Abstract
Experiments were conducted to investigate the roles of Marek's disease virus serotype 1 (MDV-1) major tegument proteins VP11/12, VP13/14, VP16, and VP22 in viral growth in cultured cells. Based on a bacterial artificial chromosome clone of MDV-1 (BAC20), mutant viruses were constructed in which the MDV-1 homologs of UL46, UL47, UL48, or UL49 were deleted alone and in various combinations. It could be demonstrated that the UL46, UL47, and UL48 genes are dispensable for MDV-1 growth in chicken embryonic skin and quail muscle QM7 cells, although the generated virus mutants exhibited reduced plaque sizes in all cell types investigated. In contrast, a UL49-negative MDV-1 (20Δ49) and a UL48-UL49 (20Δ48-49) doubly negative mutant were not able to produce MDV-1-specific plaques on either cell type. It was confirmed that this growth restriction is dependent on the absence of VP22 expression, because growth of these mutant viruses could be partially restored on cells that were cotransfected with a UL49 expression plasmid. In addition, we were able to demonstrate that cell-to-cell spread of MDV-1 conferred by VP22 is dependent on the expression of amino acids 37 to 187 of MDV-1 VP22, because expression plasmids containing MDV-1 UL49 mutant genes with deletions of amino acids 1 to 37 or 188 to 250 were still able to restore partial growth of the 20Δ49 and 20Δ48-49 viruses. These results demonstrate for the first time that an alphaherpesvirus UL49-homologous gene is essential for virus growth in cell culture.
Marek's disease virus serotype 1 (MDV-1) is a chicken herpesvirus which causes a variety of syndromes, including generalized immunosuppression, acute neuronal symptoms and paralysis, T-cell lymphomas, and, rarely, atherosclerosis in its natural host (3). MDV-1 has been classified as an alphaherpesvirus on the basis of its genomic structure and sequence homology to other Herpesviridae (2, 5, 17, 37).
The prototype alphaherpesvirus, herpes simplex virus type 1 (HSV-1), is composed of four structural elements, a core containing the double-stranded DNA genome, an icosahedral capsid harboring the core, a proteinaceous layer, the tegument, which immediately surrounds the capsid, and an envelope containing viral membrane (glyco)proteins (35). The role of herpesvirus tegument proteins is twofold, structural and regulatory. Four of the known 15 HSV-1 tegument polypeptides are major virion components (VP1/2, VP13/14, VP16, and VP22, encoded by UL36, UL47, UL48, and UL49, respectively) (14, 35), and VP16 and VP22 were shown to form the tegument body (10).
These proteins have been assigned an important role in the formation of the virion by interacting both with capsid proteins and the envelope membrane proteins (36, 39). Furthermore, VP1/2 and VP16 were shown to play a central role in virus egress and the formation of mature virions, because deletion of these genes resulted in the absence of infectious extracellular virions (8, 22, 40). Tegument proteins have also been assigned regulatory functions in the viral life cycle, and they are delivered to the cytoplasm and nuclei of infected cells, where they may interact with cellular proteins to initiate viral replication (4). Indeed, HSV-1 VP16, through the formation of a complex with the transcriptional factors HCF and Oct-1, transactivates the five immediate-early genes, resulting in the expression of the viral proteins in a cascade-like fashion (4).
HSV-1 VP11/12, VP13/14, and VP22 have been reported to modulate the transactivating function of VP16 in virus-infected cells (10, 19, 43). Tegument also contains proteins having various functions, such as kinase activity (26), shutoff of host protein synthesis (12), interaction with ribosomes (30), and DNA packaging (31).
Homologs of HSV-1 UL46, UL47, UL48, and UL49 have been identified in the genomes of all three known MDV serotypes (MDV-1, MDV-2, and herpesvirus of turkeys [HVT]), and the four MDV-1 genes are located colinearly to their HSV-1 counterparts (1, 15, 41). Despite these similarities in genomic organization between HSV-1 and MDV-1, differences in the transcriptional organization between these two alphaherpesviruses have been reported, because no monocistronic UL48 mRNA was detected in the case of MDV-1. We reported recently that the MDV-1 VP16 protein could hardly be detected in infected chicken embryonic skin cells (CESC), although a bicistronic mRNA corresponding to the UL49-48 genes was found to direct the synthesis of VP22 and VP16 in both in vitro and in vivo expression systems (9).
MDV-1 VP22 is abundantly expressed in CESC, with a mainly nuclear localization pattern. MDV-1 VP22 is a DNA-binding protein which is able to be imported into recipient cells, similar to its HSV-1 counterpart (11). By analyzing truncated versions of VP22, the region responsible for the DNA-binding activity was located between amino acids 16 and 37 in the N-terminal part of the protein, and it was hypothesized that the intercellular transport is determined by the central portion of the protein located between amino acids 93 and 173 (9, 25).
In order to elucidate the function of the major MDV-1 tegument proteins VP11/12, VP13/14, VP16, and VP22, virus mutants carrying deletions of the UL46, UL47, UL48, and UL49 as well as a double (UL48-49) and a triple deletion mutant (UL46-48) were constructed using the recently developed MDV-1 bacterial artificial chromosome (BAC) system (32).
We were able to demonstrate that the MDV-1 VP22 protein encoded by UL49 is indispensable for virus growth, whereas VP11/12, VP13/14, and VP16 are nonessential, although the respective virus mutants exhibited significant impairments in virus growth properties on primary chicken embryonic skin and quail QM7 cells.
(Part of this work was included in the Ph.D. thesis of F. Dorange, University of Tours, Tours, France.)
MATERIALS AND METHODS
Viruses and cells.
Chicken embryonic skin cells (CESC) were obtained as previously described (33) with slight modifications (9). Quail muscle cells QM7 (ATCC CRL-1962), which have been shown to support growth of MDV-1 (32), were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS).
BAC20, the bacterial artificial chromosome vector described earlier (32), was used to obtain all mutant viruses. BAC20 contains the complete genome of the attenuated MDV-1 strain 584Ap80C, in which BAC vector sequences (released from plasmid pHA1 [20]) were introduced into the US2 locus by homologous recombination (32).
Construction of mutant BAC clones.
Mutagenesis of MDV-1 BAC20 was performed essentially as described (32). Escherichia coli DH10B cells harboring the BAC20 DNA and the pGETrec vector containing the recE, recT, and bacteriophage λ gam gene under the control of the arabinose promoter (23) were induced by addition of arabinose (final concentration of 0.4%) and prepared for electroporation.
