Abstract
We have recently reported an increased heterogeneity in the human immunodeficiency virus type 1 (HIV-1) envelope gene (env) in HIV-1-infected patients with pulmonary tuberculosis (TB) compared to patients with HIV-1 alone. This increase may be a result of dissemination of lung-derived HIV-1 isolates from sites of Mycobacterium tuberculosis infection and/or the systemic activation of the immune system in response to TB. To distinguish between these two mechanisms, blood and pleural fluid samples were obtained from HIV-1-infected patients with active pleural TB in Kampala, Uganda (CD4 cell counts of 34 to 705 cells/μl, HIV-1 plasma loads of 2,400 to 280,000 RNA copies/ml, and HIV-1 pleural loads of 7,600 to 4,500,000 RNA copies/ml). The C2-C3 coding region of HIV-1 env was PCR amplified from lysed peripheral blood mononuclear cells and pleural fluid mononuclear cells and reverse transcriptase-PCR amplified from plasma and pleural fluid HIV-1 virions of eight HIV-1 patients with pleural TB. Phylogenetic and phenetic analyses revealed a compartmentalization of HIV-1 quasispecies between blood and pleural space in four of eight patients, with migration events between the compartments. There was a trend for a greater genetic heterogeneity in the pleural space, which may be the result of an M. tuberculosis-mediated increase in HIV-1 replication and/or selection pressure at the site of infection. Collectively, these findings suggest that HIV-1 quasispecies in the M. tuberculosis-infected pleural space may leak into the systemic circulation and lead to increased systemic HIV-1 heterogeneity during TB.
A diverse population of human immunodeficiency virus type 1 (HIV-1) is rapidly established in an infected individual due to high rates of viral replication and mutations (15, 56). An HIV-1-specific immune response may lead to selection of HIV-1 escape variants (22), whereas the release of cytokines and chemokines, due to a more general immune activation, appears to stimulate HIV-1 replication and increase virus levels (9, 11).
In addition to these systemic observations, HIV-1 dissemination followed by compartmentalization within various organs can lead to divergent evolution of quasispecies subpopulations within an infected individual. Tissue compartmentalization is evident in the lung (1, 16, 25, 41), genital tract (6, 7, 12, 17, 60), and lymph node (1, 13, 58) but is most notable in the brain (13, 20, 29, 58). Some patients have distinct HIV-1 quasispecies phylogenies between the brain and blood, while others have quasispecies that migrate readily between these compartments (20). Divergent evolution of HIV-1 quasispecies coupled with intermittent migration events of these quasispecies between compartments has also been shown in the blood and semen of patients (12), indicating that the semen is also an anatomical reservoir for the virus. Similarly, HIV-1 compartmentalization is observed in the genital tract of women (31). Identification of new HIV-1 reservoirs and a thorough analysis of compartmentalization are necessary to understand intrapatient HIV-1 evolution and to assess the effects of antiretroviral treatment on remote regions of HIV-1 replication.
Secondary infections such as tuberculosis (TB) can affect HIV-1 evolution and compartmentalization. Nakata et al. demonstrated that pulmonary TB may enhance viral compartmentalization as well as HIV-1 replication and heterogeneity in the lung. (24). Unlike other secondary or opportunistic infections, TB can lead to chronic immune stimulation at the sites of Mycobacterium tuberculosis coinfection (52) and may therefore affect both local and systemic viral replication for prolonged periods of time. In fact, TB is associated with increased systemic viral replication (49) and heterogeneity (5), decreased CD4 cell counts, a more rapid progression to AIDS, and increased mortality (57). In vitro, M. tuberculosis infection or purified protein derivative stimulation of primary cell populations results in potent activation of HIV-1 replication and may explain the increase in HIV-1 loads following the diagnosis of TB (11).
We have previously described an increased systemic HIV-1 heterogeneity in patients with active pulmonary TB (5). In a CD4-matched cohort, the mutation frequency of HIV-1 quasispecies in HIV-1-infected adults with TB (HIV/TB patients) was at least two- to threefold greater than in HIV-1 patients without TB. We hypothesized that this increase in systemic viral heterogeneity may be due to stimulation of HIV-1 replication at sites of M. tuberculosis infection (e.g., lung and pleural space) that could lead to a significant migration of genetically distinct lung-derived HIV-1 quasispecies into the blood. This hypothesis was supported by a greater frequency of distinct quasispecies lineages in HIV/TB patients compared to HIV-1 patients.
