Abstract
Astrovirus contains three open reading frames (ORF) on its genomic RNA, ORF1a, ORF1b, and ORF2. ORF1a encodes a 920-amino-acid (aa) nonstructural protein, nsP1a, which displays a 3C-like serine protease motif. Little is known about the processing of nsP1a or whether the protease it contains is active and involved in autocatalytic processing. Here we address both of these matters. Intact and N-terminally deleted forms of ORF1a from human astrovirus serotype 1 were expressed in BHK cells, and nsP1a-derived processing products were immunoprecipitated with an nsP1a-specific antibody or an antibody specific for an N-terminally linked epitope tag. The mapping of the main processing products, p20 and p27, suggests cleavage sites near aa 170, 410, and 655 of nsP1a. Cleavages at around aa 410 and 655, but not aa 170, were abolished when a 9-aa substitution was introduced into the protease motif in nsP1a. The p27 processing product was also found in Caco-2 cells that had been infected with human astrovirus serotype 1, confirming the presence of the cleavage sites at approximately aa 410 and 655.
Human astroviruses (HAstV) are nonenveloped, positive-strand RNA viruses which recent epidemiological studies have identified as an important cause of acute gastroenteritis among young children worldwide (15). To date, eight serotypes of HAstV have been reported in the literature, among which serotype 1 (HAstV-1) is the most prevalent (15). For two strains of HAstV-1 (12, 22), one strain of HAstV-2 (8), one strain of HAstV-3 (18), and one strain of HAstV-8 (16), the complete genome sequences have been determined. The viral genome of HAstV-1 (Oxford strain) consists of 6,771 nucleotides (nt), excluding the 3"-terminal poly(A) tail, and contains three open reading frames (ORF), ORF1a, ORF1b, and ORF2. Both ORF1a and ORF1b, which contain the conserved motifs for a 3C-like serine protease and an RNA-dependent RNA polymerase, respectively, are believed to be translated directly from the genomic viral RNA. The translation product of ORF1a is designated nonstructural protein 1a (nsP1a). Expression of ORF1b, which is in a −1 position relative to ORF1a, depends on the occurrence of a −1 ribosomal frameshift during the translation of ORF1a (5 to 7% frequency in a cell-free, uncoupled transcription-translation system [13]), leading to an nsP1a/1b fusion protein (13, 14). ORF2 encodes the viral capsid protein and is expressed from a subgenomic RNA colinear with the 3" one-third of the genomic RNA (12, 17).
Little is known about the processing of the astroviral nonstructural proteins nsP1a and nsP1a/1b translated from ORF1a and ORF1b or about the role of the presumed 3C-like serine protease in nsP1a. Gibson et al. reported the in vitro expression of the full-length nsP1a and nsP1a/1b (4) but presented no data on processing. In another study (9), a single autocatalytic processing event was observed in nsP1a in a cell-free expression system. In contrast, Willcocks et al. found multiple nsP1a-derived fragments in infected Caco-2 cells (21) but did not define any cleavage sites. Here we present a map for the processing of nsP1a of HAstV-1 in tissue culture cells as well as evidence for autocatalytic cleavage.
We constructed a series of plasmids containing ORF1a and ORF1b sequences derived from a full-length cDNA clone of HAstV-1 (pAVIC) (2). The initial construct, which contained the complete ORF1a as well as the first 367 nt of ORF1b, was modified in one or more of the following ways: the coding sequence for a 9-amino-acid (aa) Arg-Gly-Ser-His6 epitope (His tag) was fused to the 5" end of the intact ORF1a (His-nsP1a) or the 5" end of a truncated ORF1a lacking the 5" 1,179 nt coding for aa 1 to 393 of nsP1a (His-394-nsP1a), or the protease domain in nsP1a was disrupted by replacing aa 546 to 554 with an unrelated sequence (Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala), thereby deleting the proposed catalytic serine (Ser551) and two amino acids implicated in substrate binding, Thr546 and Gln547 (nsP1aΔ, His-nsP1aΔ, and His-394-nsP1aΔ). All cloning operations were performed according to standard protocols (19). Deletions, insertions, and mutations were introduced by using a PCR-based strategy (6, 7) and verified by sequencing. The construct coding for 421-nsP1a, which was kindly provided by D. Kiang, contained ORF1a-derived sequences coding for aa 421 through 920 of nsP1a and no ORF1b sequences.