The mutagenesis strategy was to replace the targeted gene with the kanamycin resistance (Kanr) gene by homologous recombination. The Kanr gene of pACYC177 (6) was amplified by PCR using PCR primers of 70 nucleotides in length that contained 50-nucleotide homology arms at their 5" ends and 20 nucleotides at their 3" ends for amplification of the Kanr gene. The PCR primers, selected on the basis of the published sequence of the MDV-1 strain Md5, are given in Table 1. pGETrec-containing BAC20 cells were electroporated with the various PCR fragments and plated on LB agar containing 50 μg of kanamycin per ml of agar.
TABLE 1.
Primers used to delete MDV-1 genes and to generate pGEM-T48-49.5a
| Primer | Sequence | Description |
|---|---|---|
| BACdel49f | GCCACACGGTACAATAGAAGGTGCACTTGTTCATATCTTACTGTTTAATACGATTTATTCAACAAAGCCACG | Kanr gene for UL49 and UL49-48 deletions |
| BACdel49r | GTCTATAAAAGACGACTTACTTGCAGTAGTAGGGCTGTTCCTATGTTACAGCCAGTGTTACAACCAATTAACC | Kanr gene for UL49 deletion |
| BACdel48f | CTACTACTGCAAGTAAGTCGTCTTTTATAGACATCCGAATTAAAAACTAGCGATTTATTCAACAAAGCCACG | Kanr gene for UL48 and UL48-46 deletions |
| BACdel48r | TATAGTTTCGTTCTGCCAACTCACCATCATACTAATAGACAAATACCTCCGCCAGTGTTACAACCAATTAACC | Kanr gene for UL48 and UL49-48 deletions |
| BACdel47f | ACATTATAAAACGGCGTAGTTTTGATAACAGTATGCTGGTAGCACATTCCCGATTTATTCAACAAAGCCACG | Kanr gene for UL47 deletion |
| BACdel47r | CCCGGTTTTAAACATGTACCACTCTTAAATGCCATTATTCTACATCCGGAGCCAGTGTTACAACCAATTAACC | Kanr gene for UL47 deletion |
| BACdel46f | CCTGTACTATATTTCAGTTTTCTCTGTAGTGTGTCCATCCATATGGGTTACGATTTATTCAACAAAGCCACG | Kanr gene for UL46 deletion |
| BACdel46r | TGAAAGGGTATACATGTTTGGGACAAATCGTTACGCTAAAGATAAAAACAGCCAGTGTTACAACCAATTAACC | Kanr gene for UL46 deletion |
| BACdel46r2 | GGGACAAATCGTTACGCTAAAGATAAAAACATATATGTTTGAACATAGTAGCCAGTGTTACAACCAATTAACC | Kanr gene for UL48-46 deletion |
| Kan1 | CCTGATATGAATAAATTGC | Sequence of recombination site in deleted BACs |
| Kan2 | GTGCAATGTAACATCAGAG | |
| UL49,5F | GGATCCGAATTCATGGGACTCATGGACA | Obtaining of UL48-49.5 sequence |
| UL48R | GTCGACTGCAGGTTATAAAGTACTGATGGTAGTGGC |
For the BAC primers, bold sequences, homologous to the pACYC177 vector sequences, were used to amplify the Kanr gene, and underlined sequences are homologous to sequences bordering the UL46, UL47, UL48, or UL49 gene. For primers UL49.5F and UL48R, used to generate the UL48-49.5 sequence, the bold and underlined sequences indicate restriction enzyme sites.
DNA analyses.
DNA from kanamycin-resistant colonies was prepared, digested with restriction endonuclease HindIII, and analyzed by agarose gel electrophoresis. DNA from clones exhibiting the predicted restriction profiles was used to transform electrocompetent DH10B or Top10 (Invitrogen) cells, in order to remove the pGETrec vector (23). Mutant BAC DNA was isolated again, digested with HindIII, and separated by 0.8% agarose gel electrophoresis, and DNA fragments were transferred to a positively charged nylon membrane (Ambion). Southern blot hybridization was performed using standard procedures and a nonradioactive detection kit (ECL direct nucleic acid labeling and detection system; Amersham-Pharmacia Biotech). Gene-specific DNA probes for UL46, UL47, UL48, UL49, and Kanr were obtained from existing pGEM-T plasmids (9) or from plasmid pACYC177.
Nucleotide sequencing of the recombination sites was performed using primers Kan1 and Kan2 (Table 1) on a Perkin Elmer automatic sequencer according to the supplier's instructions.
Construction of plasmids.
The recombinant plasmids pGEM-T UL49N1, pGEM-T UL49N2, and pGEM-T UL49s-48 were described earlier (9). The mutant genes contained within these plasmids encode the MDV-1 proteins VP22N1, VP22N2, and VP22MS, respectively. VP22N1 lacks amino acids 1 to 15, VP22N2 lacks amino acids 1 to 37, and VP22MS lacks amino acids 188 to 250 of VP22. The mutant UL49 genes contained in these clones were released from the pGEM-T vectors and cloned under the control of the human cytomegalovirus immediate-early promoter in the pcDNA3 vector (Invitrogen) to obtain recombinant expression plasmids pUL49N1, pUL49N2, and pUL49s-48.
The region encompassing the UL48 to UL49.5 sequence of MDV-1 strain RB1B was amplified by PCR and cloned in the T-tailed vector pGEM-T (Promega). Primers were selected on the basis of the published sequence of the MDV-1 GA strain (Table 1) (41). The resulting plasmid was termed pGEM-T48-49.5 and used for the generation of a UL49 rescuant virus.
DNA transfections.
One microgram of BAC20 or mutant BAC20 DNA was transfected into either CESC or QM7 cells using the calcium phosphate precipitation method (21). trans-Complementation of the deleted genes in mutant BAC20 clones was achieved by cotransfection of 1 × 105 QM7 cells seeded in 24-well plates with 1 μg of pcDNA3 vector carrying the complementary gene for the respective deficient virus together with the mutant BAC20 genome.
Construction of QM7 cell lines expressing full-length or truncated VP22.
In order to obtain QM7 cell lines expressing either full-length or truncated VP22 (VP22N1 and VP22N2, respectively), plasmids pUL49, pUL49N1, pUL49N2, and pT+ (pcDNA3 for generation of a control cell line) harboring the neomycin resistance gene were transfected into cells by using the Lipofectin reagent (Gibco-BRL). Recombinant QM7 cell lines were selected in the presence of 0.8 mg of geneticin (G418) per ml. Cell lines expressing VP22MS were obtained by transfecting plasmids pTKhyg (harboring the hygromycin resistance gene) and pUL49s-48 at a ratio of 0.25 μg of pTKhyg per 10 μg of pUL49s-48 and subsequent selection in the presence of 0.2 mg/ml of hygromycin. VP22-, VP22N1-, VP22N2-, and VP22MS-expressing clones were identified by using anti-VP22 monoclonal antibody (AcMo) (9) and were grown in the presence of 0.4 mg of G418 per ml.