In the present study, HIV-1-infected adults with pleural TB were enrolled to assess the effects of TB infection on local and systemic HIV-1 heterogeneity. Pleural TB is caused by spread of M. tuberculosis into the pleural space and subsequent local inflammation and migration of activated leukocytes into the pleural space (3). In this study, we obtained blood and pleural samples from HIV-1-infected individuals with pleural TB, compared the HIV-1 heterogeneity in pleural and blood compartments, and developed a model for HIV-1 diversity at the site of M. tuberculosis infection.
The C2-C3 region of the HIV-1 envelope (env) gene was sequenced to estimate the compartmental (blood or pleura) heterogeneity. This analysis revealed variable amounts of pleural and blood quasispecies compartmentalization and a slightly greater quasispecies heterogeneity in the pleural space. More pleura-derived quasispecies were present in the blood than vice versa. These results suggest that increased viral replication in the pleural space in patients with pleural TB may be associated with a migration of pleura-derived isolates into the blood and increased HIV-1 heterogeneity in the circulation. However, it should be noted that in contrast to the pulmonary HIV/TB model, HIV-1-infected cells did not exist in the pleural space prior to M. tuberculosis invasion and active replication. Recruitment and activation of HIV-1-infected and uninfected leukocytes to the pleural space (2) have led to stimulation of HIV-1 replication (51, 59) and divergent evolution of quasispecies.
MATERIALS AND METHODS
Study subjects and clinical assessments.
Blood and pleural fluid samples were obtained from eight HIV-1-infected adults with newly diagnosed pleural TB enrolled in a study of the immunopathogenesis of pleural TB at the National Tuberculosis Treatment Centre, Mulago Hospital, Kampala, Uganda (14). All patients gave informed written consent for study participation and HIV-1 testing. They also received pre- and post-HIV-1 test counseling. The study protocol was approved by the institutional review boards at Case Western Reserve University and the Ugandan National AIDS Research Subcommittee. Patients underwent phlebotomy for collection of venous blood in addition to diagnostic thoracocentesis and closed pleural biopsy. Following diagnosis, all patients received anti-TB therapy consisting of 2 months of daily isoniazid, rifampin, ethambutol, and pyrazinamide, followed by 4 months of daily isoniazid and rifampin (14). None of the patients received antiretroviral therapy. The diagnosis of pleural TB was established by positive pleural fluid culture (six of eight) and/or pleural biopsy (histology or culture, four of eight). The HIV-1 load in plasma and pleural fluid was measured using the Amplicor HIV-1 monitor assay (Roche Molecular Systems, Branchburg, N.J.) according to the manufacturer's instructions.
Proviral DNA purification, RT-PCR, PCR, and molecular cloning.
Peripheral blood mononuclear cells (PBMC) and pleural fluid mononuclear cells (PFMC) were isolated by Ficoll-Paque density gradient centrifugation from whole heparinized blood and drained pleural fluids, respectively (Pharmacia Biotech, Piscataway, N.J.) (4). Proviral DNA was extracted from lysed PBMC and PFMC as described previously (33). Briefly, PBMC and PFMC were pelleted and then resuspended in gel lysis buffer (0.01% gelatin, 50 mM NaCl, 10 mM Tris [pH 8.3], 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20). Proteinase K (1 mg/ml) was added, and samples were incubated at 60°C for 1 h and then at 95°C for 10 min. Samples were then extracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by two chloroform-isoamyl alcohol (24:1) extractions and ethanol precipitation of DNA.
Pleural fluid and plasma were centrifuged at 4°C at 32,000 × g for 40 min to pellet virus. Viral RNA was isolated from the pellets using the Qiagen RNeasy kit and QIAshredder spin columns (Qiagen, Valencia, Calif.), (32). HIV-1 env RNA was reverse transcribed from viral RNA using the primer E105 (38) (positions 7522 to 7502 of HXB2) and murine leukemia virus reverse transcriptase (RT; Roche Molecular Systems Inc., Branchburg, N.J.). HIV-1 genomic regions in the C2-C3 regions of the gp120 coding region were PCR amplified from the proviral DNA and cDNA samples (1 μg) using primers E80 (38) (positions 6858 to 6879 in HXB2) and E105 (38) (positions 7522 to 7502 of HXB2). These external PCRs were carried out in a 100-μl reaction mixture as described previously (5, 32, 34).
A nested PCR amplification of the C2-C3 region of gp120 was performed on the external PCR products using primers E110 (38) (positions 7002 to 7025 in HXB2) and E125 (38) (positions 7338 to 7315 in HXB2) (5, 32, 34). Nested PCR products were purified with the Qiagen PCR purification kit (Qiagen, Valencia, Calif.) and then cloned into the pCR2.1-Topo vector (Invitrogen Corp., San Diego, Calif.). Bacterial transformants were selected from Luria-Bertani (LB)-ampicillin-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates and then used as templates for colony PCR amplifications. Bacterial colonies were added directly into a PCR mixture containing the M13 forward and reverse primers, i.e., flanking the env insert. This PCR protocol was repeated for 9 to 10 colonies from each patient sample (plasma, pleural fluid, PBMC, and PFMC) as described for the external PCRs.