The vaccinia virus-driven infection-transfection system (1) was used to express the series of astrovirus ORF1a- and ORF1b-derived sequences in BHK cells. After metabolic labeling with [35S]methionine-cysteine, 35S-labeled nsP1a-specific products were immunoprecipitated from cell lysates with either a monoclonal antibody against the N-terminal His tag (MAb αHis; Qiagen, Valencia, Calif.) or an nsP1a-specific polyclonal antibody (pAb 5-6). The latter was produced by mice immunized with a fragment of nsP1a spanning aa 445 to 688 that was expressed in bacteria as a fusion protein with glutathione S-transferase (20). Immunoprecipitates were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and detected by autoradiography. Differences in the intensities of the 35S bands among the lanes can be attributed to the levels of expression of the individual ORF1a-derived sequences as well as to the stabilities of the translation products. However, within one lane, the intensities of the bands reflect the relative amounts of cleavage products for a given translation product.
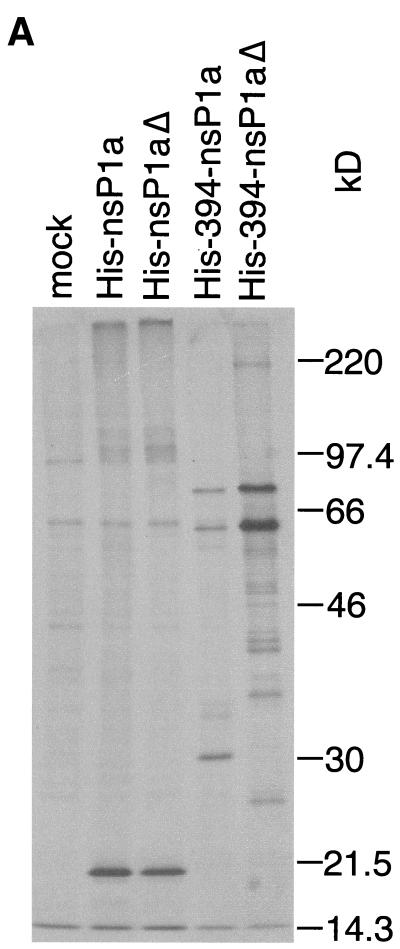
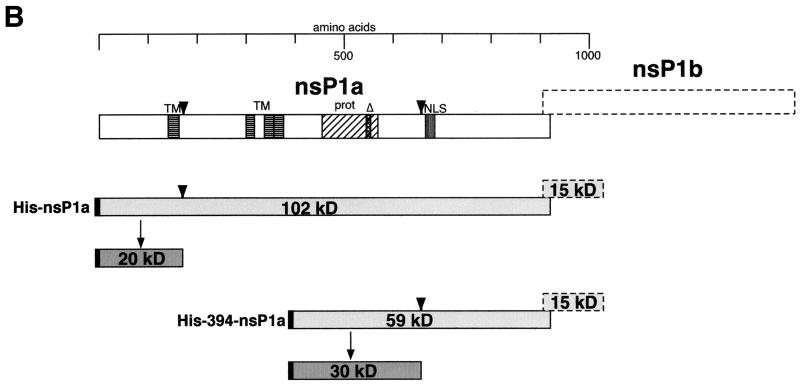
When the full-length nsP1a linked to an N-terminal His tag (His-nsP1a) was expressed in BHK cells as described above, a 20-kDa fragment was immunoprecipitated with MAb αHis (Fig. 1A). The 20-kDa fragment has to extend from the N terminus of nsP1a, as it contains the N-terminally linked His tag. Therefore, the 20-kDa band suggests cleavage at approximately aa 180 of His-nsP1a (or at aa 170 of nsP1a) (Fig. 1B). A faint band of about 101 kDa probably represents the unprocessed, full-length His-nsP1a translation product (expected molecular mass, 102 kDa). Disruption of the protease motif in nsP1a (His-nsP1aΔ) did not diminish immunoprecipitation of the 20-kDa fragment (Fig. 1A), indicating that cleavage at around aa 170 of nsP1a does not depend on an intact ORF1a-encoded protease.
FIG. 1.