Virus growth curves.
Virus growth curves were determined as described (27). Briefly, 2 × 106 CESC were infected with approximately 100 PFU of BAC20 or mutant viruses. At 0, 6, 16, 24, 48, 72, 96, 120, 144, and 168 h postinfection (p.i.), infected cells were harvested and titrated on fresh CESC. The PFU-per-plaque yield was calculated by dividing the number of plaques obtained after trypsinization by the number of plaques at day 0 (approximately 100).
MAbs and IIF analyses.
Monoclonal antibodies (MAbs) were raised against baculovirus-expressed VP5 (major capsid protein, [17, 24]) and VP11/12 and VP13/14 (F. Dorange and J.-F. Vautherot, unpublished data) by using a protocol described earlier (9, 38). The antibodies were termed F19 (anti-VP5), J3 (anti-VP11/12), and L13B (anti-VP13/14).
Infected or transfected cell monolayers were fixed and permeabilized at 4 or 5 days p.i. (or posttransfection) by addition of ethanol-acetone (75:25) at −20°C for 30 min, and indirect immunofluorescence (IIF) was performed as previously described (9), except that Oregon Green 488-labeled anti-VP5 MAb F19 and Texas Red-X labeled anti-VP22 MAb D18 (9) were used for colocalization studies.
To perform double immunostaining, MAbs were purified on protein A columns (Immunopure Plus immobilized protein A IgG purification kit; Pierce) and conjugated to Oregon Green or Texas Red-X by using the FluoReporter Oregon Green 488 or Texas Red-X protein labeling kits (Molecular Probes).
RESULTS
Construction of MDV-1 mutant viruses.
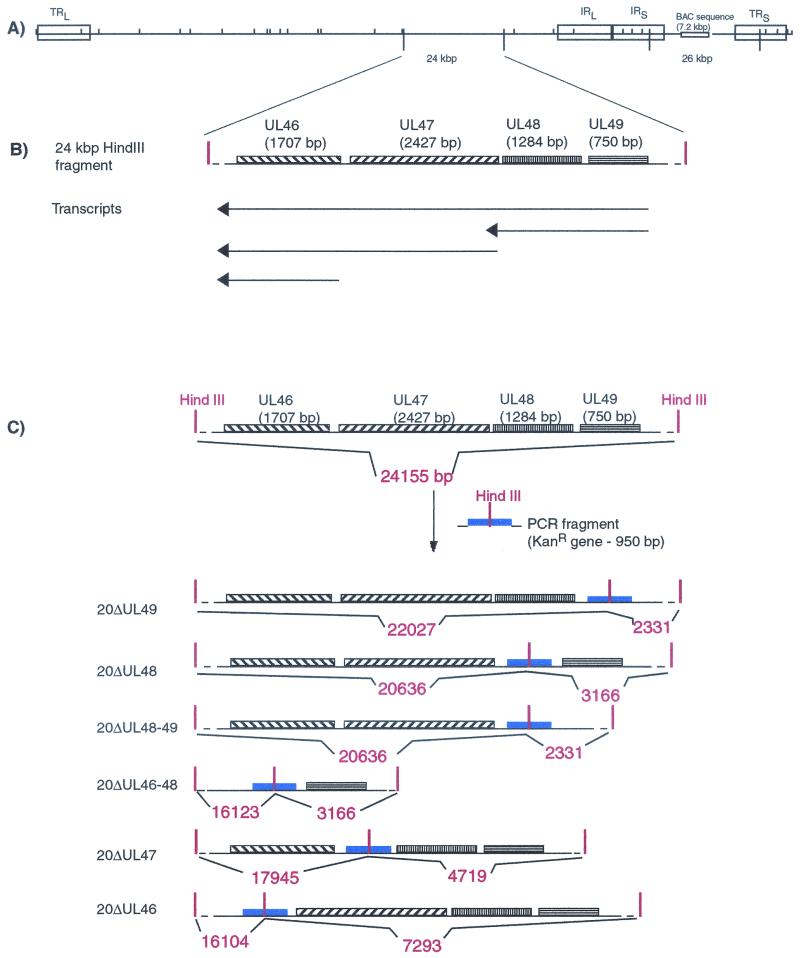
In the BAC20 genome, the UL46 to UL49 genes are present on a 24-kbp HindIII fragment (Fig. 1A). In mutant BAC20 clones, insertion of the Kanr gene by homologous recombination within this fragment introduced an additional HindIII restriction site, dividing the 24-kbp fragment into two smaller fragments (Fig. 1C).
FIG. 1.
(A) Schematic diagram of the genomic organization and HindIII restriction map of BAC20 virus. (B) Transcriptional organization in the UL46 to UL49 gene region of MDV-1 (41). (C) Schematic illustration of the BAC20 mutagenesis and the construction of mutant BAC20 clones 20Δ46, 20Δ47, 20Δ48, 20Δ46-48, 20Δ49, and 20Δ48-49. Digestion of BAC20 with HindIII leads to a 24-kb fragment which contains the UL46 to UL49 genes. The insertion of the Kanr gene by homologous recombination generated a new HindIII restriction site in all mutant BAC20 clones, which results in generation of two fragments. Fragment sizes are given in base pairs and were calculated on the basis of the MDV-1 strain Md5 sequence.