Sequencing and analysis.
Qiagen-purified PCR products were sequenced using the E110 (for the nested PCR amplifications) or M13 reverse (for the colony PCR amplifications) primer and the ABI automated sequencer at the Davis sequencing facility (Davis, Calif.). The chromatogram files were read using the Chromas 1.6 program (Helensvale, Australia). The C2-C3 env sequences were aligned, phylogenetic trees including reference env sequences were constructed, and bootstrap resampling (1,000 data sets) of multiple alignments to test the statistical robustness of the trees was performed using the Clustal X version 1.64b program (48). Phylogenetic trees were drawn using Treeview 1.6.1 (28). Kimura two-parameter distances were calculated with the DNADIST program in the PHYLIP package (35). Synonymous/nonsynonymous ratios (ds/dn) were calculated using the SNAP program of the HIV sequence database (http://hiv-web.lanl.gov) (18, 26).
Phylogenetic compartmental analysis.
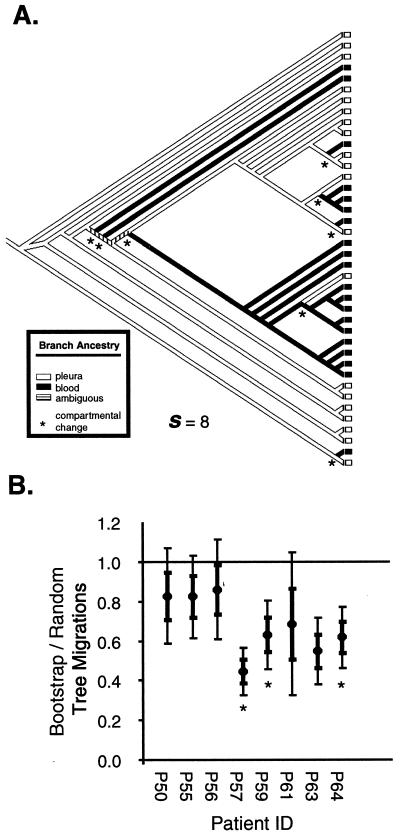
We used a variant of the Swofford-Maddison (47) and Slatkin-Maddison tests (31, 42, 43) to determine if HIV-1 env sequences obtained from one compartment (i.e., blood or pleural space) were more related to each other than to sequences derived from the other compartment (31). The null hypothesis was that the arrangements of sequences were due to chance events and not to tissue compartmentalization of the isolates. The sequences were plotted on each of 1,000 bootstrap phylogenetic trees (constructed using the SEQBOOT program in the PHYLIP package [35]). A compartmental change was defined when the branch ancestry changed from one compartmental source to another (see Fig. 4A).
FIG. 4.
Phylogenetic and phenetic evaluation of blood and pleural compartmentalization in HIV-1-infected Ugandans with pleural TB. (A) Determination of compartmental changes. The source of each sequence is determined by the compartment from which it was isolated (blood or pleural space). The ancestry of each branch was determined by the sources of the terminal quasispecies so as to minimize compartmental changes, which occur when the ancestry of a branch changes. The sum total of compartmental changes (s) was calculated for each tree. (B) Migration events between compartments for the 1,000 actual bootstrap trees and 1,000 randomly constructed trees were constructed using the neighbor-joining method. Tree topology for the actual bootstrap trees was based on nucleotide sequence data for the C2-C3 region of the env (see Fig. 3 and Materials and Methods). Thin and thick error bars indicate 1 and 2 standard deviations of s ratios of actual to random trees, respectively. s ratios that fall 2 standard deviations below 1 are considered significant and suggest compartmentalization. Patients with significant blood and pleural compartmentalization of env quasispecies (P < 0.01) as determined by Mantel's test are marked by asterisks.