Immunoprecipitation of His-tagged nsP1a-derived proteins expressed in BHK cells with MAb αHis. (A) Confluent BHK cells in six-well plates were infected with a recombinant vaccinia virus (multiplicity of infection, 10) encoding T7 DNA-dependent RNA polymerase. After 45 min, the infected BHK cells were transfected with 2 to 3 μg of plasmid DNA encoding nsP1a/1b-derived sequences under the control of a T7 DNA-dependent RNA polymerase promoter, or a control plasmid, by using Lipofectamine in accordance with the manufacturer's directions (Gibco-BRL, Gaithersburg, Md.). At 4 h posttransfection, the medium was removed, and the cells were starved in Met-Cys-free medium for 30 min and then labeled with 30 to 50 μCi of [35S]Met-Cys (Tran35S-label; ICN Biomedicals, Irvine, Calif.) per well for 15 min. Cell lysates were prepared, and nsP1a-specific products were immunoprecipitated with MAb αHis (Qiagen) as described previously (5). The immunoprecipitated products were separated on an SDS-10% polyacrylamide gel (11), and the radiolabeled bands were visualized by autoradiography. Prestained molecular mass standards (Rainbow Marker; Amersham Pharmacia Biotech, Piscataway, N.J.) were run in parallel, and their positions on the gel were transferred to the autoradiogram. Mock, mock-infected cells. (B) Interpretation of the bands seen in panel A. Open boxes represent the generic nsP1a and nsP1b encoded by ORF1a and ORF1b, respectively. nsP1b is translated as a fusion protein with nsP1a only after the occurrence of a −1 ribosomal frameshift. Conserved motifs are indicated by horizontal stripes (TM, transmembrane helices), vertical stripes (NLS, nuclear localization signal), diagonal stripes (prot, protease), and cross-hatching (Δ, 9-aa substitution in protease; see text). nsP1a-derived translation products expressed in BHK cells are depicted as lightly shaded boxes. Processing products identified on the gel are shown as darkly shaded boxes. The N-terminally linked His tag is indicated in black. Suggested cleavage sites are indicated by arrowheads.
An ORF1a-derived translation product lacking the N-terminal 393 aa of nsP1a and containing a His tag coupled to aa 394 (His-394-nsP1a) (Fig. 1A) gave rise to three His-tagged fragments, measuring 30, 61, and 76 kDa in size. The 30-kDa band suggests cleavage 260 aa from the His tag, at approximately aa 655 of nsP1a (Fig. 1B), while the 61-kDa band probably reflects the full-length His-394-nsP1a translation product (calculated molecular mass, 59 kDa). The 76-kDa band may be explained by the occurrence of a frameshift after aa 903 of nsP1a that led to the translation of the 122 ORF1b-derived amino acids followed by 11 vector-encoded amino acids (expected molecular mass of the fusion protein, 72 kDa). The prominence of the 76-kDa band suggests that during translation of His-394-nsP1a, frameshifting occurs at a frequency higher than the 5 to 7% reported previously (13). This may be due to an altered conformation of the nascent His-394-nsP1a during translation compared to that of the full-length nsP1a.
When the protease motif was disrupted in His-394-nsP1aΔ, immunoprecipitation with MAb αHis gave rise to the same 61- and 76-kDa bands (Fig. 1A). A series of much fainter bands with smaller molecular masses was also seen and may represent nonspecific degradation products. However, the specific 30-kDa band seen in the case of His-394-nsP1a was no longer detected, suggesting that cleavage at approximately aa 655 of nsP1a is dependent on the protease in nsP1a.