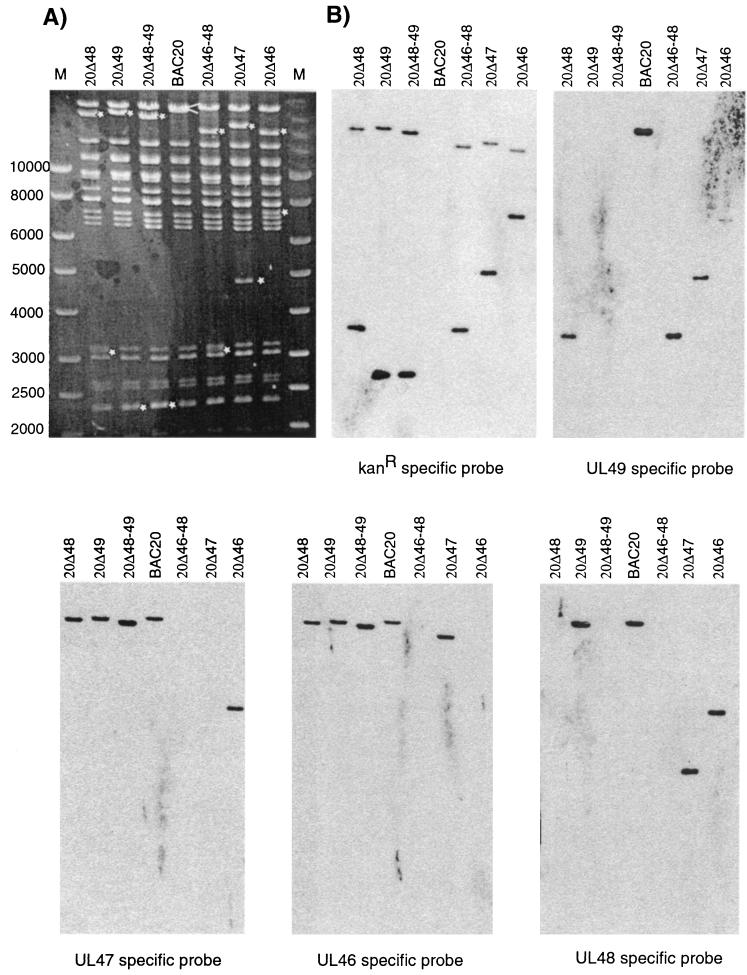
In an ethidium bromide-stained agarose gel (Fig. 2A), a double band was visualized in digested BAC20 DNA (arrowhead) which corresponds to the 24- and 26-kbp HindIII fragments present in the BAC20 genome (Fig. 1A). In mutant BAC20 DNA, the 24-kbp fragment disappeared, and two smaller fragments were clearly visible (Fig. 2A, asterisks).
FIG. 2.
HindIII digestion and Southern blot analysis of mutant BAC20 clones. (A) BAC DNA was digested with HindIII, and the resulting fragments were separated on an ethidium bromide-stained 0.8% agarose gel. Arrowhead and asterisks indicate the 24- and 26-kb doublet band in BAC20 and the new fragments generated by the insertion of the Kanr gene in mutant BAC clones, respectively. (B) DNA fragments were transferred to a nylon membrane, and hybridizations were performed using Kanr-, UL49-, UL47-, UL46-, and UL48-specific probes. The DNA separated on the individual lanes is given. The sizes of the DNA molecular weight markers in lanes M (Smartladder; Eurogentec) are indicated.
Separated DNA fragments were transferred to a nylon membrane, and hybridizations were carried out using Kanr or gene-specific probes (Fig. 2B). As expected, the Kanr-specific probe detected two fragments in DNA of all mutant BAC20 clones and did not hybridize with BAC20 DNA (Fig. 2B). These fragments exhibited the calculated sizes after introduction of the Kanr gene. The gene-specific probes detected a single fragment of the calculated size in all mutant BAC20 clones except for the clone in which the respective gene(s) had been deleted, confirming the deletion of the gene(s) of interest in the individual mutant BAC20 clones. In all cases, the gene-specific probes detected the 24-kbp HindIII fragment of BAC20 (Fig. 2B). In addition, determination of the nucleotide sequences of the recombination sites in all mutant genomes confirmed the correct insertion of the Kanr fragment into the targeted locus (data not shown).
VP11/12, VP13/14, and VP16 are nonessential for growth of MDV-1.
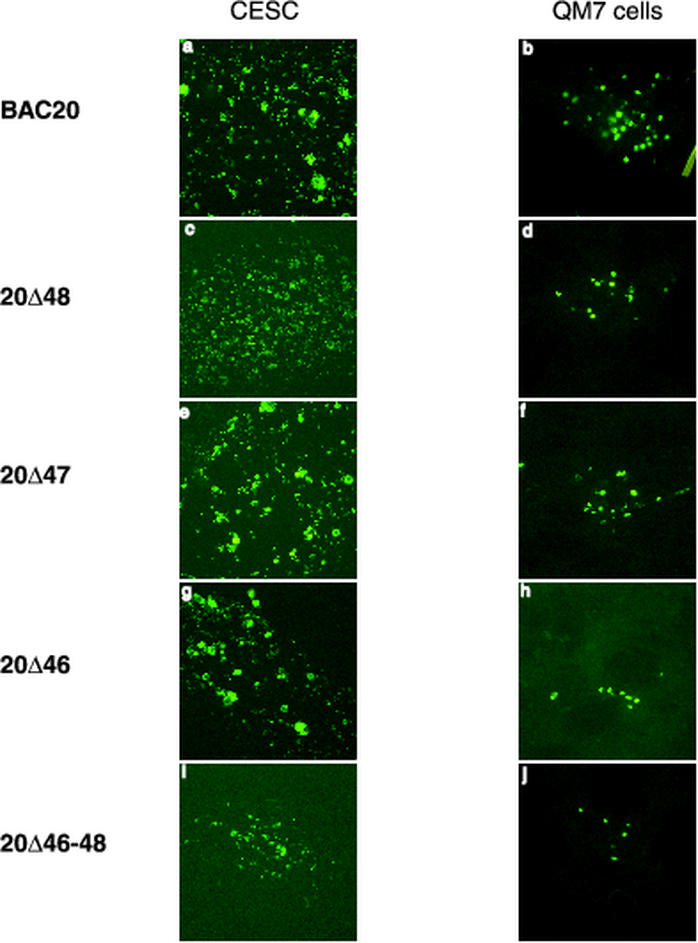
DNA of mutant BAC clones 20Δ46, 20Δ47, and 20Δ48, which are unable to express VP11/12, VP13/14, and VP16, respectively, were transfected into either CESC or QM7 cells to analyze the growth properties of the mutant viruses. CESC are efficient targets for propagation of MDV-1 strains because plaques can easily be visualized from day 2 after infection or transfection. Plaque formation was readily observed after transfection of BAC20 or any of the mutant DNAs mentioned above, and viral antigen was detected by IIF by using the anti-VP5 MAb F19 from day 4 after transfection (Fig. 3). Similarly, fluorescent foci were observed in QM7 cells transfected with the mutant BAC20 DNAs (Fig. 3). All tested mutant viruses, however, formed smaller plaques in both cell types when compared to those of parental BAC20 virus (Fig. 3). Moreover, the sizes of the plaques formed by the 20Δ46 and 20Δ47 viruses appeared to become smaller by day 5, and no 20Δ46 virus plaques could be seen at day 7 after transfection under the light microscope.
FIG. 3.
Digital images of immunofluorescence analysis of BAC20 virus and mutant 20Δ46, 20Δ47, 20Δ48, and 20Δ46-48 viruses. The respective BAC DNA was transfected into CESC and QM7 cells. Cells were fixed on days 5 and 4 after transfection, respectively, and reacted with an Oregon Green-labeled anti-VP5 MAb F19. Cells were examined with a 10× objective (a to i) or a 20× objective (b to j) using a Nikon fluorescent microscope equipped with a TV Lens Nikon digital camera.