The ancestral origin of each branch was determined to minimize the number of putative compartmental changes (s) for each tree (47) using the trace character feature of MacClade 4.0 (23). A sequence that maps to a cluster with a single ancestry (e.g., blood from origin to end of branch) would have an s of 0. An s of greater than 0 suggests that an ancestor of the viral particle migrated between compartments. In the latter case, a viral particle may have been isolated in the blood but have the pleural space as the ancestral origin. The number of s required to fit all sequences to each phylogenetic tree was computed using the treelength feature of MacClade 4.0 (23), counting each node or branch point in the tree representing a compartmental change only once (see Fig. 4A). To determine if lower s values represented compartmentalization, the 1,000 bootstrap trees based on genetic distances between the env sequences obtained from pleural space and blood were compared to 1,000 random trees generated by the random joining-splitting and treelength options in MacClade 4.0 (23). The standard deviation of the ratio was calculated as described previously (36). P values were obtained by comparing the mean of s for all bootstrap trees to the null distribution of s(srand). The P value is the number of times the srand is less than the average bootstrap s divided by 1,000 (for the number of random trees). Thus, the ratio of s values from the random trees and the bootstrap trees provides a measure of compartmentalization.
Phenetic compartmental analysis.
In addition to testing the ancestral relationships between sequences, we used phenetic analysis to determine the degree of genetic similarity among sequences (31). Mantel's test (44, 54) determined if sequences from the blood compartment were more genetically similar to each other than to sequences from the pleural space. This test involves the construction and comparison of two matrices based on the genetic distances between env sequences of the two compartments. A generalized regression permutation procedure comparing two distance matrices was performed for each sample. The first distance matrix, Mk, contains pairwise Kimura two-parameter distances of sequences from both compartments. The second matrix, Mc, contains distances of 0, for sequences in the same compartment, and 1, for sequences in different compartments. Each value of Mc was plotted against Mk, and the square of the Pearson product-moment correlation (Pearson), r2, was computed for all pairs of elements.
To determine a P value and the robustness of compartmentalization, the rows and columns of the Mc were permutated 1,000 times, and r2 was calculated for each permutation. The hypothesis that there is compartmental phenetic structure is rejected if more than 5% of the r2 values from the random permutations exceed the r2 values from Mc and Mk.
Nucleotide sequence accession numbers.
All sequences in this paper were deposited into the GenBank database, with accession numbers AF383297 to AF383304 for the average env sequence of each patient and accession numbers AF383305 to AF383620 for the individual clones.
RESULTS
Analysis of HIV-1-infected patients with pleural TB.
Five men and three women (mean age, 29 years; range, 22 to 35 years) diagnosed with HIV-1 and pleural TB were studied (Table 1). On chest radiograms, all subjects had moderate to massive pleural effusions, and one patient (P64) had a pulmonary infiltrate in addition. CD4 lymphocyte cell counts ranged from 34 to 705 cells/μl, with a mean of 255 cells/μl (Table 1). Four of the patients had CD4 cell counts below 200 cells/μl, whereas the other patients had CD4 cell counts above 300 cells/μl (Table 1). There was no correlation between CD4 cell counts and the pleural or plasma viral load (r < 0.5, P > 0.05, Pearson); however, the pleural fluid viral loads were significantly higher than the plasma viral loads (P < 0.01, Wilcoxon signed rank test).
TABLE 1.
Clinical and virological data on HIV-1-infected patients from Uganda
| Patient no. | Age (yr) | Gendera | CD4 (cells/μl) | Viral load (103 copies/ml)
|
Pulmonary infiltrate | |
|---|---|---|---|---|---|---|
| Plasma | Pleural | |||||
| P64 | 35 | M | 34 | 7.2 | 9.6 | Yes |
| P57 | 33 | F | 42 | 208 | 384 | None |
| P56 | 27 | M | 46 | 123.4 | 4,500 | None |
| P50 | 33 | M | 158 | 2.4 | 7.6 | None |
| P63 | 37 | M | 310 | 45 | 134 | None |
| P55 | 22 | F | 372 | 25 | 160 | None |
| P61 | 24 | M | 374 | 63.3 | 257 | None |
| P59 | 23 | F | 705 | 279.8 | 750 | None |
M, male; F, female.
Genetic characterization of HIV-1 isolates from patients with pleural TB.
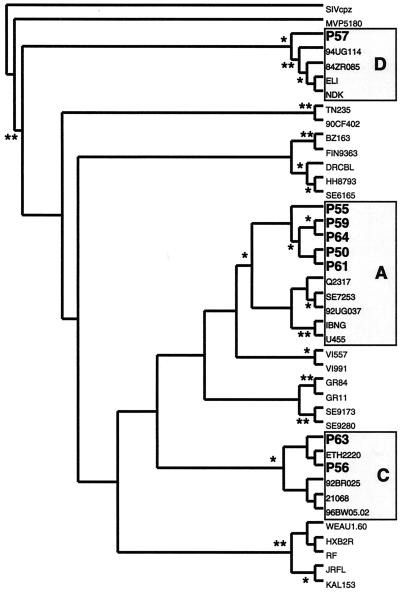
Total cell DNA was isolated from PBMC and PFMC from eight HIV-1 patients with pleural TB. Viral RNA was also isolated from the pleural fluid and plasma from each patient and then reverse transcribed with a primer annealing to the C3 region in env. A region of the HIV-1 env (C2-C3) gene was then PCR amplified and sequenced from the DNA or cDNA from each source. Average HIV-1 env sequences were then compared to the env sequences of known HIV-1 clades using a neighbor-joining method to construct phylogenetic trees and determine clade identity (Fig. 1).