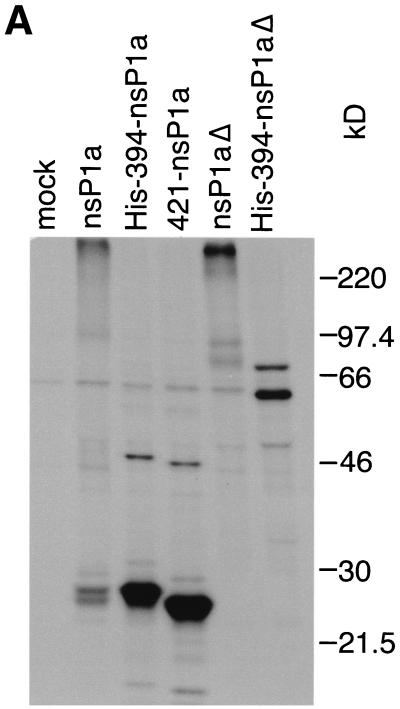
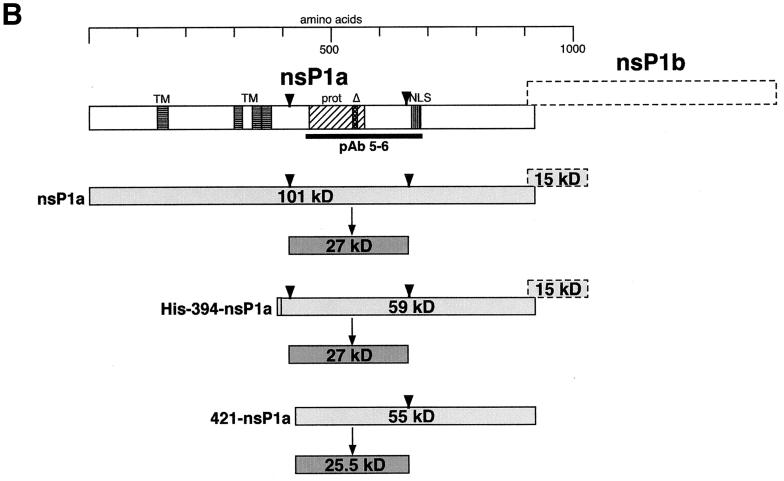
To confirm the locations of the proposed cleavage sites and to identify others, we examined the processing of nsP1a and His-394-nsP1a by using an antibody that recognizes an internal region of nsP1a (pAb 5-6; epitope contained in aa 445 to 688). Lysates of BHK cells expressing nsP1a or His-394-nsP1a were immunoprecipitated with pAb 5-6, and the immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis as described above (Fig. 2A). For nsP1a, two bands, of 26 and 27 kDa, were detected. His-394-nsP1a gave rise to a very strong signal at 27 kDa and a much fainter one at 47 kDa. The presence of the 27-kDa fragment for both nsP1a and His-394-nsP1a indicates that it is derived from a location downstream of aa 393. At the same time, this fragment has to overlap at least part of the region from aa 445 to 688, which contains the pAb 5-6 epitope. Immunoprecipitation of His-394-nsP1a with MAb αHis did not yield a 27-kDa band but did yield a slightly larger, 30-kDa fragment, which extends from aa 394 to the proposed cleavage site at about aa 655 and thus spans most of the region containing the pAb 5-6 epitope (Fig. 1A). This finding suggests that the pAb 5-6-precipitable 27-kDa processing product derived from His-394-nsP1a represents an N-terminally truncated version of the MAb αHis-precipitable 30-kDa fragment. Cleavage at around aa 410 of nsP1a would lead to the removal of a 3-kDa sequence, including the His tag, from the N terminus of the 30-kDa fragment to yield the 27-kDa fragment. To test this hypothesis, we examined the processing of an nsP1a-derived construct lacking the N-terminal 420 aa (421-nsP1a), which we expected to yield an N-terminally truncated pAb 5-6-precipitable processing product about 1 kDa smaller than 27 kDa. As predicted, a 25.5-kDa band and a much fainter 45.5-kDa band (Fig. 2A) were detected, confirming the location of the N terminus of the 27-kDa fragment (and the 47-kDa fragment) at approximately aa 410 of nsP1a (Fig. 2B). At the same time, the length of the 25.5-kDa fragment extending from aa 421 of nsP1a is consistent with the proposed cleavage site at aa 655 of nsP1a. The 45.5- and 47-kDa bands seen in Fig. 2A are likely to have resulted from aberrant processing, since they are detected only for the N-terminally truncated 421-nsP1a and His-394-nsP1a constructs, respectively, and not for the full-length nsP1a.
FIG. 2.
Immunoprecipitation of nsP1a-derived proteins expressed in BHK cells with pAb 5-6. (A) nsP1a/1b-derived sequences were expressed in BHK cells, and processing products were analyzed as described in the legend to Fig. 1A, except that labeling was done for 90 min and pAb 5-6 was used for immunoprecipitation. (B) Interpretation of the bands seen in panel A. Representation is as described in the legend to Fig. 1B. The epitope for pAb 5-6 is indicated by a thick black line.