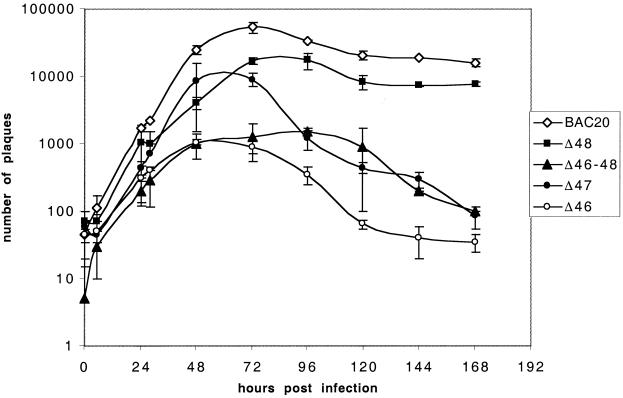
These results prompted us to monitor a possible growth impairment in the mutant viruses by analyzing their growth kinetics on CESC (Fig. 4). BAC20 virus titers reached a maximum at 72 h p.i. (6 × 104 PFU/2 × 106 CESC), and slightly decreased until day 7 (2 × 104 PFU/2 × 106 cells). All four mutant viruses, 20Δ46, 20Δ47, 20Δ48, and 20Δ46-48, exhibited reduced maximal virus titers compared to BAC20. The 20Δ48 virus reached maximal titers of 2 × 104 PFU/2 × 106 cells at 72 h p.i. Mutant virus 20Δ47 reached a maximal titer of 9 × 103 PFU/2 × 106 cells at 72 h p.i., but afterwards virus titers continuously decreased to 1 × 102 PFU/2 × 106 cells on day 7 p.i.
FIG. 4.
Growth curves of mutant MDV-1. After infection of primary CESC with approximately 100 PFU of the individual mutant viruses, virus titers were determined at the indicated time p.i. by inoculating virus dilutions onto fresh CESC. Average values and standard deviations of data from two independent experiments are shown.
Mutant virus 20Δ46 was significantly impaired in virus growth, and the maximal titer reached was 1 × 103 PFU/2 × 106 cells at 72 h. p.i. The virus carrying a deletion of UL46, UL47, and UL48 (20Δ46-48) was severely impaired early in infection even compared to the 20Δ46 virus, but reached maximal titers which were comparable to those of 20Δ46 at 72 h p.i.
Taken together, the results of the virus growth curves for the individual MDV-1 mutants corroborated our initial observations, which had indicated a marked reduction of plaque sizes for the 20Δ46, 20Δ47, and 20Δ46-48 viruses, but it could also be demonstrated that each of the major tegument proteins alone or in combination is nonessential for cell-to-cell spread and growth of MDV-1, although a moderate additive effect after deletion of all three major tegument was readily observed.
VP22 is essential for replication for MDV-1.
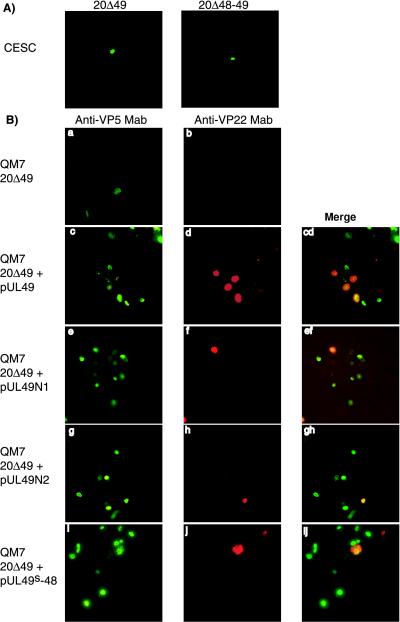
The next series of experiments addressed the effect of a deletion of UL48 and UL49 from the MDV-1 genome either individually or in combination. As described above, DNA of mutant BAC clones 20Δ49 and 20Δ48-49 was transfected into either CESC or QM7 cells to analyze the growth properties of mutant MDV-1 harboring deletions in VP16 and/or VP22. After transfection of mutant 20Δ49 or 20Δ48-49 DNA into CESC or QM7 cells, only single cells expressing the major capsid protein VP5 could be detected (Fig. 5A and 5Ba), suggesting that the reconstituted mutant viruses were not able to replicate in cultured cells. Fluorescent foci, however, were observed when 20Δ49 and 20Δ48-49 were cotransfected into QM7 cells together with pUL49, from which the MDV-1 homolog of VP22 is expressed (Fig. 5Bc, 5Bd, and 5Bcd). These results demonstrated that growth of the 20Δ49 and 20Δ48-49 mutant viruses could be at least partially complemented in trans by the transient expression of UL49 in QM7 cells. These results also demonstrated that VP22 is essential for growth of the highly cell-associated MDV-1 in cultured cells.
FIG. 5.
(A) Immunofluorescence analysis of 20Δ49 and 20Δ48-49 replication in CESC and (B) trans-complementation of 20Δ49 in QM7 cells. (A) CESC were transfected with 20Δ49 or 20Δ48-49 DNA, fixed 5 days after transfection, and reacted with an Oregon Green-labeled anti-VP5 MAb F19. (B) QM7 cells (a to j) were transfected with 20Δ49 DNA (a and b), 20Δ49 and pUL49 DNA (c and d), 20Δ49 and pUL49N1 DNA (e and f), 20Δ49 and pUL49N2 DNA (g and h), or 20Δ49 and pUL49s-48 DNA (i and j) and fixed at 4 days after transfection. Cells were reacted with Oregon Green-labeled anti-VP5 MAb F19 and Texas Red-X-labeled anti-VP22 MAb D18. Single-color images were recorded separately and are shown in the left and middle columns. The merged images are shown in the right column. Cells were examined with a 40× objective using a Nikon fluorescent microscope equipped with a TV Lens Nikon digital camera.
To further examine which region of VP22 is required for MDV-1 growth, cotransfection experiments of 20Δ49 DNA and pcDNA3 constructs, which encode truncated forms of VP22, were performed. The N- and C-terminally truncated forms of VP22, VP22N1, VP22N2, and VP22MS, were expressed in QM7 cells at levels that were comparable to that of VP22 (data not shown). It could be clearly demonstrated that all mutant VP22 constructs were able to trans-complement for growth of the UL49-negative MDV-1, as foci of fluorescent cells were observed after cotransfection of mutant BAC20 DNA with the respective expression plasmid. All foci were associated with or close to individual cells expressing VP22 or its deleted forms from pcDNA3 constructs (Fig. 5Bc to Bj). These observations indicated restored cell-to-cell spread after cotransfection of UL49 expression plasmids (Fig. 5), whereas only single infected cells were observed after transfection of QM7 cells with the UL49- or UL48-49-negative BAC20 DNA alone (Fig. 5Ba and Bb).