FIG. 1.
Phylogenetic tree of the PBMC average sequence patient samples for subtyping. Bootstrap resampling values of 90 to 100% and 70 to 90% are represented by ∗∗ and ∗, respectively. A 277-nt segment of the C2-C3 region was used to construct these trees by the neighbor-joining method. env genetic subtypes are indicated. GenBank accession numbers for the reference sequences are: SIVcpz, AF115393; group O isolate MVP5180, E16837; subtype C isolates 94UG114 (U88824), 84ZR085 (U88822), ELI (K03454), and NDK (A34828); subtype AE isolates TN235 (L03P55) and 90CF402 (U51188); subtype F isolates BZ163 (L22085) and FIN9363 (AF075703); subtype G isolates DRCBL (AF084936), HH8793 (AF061640), and SE6165 (AF061642); subtype A isolates Q2317 (AF004885), SE7253 (AF069670), 92UG037 (U51190), IBNG (L39106), and U455 (M62320); subtype H isolates VI557 (U09666) and VI991 (AF190127); subtype cpx isolates GR84 (AF119819) and GR11 (AF119820); subtype J isolates SE9173 (AF082395) and SE9280 (AF082394); subtype C isolates ETH2220 (U46016), 92BR025 (U52953), 21068 (AF067155), and 96BW05.02 (AF110967); and subtype B isolates WEAU1.60 (U21135), HX2BR (K03455), RF (M17451), JRFL (U63632), and KAL153 (AF193276).
env sequences from the HIV/TB subjects in this study clustered into several clades. Isolates P56 and P63 clustered with clade C, isolate P57 clustered with clade D, and isolates P50, P55, P59, P61, and P64 clustered with clade A (Fig. 1). Amino acids at positions 306 (HXB2 nucleotide [nt] positions 7140 to 7142 [19]) and 322 (at positions 7188 to 7190) in the V3 domain of env were used to predict the biological phenotype and coreceptor usage. A positively charged amino acid at either of these positions in the hypervariable, immunodominant V3 loop of gp120 is highly predictive of a syncytium-inducing, CXCR4-tropic phenotype (8). An analysis of the env V3 loop sequences from all samples revealed only negatively charged or neutral amino acids at amino acid residues 306 and 322, suggesting that all patients harbored non-syncytium-inducing CCR5-tropic HIV-1 isolates in both the blood and pleural compartments.
For detailed analyses of HIV-1 heterogeneity, we cloned and sequenced the HIV-1 env region from each source (PBMC, plasma, PFMC, and pleural fluid) of each patient. A total of 316 (9 or 10 per source) HIV-1 env clones were isolated from average env PCR products and sequenced (see Materials and Methods). All clones had V3 sequences predictive of a non-syncytium-inducing, CCR5-tropic viral phenotype (see above). Potentially inactivating mutations (e.g., stop codons or frameshift mutations) were observed in just 2 of 316 molecular clones.
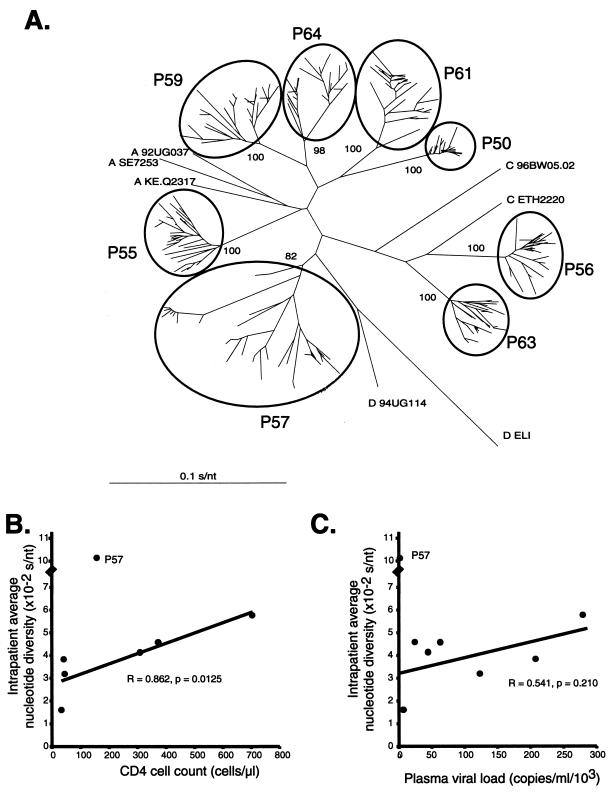
Neighbor-joining trees containing every clone from each patient were constructed to compare the phylogenetic relationships of all env sequences and confirm the presence of intrapatient clusters (Fig. 2A). Bootstrap values indicated that sequences from each patient formed a monophyletic group with no aberrant, contaminating sequences. env quasispecies diversity within each patient was estimated by the size of the intrapatient cluster. For example, patient P57 harbored a more diverse population of HIV-1 env quasispecies than patient P50. A more accurate method of determining intrapatient diversity is to compare the nucleotide diversity between quasispecies. Upon removal of sample P57, which had an extremely high env genetic heterogeneity (0.1011 substitution per nucleotide [s/nt], 2.5-fold greater than the average for the other sequences), there was a direct correlation between the CD4 cell count and intrapatient genetic diversity at the time of sample collection (r = 0.862, P = 0.0125, Pearson; Fig. 2B). The correlation between env sequence diversity and pleural or plasma viral load was not significant (data not shown and Fig. 2C, respectively).
