidase protein are resistant to Newcastle disease virus infection. Virology 171:10-17. [DOI] [PubMed] [Google Scholar]
- 19.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 21.Potier, M., L. Maneli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-D-N-acertylneuraminate) substrate. Analyt. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 22.Portner, A., R. G. Webster, and W. J. Bean. 1980. Similar frequencies of antigenic variants in Sendai, vesicular stomatitis, and influenza A viruses. Virology 104:235-238. [DOI] [PubMed] [Google Scholar]
- 23.Scheid, A., and P. Choppin. 1973. Isolation and purification of the envelope proteins of Newcastle disease virus. J. Virol. 11:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:470-490. [DOI] [PubMed] [Google Scholar]
- 25.Sergel, T., L. McGinnes, and T. Morrison. 1993. Role of a conserved sequence in the maturation and function of the Ndv Hn glycoprotein. Virus Res. 30:281-294. [DOI] [PubMed] [Google Scholar]
- 26.Sergel, T., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717-726. [DOI] [PubMed] [Google Scholar]
- 27.Sheehan, J. P., and R. M. Iorio. 1992. A single amino acid substitution in the hemagglutinin-neuraminidase of Newcastle disease virus results in a protein deficient in both functions. Virology 189:778-781. [DOI] [PubMed] [Google Scholar]
- 28.Stehle, T., and S. C. Harrison. 1996. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 4:183-194. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi, T., K. W. Ryan, and A. Portner. 1992. A plasmid that improves the efficiency of foreign gene expression by intracellular T7 RNA polymerase. Genet. Anal. Tech. Appl. 9:91-95. [DOI] [PubMed] [Google Scholar]
- 30.Takimoto, T., G. L. Taylor, S. J. Crennell, R. A. Scroggs, and A. Portner. 2000. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology 270:208-214. [DOI] [PubMed] [Google Scholar]
- 31.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taroni, C., S. Jones, and J. M. Thornton. 2000. Analysis and prediction of carbohydrate binding sites. Protein Eng. 13:89-98. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, G. 1996. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 6:830-837. [DOI] [PubMed] [Google Scholar]
- 34.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 35.Varghese, J. N., W. G. Laver, and P. M. Colman. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303:35-40. [DOI] [PubMed] [Google Scholar]
- 36.Weis, W., J. H. Brown, S. Cusack, J. C. Paulson, J. J. Skehel, and D. C. Wiley. 1988. Structure of the influenza virus hemagglutinin complexed with its receptor, sialic acid. Nature 333:426-431. [DOI] [PubMed] [Google Scholar]