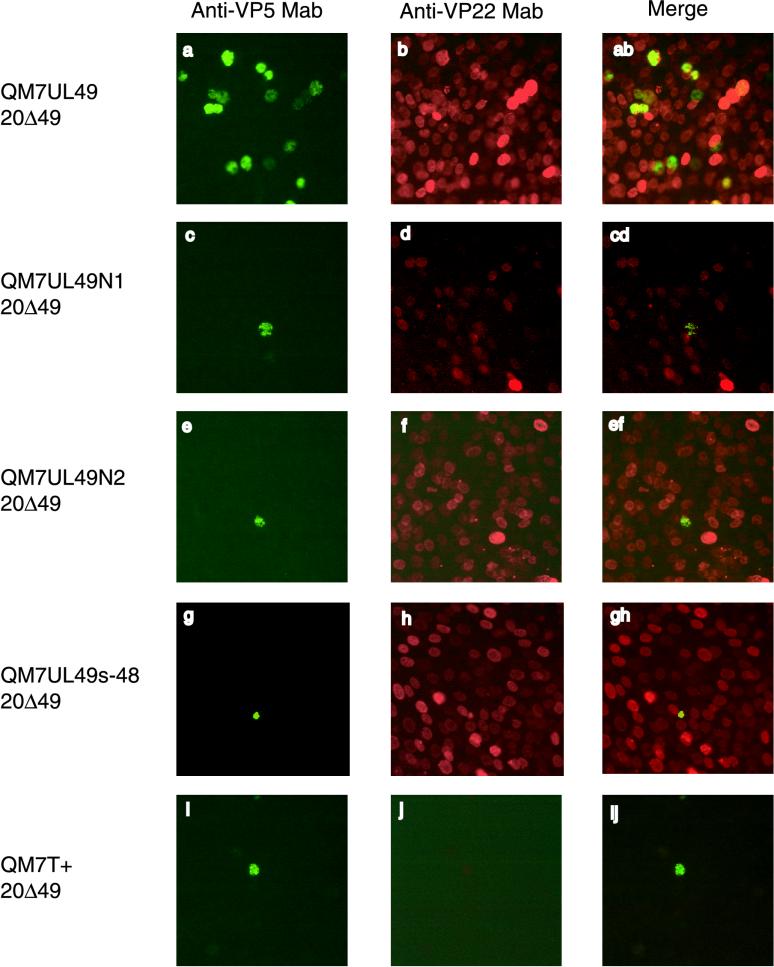
Because transient transfection using pcDNA3 vectors led to expression of full-length and truncated VP22 in a variable number of cells, we selected QM7 cell lines which constitutively expressed VP22, VP22N1, VP22N2, and VP22MS. These cell lines were named QM7UL49, QM7UL49N1, QM7UL49N2, and QM7UL49s-48, respectively, and were used to investigate whether they would more efficiently trans-complement growth of the 20Δ49 and 20Δ48-49 mutant viruses. Full-length and truncated VP22 were detected at similar levels except for the QM7UL49N1 cell line, in which VP22N1 appeared to be expressed at a lower level (Fig. 6d). In the QM7UL49s-48 cell line, VP22MS was readily detectable (Fig. 6h), whereas VP16 was not detected by IIF assays (data not shown).
FIG. 6.
Immunofluorescence analysis of trans-complementation assays of 20Δ49 in QM7 cell lines constitutively expressing full-length or truncated VP22. 20Δ49 DNA was transfected into QM7UL49 (a and b), QM7UL49N1 (c and d), QM7UL49N2 (e and f), QM7UL49s-48 (g and h), or QM7T+ cell lines (i and j). Four days after transfection, cells were fixed and stained with Oregon Green-labeled anti-VP5 MAb F19 and Texas Red-X-labeled anti-VP22 MAb D18. Single-color images were recorded separately and are shown in the left and middle columns. The merged images are shown in the right column. Cells were examined with a 40× objective using a Nikon fluorescent microscope equipped with a TV Lens Nikon digital camera.
Surprisingly, none of the cell lines constitutively expressing truncated VP22 was able to trans-complement the growth defect of 20Δ49 and 20Δ48-49 (Fig. 6c to 6h). In the QM7UL49 cell line expressing full-length VP22, however, foci of infected cells indicating trans-complementation could be observed (Fig. 6a, 6b, and 6ab). It must be noted that the average number of foci per 20Δ49- or 20Δ48-49-transfected QM7UL49 cells was rather low, as only 1 focus per 15 transfected cells was observed after transfection. In contrast, formation of foci was observed in all cell lines expressing full-length or truncated VP22 after transfection of BAC20 DNA, suggesting that the inability of mutant MDV-1 to grow on VP22-expressing cell lines was not caused by an interference phenomenon (data not shown), but rather by an inability of constitutively expressed VP22 mutant proteins to trans-complement growth of UL49-negative MDV-1.
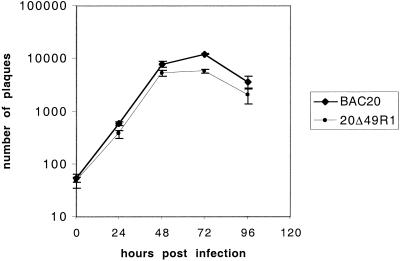
Because both the 20Δ49 and 20Δ48-49 viruses exhibited a major growth defect which could not be totally complemented by QM7UL49 cells, a rescuant virus was generated to ascertain that this lethal mutation was associated with the deletion of UL49 and did not result from a secondary site mutation. The rescuant virus, 20Δ49R1, was obtained by cotransfecting 20Δ49 DNA and pGEM-T48-49.5 in CESC. Monolayers were examined for visible plaques from day 3 posttransfection, and a single plaque was picked and amplified on fresh CESC. Both BAC20 and 20Δ49R1 replicated at comparable levels in CESC (Fig. 7), suggesting that the repair of the deletion in UL49 of the 20Δ49 virus was able to totally rescue virus growth in cultured cells and that the UL49 gene is indeed essential for MDV-1 growth.
FIG. 7.
Growth characteristics of the 20Δ49R1 rescuant virus. Primary CESC were infected with 100 PFU of either 20Δ49R1 or BAC20 virus per 2 × 106 CESC, and virus titers were determined at various times p.i. by inoculating dilutions of infected cells with fresh CESC.
DISCUSSION
By using a recently established MDV-1 BAC clone (32), we have constructed a series of MDV-1 mutants with deletions of the UL46 to UL49 genes, which encode the MDV-1 homologs of HSV-1 major tegument proteins.