FIG. 2.
Analysis of HIV-1 env quasispecies from the blood and pleural space of HIV-1-infected individuals with pleural TB. (A) Phylogenetic tree of all quasispecies. The letters preceding the name of the reference sequence indicate the subtype. The bootstrap values (percentages) separating each set of patient samples are indicated. The scale bar represents 0.1 substitution per nucleotide position (s/nt). The average nucleotide diversity of all HIV-1 env quasispecies within an individual was plotted against CD4 cell count (B) and plasma viral load (C) obtained from the same sample and time point.
Evidence of compartmentalization and migrations between the blood and pleural space.
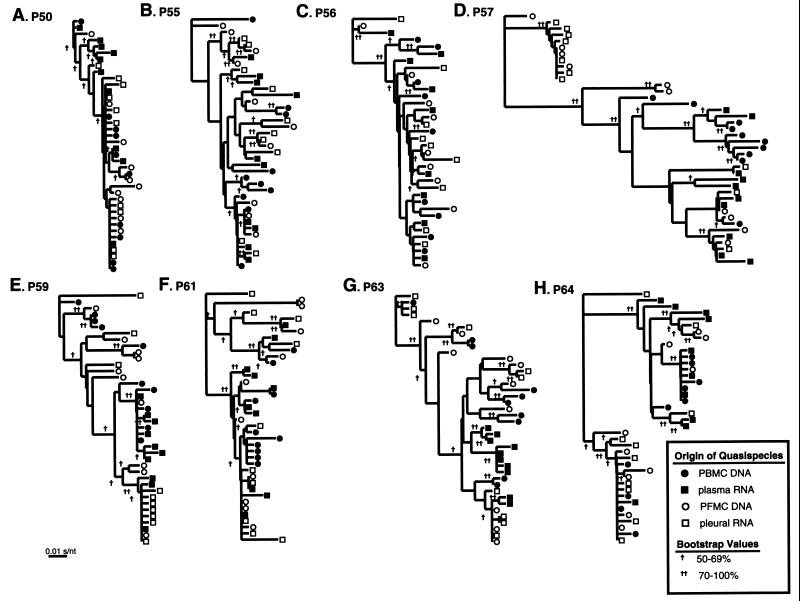
env quasispecies from blood (PBMC and plasma) and pleural space (PFMC and pleural fluid) of each HIV-1-infected patient with pleural TB were used to construct eight cladograms using the neighbor-joining method (Fig. 3). Clusters were comprised mainly of quasispecies from a single anatomical compartment (pleural space or blood), with isolates appearing to migrate frequently between compartments. Patients P50, P57, P59, and P64 showed the strongest evidence of phylogenetic compartmentalization of the blood and pleural isolates (Fig. 3A, D, E, and H, respectively). However, subjects P55, P56, and P63 had few compartment-specific clusters (Fig. 3B, C, and G, respectively). Subject P61 seemed to fall between the groups, with some compartmentalization and migrations of the quasispecies from the four different sources (Fig. 3F). There was no relationship between the degree of compartmentalization and viral load or CD4 cell count (data not shown). This analysis indicates that in some patients, pleural TB may lead to anatomic separation of HIV-1 quasispecies in the pleural space from those in blood.
FIG. 3.