When the protease motif was disrupted in nsP1aΔ or His-394-nsP1aΔ (Fig. 2A), the 27-kDa fragment could no longer be detected after immunoprecipitation with pAb 5-6. This finding indicates that cleavage at approximately aa 410 and 655 is dependent on an intact protease motif in nsP1a. In the case of His-394-nsP1aΔ, the same pattern of bands representing the full-length translation product and the frameshifted His-394-nsP1aΔ/1b fusion protein could be seen after immunoprecipitation with pAb 5-6 (Fig. 2A, 60 and 72 kDa) as had been observed with MAb αHis (Fig. 1A, 61 and 76 kDa). For nsP1aΔ, a specific band of about 76 kDa was detected (Fig. 2A). This likely reflects the C-terminal cleavage product obtained after processing at approximately aa 170 (expected molecular mass, 82 kDa), which was shown to be independent of the protease contained in nsP1a. It is noteworthy that in the case of nsP1aΔ, much of the radioactivity remains at the top of the gel. To a lesser degree, this is also true for the other full-length constructs, nsP1a (Fig. 2A), His-nsP1a, and His-nsP1aΔ (Fig. 1A). In contrast, for the N-terminally truncated constructs, His-394-nsP1a, His-394-nsP1aΔ, and 421-nsP1a (Fig. 2A), none of the radioactivity can be found at the top of the gel. This observation suggests that sequences between aa 394 and the proposed cleavage site at approximately aa 170 promote the formation of large complexes.
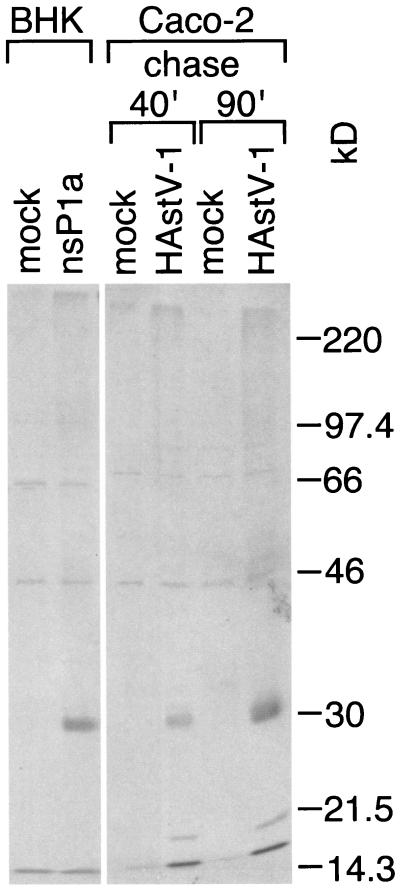
While the expression of ORF1a in BHK cells through a vaccinia virus-driven infection-transfection sytem allows us to study the processing of deleted and mutated forms of nsP1a, the astrovirus replication-independent expression in vaccinia virus-infected cells and the secondary conformational effects of the deletions and mutations may also lead to artificial processing events. We therefore used immunoprecipitation with pAb 5-6 to detect nsP1a processing products present in Caco-2 cells that had been infected with wild-type HAstV-1 and compared them with those found in BHK cells expressing ORF1a by means of the vaccinia virus-based expression system. The results are shown in Fig. 3. The same specific band (28 kDa) can be detected in both astrovirus-infected cells and BHK cells expressing ORF1a through the vaccinia virus-based expression system. This result clearly validates the findings on nsP1a processing in BHK cells. In particular, it demonstrates the presence of the 27- to 28-kDa nsP1a fragment believed to result from cleavage at approximately aa 410 and 655 of nsP1a in infected Caco-2 cells, where nsP1a is expressed and processed in the natural context of astroviral replication.
FIG. 3.
pAb 5-6-precipitable processing products derived from nsP1a expressed in BHK cells or Caco-2 cells. nsP1a was expressed in BHK cells as described in the legend to Fig. 1A, except that labeling was followed by an 85-min cold chase. Caco-2 cells in six-well plates were infected with astrovirus at a multiplicity of infection of 4 or were mock infected. At 4 h postinfection, the cells were starved for 30 min in medium without Cys or Met and then metabolically labeled with 50 μCi of [35S]Met-Cys for 30 min. Medium containing 35S label was replaced with cold medium, and cell lysates were prepared after either a 40- or a 90-min chase. nsP1a-specific products were immunoprecipitated with pAb 5-6 and analyzed as described in the legend to Fig. 1A.