We could demonstrate that MDV-1 VP16, encoded by the UL48 gene, is nonessential for virus replication in cell culture, because a virus carrying a deletion of UL48 was able to form plaques in CESC and QM7. This result supported our initial IIF studies, in which we were not able to detect expression of the VP16 protein in CESC infected by MDV-1 strain RB1B (9). However, the UL48-negative virus exhibited a slight growth impairment in CESC compared to the BAC20, and PFU-per-plaque yield was shown to be reduced to 240 versus 1,100 in the BAC20 virus.
In recent IIF analyzes on CESC infected with either MDV-1 strain RB1B or BAC20 virus, discrete but very weak immunofluorescent staining was detected with a VP16-specific MAb in enlarged cells corresponding to fused infected cells, especially when paraformaldehyde was used as the fixation agent (Dorange and Vautherot, unpublished). This may suggest low-level expression and accumulation of VP16 in later stages of infection and in dying cells and would explain the moderate but detectable impairment of 20Δ48 in virus growth.
Another possibility for the reduced growth properties of 20Δ48 may be that a short open reading frame (ORF91), which encodes a polypeptide of 91 amino acids in length, and overlaps the UL48 ORF (41) could be functional and may encode a peptide playing a role in virus replication in cultured cells. As UL49.5, UL49, and UL48 seem to have the same polyadenylation signal located downstream of UL48 (13, 42), the replacement of UL48 by the Kanr gene could also have a deleterious effect on the transcription of the bi- and tricistronic mRNAs. However, this poly(A) signal was not affected by the mutagenesis performed, and VP22 appeared to be expressed at levels similar to those in 20Δ48- or BAC20-infected CESC (data not shown). The UL47 promoter, located between the UL48 and UL47 genes in the intergenic region, was also not affected by the mutagenesis, and VP13/14 was readily detected in IIF assays (data not shown), indicating that the overall transcriptional organization of this region is still intact after insertion of the Kanr gene.
Comparison of MDV-1 VP16 with its homologs of other alphaherpesviruses suggests that VP16 expression is regulated differently in MDV-1, and the exact role of VP16 in infection still remains to be determined. For example, varicella-zoster virus (VZV) VP16 (ORF10) is abundantly expressed in infected cells but is dispensable for virus growth (7). In contrast, HSV-1 VP16 is absolutely required for virus growth, as deletion of VP16 abolishes the ability of HSV-1 to grow in cell culture (40). HSV-1 VP16 has been demonstrated to perform several roles in virus replication through transactivation of immediate-early gene expression (4), interaction with virion host shutoff protein VHS (34), and involvement in virus egress (22). Whether MDV-1 VP16 is involved in viral replication in the early events of infection or in virus maturation and cell-to-cell spread in the late stages of infection or in both will be determined in future experiments, but deletion of this gene had only a minor impact on MDV-1 growth in cultured cells. Moreover, the study of the expression and function of the MDV-1 VHS homolog will be of major interest for an understanding of the role of VP16 in MDV-1 infection.
MDV-1 strains carrying deletions of the UL46 and UL47 genes were able to form plaques on CESC. Both viruses, however, showed a severe growth impairment compared to BAC20 virus and reached a maximal PFU-per-plaque yield of 130 and 17, respectively, compared to 1,100 for the BAC20 virus. Moreover, both viruses also exhibited a significant decrease in plaque size and in PFU-per-plaque yield between days 5 and 7 p.i. When considering the growth curves, the growth impairment of 20Δ46 and 20Δ47 became obvious at day 3 p.i.
A decrease in plaque size was reported for an MDV-1 mutant with a deletion of the US1 gene, coding for the ICP22 homolog (27). The growth curve of the US1-negative virus was very similar to that of the parent virus until day 4, but then a decrease in plaque size and virus yield by day 5 to 7 with a PFU-per-plaque yield of 100 versus 200 for the parent virus by day 7 was observed. The authors demonstrated that this decrease correlated with the survival of cells of the CEF monolayer (27). Although the cell system used in our study is different, this difference in growth titer may also be related to aging of cells. However, we cannot exclude that VP11/12 and VP13/14 play an important role at late stages of infection. Indeed, both proteins were detected mainly in highly infected cells in the center of viral plaques (Dorange and Vautherot, unpublished).
In the case of HSV-1, VP11/12 and VP13/14 have been suggested to be involved in modulation of the activity of VP16 (42, 43). However, both genes are dispensable for virus growth, and deletion of UL46 and UL47 genes either separately or in combination showed little impact on virus growth (42). In the case of HSV-1, however, cells can be infected with mutant viruses harboring deletions of UL46, UL47, and UL46-47 at high multiplicities of infection (5 PFU/cell), which is not possible in the case of MDV-1 due to the high cell association of the agent. The virus harboring a deletion of the UL46, UL47, and UL48 genes, 20Δ46-48, was able to grow on CESC and QM7 cells, but also showed a major growth impairment. We concluded that neither of these genes and their products, either separately or in combination, are essential for MDV-1 growth in cultured cells.
By a series of experiments, it was shown that BAC20 mutant viruses with a deletion of UL49 alone or in combination with UL48 were unable to grow in cultured cells. By generating a rescuant virus, 20Δ49R1, we were able to definitely confirm the essential role of the UL49 gene product in viral replication. We have previously shown that MDV-1 VP22 is an abundantly expressed protein in MDV-1-infected cells, with a mainly nuclear localization (9). The rationale for our trans-complementation using various VP22 constructs was related to our initial observation that upon transfection of the gene into cells, the protein was entering neighboring cells of the monolayer. These results suggested the possibility of interaction between the protein and the maturing mutant viruses.
Both the 20Δ49 and the 20Δ48-49 virus were rescued by cotransfection of the pUL49 expression plasmid, suggesting that VP22 can complement the defect of UL49-negative MDV-1 in trans. VP22 expressed in the baculovirus-insect cell system, although taken up by CESC and QM7 cells in the monolayer, was not able to complement the deficient viruses (data not shown). As expected, MDV-1 VP16, which is also absent in 20Δ48-49, was not required for virus replication, because the defect of this mutant virus could be complemented by expression of VP22 alone. Both N- and C-terminally truncated forms of the MDV-1 VP22 proteins, VP22N1, VP22N2, and VP22MS, also complemented the 20Δ49 virus in trans, suggesting that the DNA-binding region, which is absent in VP22N2, is not essential for virus growth in cell culture. The three truncated proteins contained the region involved in intercellular spread, suggesting an important role of this VP22 property in MDV-1 infection. To our knowledge, this is the first report on an alphaherpesvirus for which VP22 plays an essential role in virus growth and cell-to-cell spread.