Analysis of HIV-1 env quasispecies from two different tissue compartments of individual patients with pleural TB. Phylogenetic trees were derived from individual env sequences from the blood and pleural space of patients P50, P55, P56, P57, P59, P61, P63, and P64 (A to H, respectively). Branch lengths are drawn to scale. The scale bar represents 0.01 substitution per nucleotide position. The legend indicates bootstrap values greater than 50%. HIV-1 env sequences were obtained from 9 or 10 clones from each sample of PBMC, plasma, PFMC, and pleural fluids. HIV-1 env PCR products were cloned into the pCR2.1-Topo vector (Invitrogen) to select the quasispecies for sequencing and subsequent analyses.
To obtain a more accurate analysis of HIV-1 compartmentalization, we compared the phylogenetic relationships and source of quasispecies using the Swofford-Maddison and Slatkin-Maddison tests (31, 42, 43). These tests determined if HIV-1 env sequences obtained from one compartment (blood or pleural space) were more related to each other than to sequences from the other compartment. Compartmentalization is evident in a patient if the ratio of the s values from the actual to random trees was significantly less than 1, i.e., the mean of this ratio plus 2 standard deviations (Materials and Methods and Fig. 4B).
Using the Slatkin-Maddison test, patients P57, P59, P63, and P64 clearly have HIV-1 quasispecies that separate into two compartments (P ≤ 0.001, Fig. 4B). In contrast, patients P50, P55, and P56, had quasispecies that did not compartmentalize, that is, s ratios of actual to random trees that were not significantly less than 1 (P ≥ 0.05, Fig. 4B). Interestingly, sample P61 had a ratio with 2 standard deviations exceeding 1 but still maintained a P of less than 0.001, suggesting a weak blood and pleural compartmentalization.
Compartmentalization was also assessed using an analysis of phenetic structure. Mantel's test involves the construction and comparison of two matrices based on (i) genetic distances between pairs of env sequences and (ii) the sources (blood or pleural space) of these pairs of env sequences. Evidence of compartmentalization is depicted by greater genetic distances separating quasispecies of different sources and smaller distances separating those of the same source (see Materials and Methods). Similar or small differences in genetic distances separating intra- and intercompartmental quasispecies suggest a lack of compartmentalization. Although this analysis provides a more narrow definition of compartmentalization than the Slatkin-Maddison test, quasispecies from patients P57, P59, and P64 still had phenetic structure (Fig. 4B, P < 0.01) and clear evidence of separate blood and pleural compartments.
Analysis of quasispecies in the pleural space and blood.
To determine the local effects of pleural TB on the HIV-1 quasispecies population, we compared the pairwise genetic distances of env quasispecies from the pleural space and blood (Table 2). There was a significant increase in the quasispecies heterogeneity within the pleural space compared to blood of patients P57, P59, and P61 (Table 2). However, sample P50 had a much lower heterogeneity in the pleural space, while the samples from the other four patients had no difference in the heterogeneity between compartments. Overall, there was a slight trend for increased heterogeneity in the pleural space.
TABLE 2.
Heterogeneity and ds/dn values
| Patient no. | Avg genetic distancea (10−2 s/nt ± SE)
|
ds/dn ± SEb
|
||
|---|---|---|---|---|
| Blood | Pleura | Blood | Pleura | |
| P50 | 1.69 ± 0.07 | 1.42 ± 0.05 | 2.82 ± 1.14 | 3.78 ± 0.16 |
| P55 | 3.77 ± 0.12 | 3.81 ± 0.11 | 1.65 ± 0.43 | 2.18 ± 0.89 |
| P56 | 3.16 ± 0.11 | 3.15 ± 0.12 | 2.05 ± 0.31 | 4.61 ± 0.94 |
| P57 | 7.65 ± 0.24 | 8.90 ± 0.44 | 0.91 ± 0.08 | 1.51 ± 0.17 |
| P59 | 3.51 ± 0.16 | 4.33 ± 0.16 | 2.55 ± 0.49 | 2.16 ± 0.40 |
| P61 | 3.79 ± 0.20 | 5.30 ± 0.26 | 2.18 ± 0.42 | 1.39 ± 0.42 |
| P63 | 4.42 ± 0.13 | 4.48 ± 0.15 | 1.32 ± 0.13 | 1.38 ± 0.16 |
| P64 | 5.24 ± 0.23 | 5.23 ± 0.25 | 1.31 ± 0.18 | 1.19 ± 0.15 |
Average distance was calculated by averaging all the Kimura two-parameter distances of all the quasispecies for each patient.
The consensus sequence was determined for each patient, and the average ratio of synonymous to nonsynonymous base substitions (ds/dn) was determined relative to the consensus sequence for each patient.