In summary, we propose the following processing sites for nsP1a. Cleavage at approximately aa 170 occurs independently of the protease contained in nsP1a, leading to an N-terminal 19-kDa fragment (20-kDa in the case of His-nsP1a). Processing at approximately aa 410 and 655 yields a 27-kDa product, which encompasses the conserved protease motif spanning aa 454 to 569. Since cleavage at around aa 655 occurs just upstream of the nuclear localization signal found between aa 666 and 679 (8), the 27-kDa fragment encompassing the protease is not expected to be located in the nucleus. In fact, when HAstV-1-infected Caco-2 cells were immunostained with pAb 5-6 and a secondary fluorescein isothiocyanate-conjugated immunoglobulin G, the fluorescent signal was detected only in the cytoplasm (3). The predicted transmembrane helices present upstream of aa 400 in nsP1a (8) are also cleaved from the 27-kDa fragment, making it unlikely that the nsP1a-derived protease is membrane anchored.
Cleavage at approximately aa 410 and 655 appears to depend on the 3C-like serine protease in nsP1a. 3C-like proteases generally require a glutamine, a glutamic acid, or occasionally a valine in the −1 position (10). The +1 position is usually occupied by glycine, alanine, serine, threonine, leucine, isoleucine, valine, or methionine. If we assume the involvement of the ORF1a-encoded 3C-like serine protease, the likely cleavage sites in nsP1a are between Val409 and Ala410 and between Glu654 and Ile655. Since the nsP1a sequence is largely conserved among the known human astrovirus sequences, sequence conservation cannot be used to confirm the proposed cleavage sites. In addition, the possibility has not been excluded that the 9-aa substitution in the protease motif affects the processing of nsP1aΔ indirectly, by distorting the substrate, rather than directly, by disabling the protease. To study the involvement of the protease contained in nsP1a in autocatalytic cleavage in more detail, site-directed mutagenesis of the presumed catalytic triad will have to be performed. The exact definitions of the cleavage sites proposed here will ultimately require N-terminal sequencing of the processing products.
Acknowledgments
We thank J. Gilbert for help with the assay system, D. Kiang for providing the 421-nsP1a construct, and S. Schlesinger, D. Feigelstock, and A. Gnirke for critical reading of the manuscript and many helpful discussions.
This work was supported by grants from the NIH (1 R21 AI43513) and the Department of Veterans Affairs (VA Merit Review) to S.M.M.
REFERENCES
- 1.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5" sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geigenmüller, U., N. H. Ginzton, and S. M. Matsui. 1997. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J. Virol. 71:1713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geigenmüller, U., and S. M. Matsui. Studies on the molecular biology of human astrovirus serotype 1. Perspect. Med. Virol. (Viral Gastroenteritis), in press.
- 4.Gibson, C. A., J. Chen, S. S. Monroe, and M. R. Denison. 1998. Expression and processing of nonstructural proteins of the human astroviruses. Adv. Exp. Med. Biol. 440:387-391. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert, J. M., and H. B. Greenberg. 1997. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J. Virol. 71:4555-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiang, D., and S. M. Matsui. Proteolytic processing of a human astrovirus nonstructural protein. J. Gen. Virol., in press. [DOI] [PubMed]
- 10.Kräusslich, H.-G., and E. Wimmer. 1988. Viral proteinases. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, T. L., H. B. Greenberg, J. E. Herrmann, L. S. Smith, and S. M. Matsui. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 68:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis, T. L., and S. M. Matsui. 1995. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch. Virol. 140:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marczinke, B., A. J. Bloys, T. D. K. Brown, M. M. Willcocks, M. J. Carter, and I. Brierley. 1994. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 68:5588-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui, S. M., and H. B. Greenberg. 2001. Astrovirus. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Mendez-Toss, M., P. Romero-Guido, M. E. Munguía, E. Mendez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 17.Monroe, S. S., S. E. Stine, L. Gorelkin, J. E. Herrmann, N. R. Blacklow, and R. I. Glass. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J. Virol. 65:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh, D., and E. Schreier. 2001. Molecular characterization of human astroviruses in Germany. Arch. Virol. 146:443-455. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 21.Willcocks, M. M., A. S. Boxall, and M. J. Carter. 1999. Processing and intracellular location of human astrovirus non-structural proteins. J. Gen. Virol. 80:2607-2611. [DOI] [PubMed] [Google Scholar]
- 22.Willcocks, M. M., T. D. K. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 75:1785-1788. [DOI] [PubMed] [Google Scholar]