In the case of HSV-1, a virus deleted of the entire UL49 gene has not yet been reported. Pomeranz and Blaho (28) report on unsuccessful trials to generate virus mutants with deletions of the entire VP22, and show that a virus coding for a C-terminally truncated form of VP22 grows to similar titers compared to parental HSV-1. However, the authors have noted a reduced plaque size compared to the parental virus and suggested that HSV-1 VP22 is needed for efficient cell-to-cell spread in cultured cells (28).
Whether MDV-1 VP22 is required for virus replication by acting as a trans-activating protein or for virus egress will be determined in future experiments. Our results showing that the MDV-1 UL49-homologous gene is essential for virus growth are in contradiction to the report that the UL49-homologous gene of bovine herpesvirus type 1 (BHV-1) is dispensable for virus growth in cultured cells (18). However, recent studies suggest that BHV-1 VP22 is functionally distinct from its HSV-1 homolog (13) and may therefore also be different from its MDV-1 counterpart.
We reported on the engineering of QM7 cell lines constitutively expressing full-length and truncated VP22. None of the cell lines, however, was able to trans-complement both UL49-negative viruses, although formation of some foci occurred in a very limited number of 20Δ49-transfected QM7 UL49 cells. It should be noted that transfection of BAC20 DNA in all four lines led to foci of infected cells, suggesting that no major cellular factors involved in the infection were damaged in these cell lines. The fundamental difference between the abilities of the cell lines permanently expressing truncated VP22 proteins versus QM7 cells transiently expressing the same proteins, where efficient trans-complementation of UL49-negative viruses was observed, needs to be clarified. Improper posttranslational modifications of HSV-1 VP22 have been suggested to exert an important effect on the distribution of VP22 and may also contribute to different function of the protein in infected cells (29).
We have previously reported that the recombinant VP22 and VP22MS proteins are phosphorylated and that two phosphorylated forms are detectable in insect cells (9). It is conceivable that incorrect phosphorylation of the truncated (and full-length) proteins in permanent cell lines prevents efficient trans-complementation of the UL49-negative MDV-1 and that a timely coordinated expression of the VP22 protein with another viral protein(s) may be required for the formation of functional VP22. Further work will be directed to elucidating whether the multiple forms of full-length and truncated VP22 are present in QM7 cells which either transiently or stably express these proteins and whether the phosphorylation pattern of VP22 has an impact on its ability to complement growth of UL49-negative MDV-1.
Acknowledgments
The technical assistance of A. Francineau for all cell culture experiments is gratefully acknowledged.
F. Dorange was supported by an INRA/Region Centre fellowship. The work was supported by grant QLK2-CT-1999-00601 from the Commission of the European Union. The laboratory of Jean-François Vautherot is a member of the IFR Biologie Comparée des Transposons et Virus.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckmaster, A. E., S. D. Scott, M. J. Sanderson, M. E. Boursnell, N. L. Ross, and M. M. Binns. 1988. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J. Gen. Virol. 69:2033-2042. [DOI] [PubMed] [Google Scholar]
- 3.Calnek, B. W., and R. L. Witter. 1991. Marek's disease, 10th ed. Iowa State University Press, Ames, Iowa.
- 4.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 5.Cebrian, J., C. Kaschka-Dierich, N. Berthelot, and P. Sheldrick. 1982. Inverted repeat nucleotide sequences in the genomes of Marek disease virus and the herpesvirus of the turkey. Proc. Natl. Acad. Sci. USA 79:555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219-2230. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 12.Fenwick, M. L., and M. J. Walker. 1978. Suppression of the synthesis of cellular macromolecules by herpes simplex virus. J. Gen. Virol. 41:37-51. [DOI] [PubMed] [Google Scholar]
- 13.Harms, J. S., X. Ren, S. C. Oliveira, and G. A. Splitter. 2000. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J. Virol. 74:3301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumiya, Y., H. K. Jang, H. Kashiwase, J. S. Cai, Y. Nishimura, Y. Tsushima, K. Kato, T. Miyazawa, C. Kai, and T. Mikami. 1998. Identification and transcriptional analysis of the homologues of the herpes simplex virus type 1 UL41 to UL51 genes in the genome of nononcogenic Marek's disease virus serotype 2. J. Gen. Virol. 79:1997-2001. [DOI] [PubMed] [Google Scholar]
- 16.Kut, E. 2001. Assemblage de la capside de l'herpèsvirus de la maladie de Marek (MDV). Mémoire présenté pour l'obtention du diplôme de l'Ecole Pratique des Hautes Etudes (EPHE), Ministère de l'Éducation Nationale, de la Recherche et de la Technologie (MENRT).
- 17.Lee, L. F., P. Wu, D. Sui, D. Ren, J. Kamil, H. J. Kung, and R. L. Witter. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc. Natl. Acad. Sci. USA 97:6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight, J. L., P. E. Pellett, F. J. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate alpha-trans-inducing factor-dependent activation of alpha genes. J. Virol. 61:992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan, R. W., J. L. Cantello, and C. H. McDermott. 1990. Transfection of chicken embryo fibroblasts with Marek's disease virus DNA. Avian Dis. 34:345-351. [PubMed] [Google Scholar]
- 22.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6:442-447. [DOI] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 25.Normand, N., H. van Leeuwen, and P. O'Hare. 2001. Particle formation by a conserved domain of the herpes simplex virus protein VP22 facilitating protein and nucleic acid delivery. J. Biol. Chem. 276:15042-15050. [DOI] [PubMed] [Google Scholar]
- 26.Overton, H. A., D. J. McMillan, L. S. Klavinskis, L. Hope, A. J. Ritchie, and P. Wong-kai-in. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 27.Parcells, M. S., A. S. Anderson, J. L. Cantello, and R. W. Morgan. 1994. Characterization of Marek's disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames. J. Virol. 68:8239-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silim, A., M. A. El Azhary, and R. S. Roy. 1982. A simple technique for preparation of chicken-embryo-skin cell cultures. Avian Dis. 26:182-185. [PubMed] [Google Scholar]
- 34.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vautherot, J. F., M. F. Madelaine, P. Boireau, and J. Laporte. 1992. Bovine coronavirus peplomer glycoproteins: detailed antigenic analyses of S1, S2 and HE. J. Gen. Virol. 73:1725-1737. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Z. H., M. D. Gershon, O. Lungu, Z. Zhu, S. Mallory, A. M. Arvin, and A. A. Gershon. 2001. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J. Virol. 75:323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanagida, N., S. Yoshida, K. Nazerian, and L. F. Lee. 1993. Nucleotide and predicted amino acid sequences of Marek's disease virus homologues of herpes simplex virus major tegument proteins. J. Gen. Virol. 74:1837-1845. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]