A ratio of synonymous (ds) to nonsynonymous (dn) substitutions equal to 1 has been associated with limited selection from a pool of random nucleotide substitutions; values above 1 suggest purifying selection against amino acid changes; and values below 1 provide evidence of selective pressure (e.g., immune response) on virus amino acid evolution (10, 40). All subjects had ds/dn values greater than or equal to 1, suggesting either no selective pressure or selection against amino acid changes (Table 2). In subject P57, the ds/dn ratio in the pleura was higher than in the blood (P = 0.025, Mann-Whitney ranked sum test), suggesting greater selection against amino acid changes in the pleural space.
The intrapatient phylogenetic trees in Fig. 3 contain quasispecies from a specific source (blood or pleural space) that cluster with quasispecies of another source or compartment. Quasispecies migrating between the blood and pleural space were first identified and then removed from subsequent pairwise genetic distance analyses. Several criteria were used to identify migrants. The Slatkin-Maddison test was used to trace the root of sequences within clusters. The compartmental origin of each cluster was defined by the majority of blood or pleural sequences. The probability that sequences within a cluster grouped together was defined by the bootstrap value of the node belonging to the cluster (value of >50%). Sequences were treated as migrants if the bootstrap value multiplied by the proportion of sequences from the other compartment was greater than 0.50. For example, pleural isolates were removed from a cluster of blood sequences when the cluster contained at least a 3:1 ratio of blood to pleural sequences and only if the bootstrap value defining a cluster was >66%.
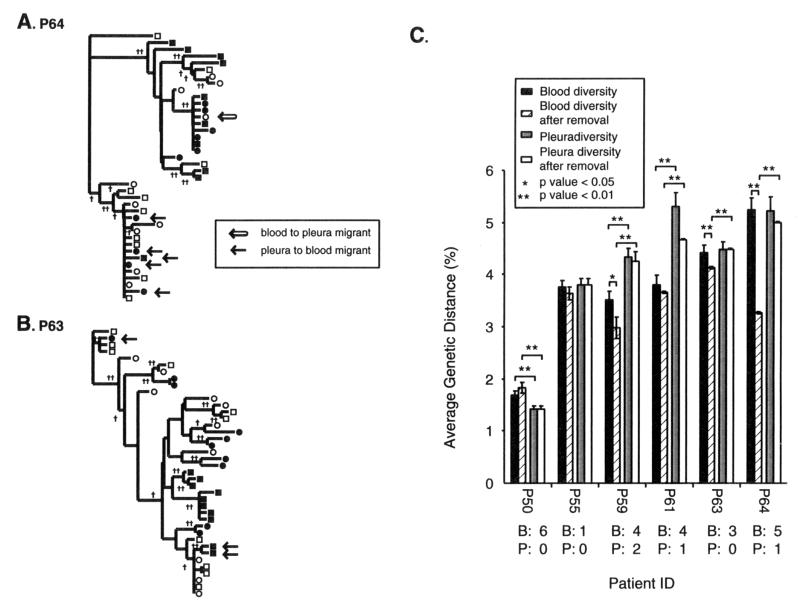
Examples of migrant removals are provided in Fig. 5A and 5B. Following the removal of these migrants from the phylogenetic clusters, we repeated the analyses on genetic distances with the remaining quasispecies of each patient. It is important to note that these uniform criteria for identification of migrants resulted in the removal of 6 sequences (of 40) from patient P50, 1 from P55, 6 from P59, 5 from P61, 3 from P63 (Fig. 5B), and 6 from P64 (Fig. 5A). The source of these env migrants is provided in Fig. 5C. No sequences were removed from patient P56 and P57 samples because they lacked defined compartmental clusters or migrants within compartmental structures (Fig. 3). The majority of env migrants removed from the analyses were physically isolated from the blood but appeared to originate from the pleural space, based on close phylogenetic relations with pleural HIV-1 quasispecies. Thus, more pleural HIV-1 quasispecies may be migrating to the blood than vice versa (P < 0.01, t test).
FIG. 5.
Analysis of env genetic diversity in blood and pleural compartments before and after removal of migrant quasispecies. Quasispecies migrating from the blood to the pleural space (open arrow) or from the pleural space to the blood (solid arrow) are highlighted in the phylogenetic trees from patients P64 (A) and P65 (B). These migrants were then removed from the group of blood or pleural env quasispecies. Intracompartmental genetic distances were then calculated before and after the removal on migrant sequences and plotted in panel C. Error bars represent 1 standard error of the mean. Below each patient number is the number of sequences removed from the blood (B) and pleural space (P). Brackets indicate significant differences in the genetic distances between blood and pleural env populations. ∗, P < 0.05; ∗∗, P < 0.01.
This migration and introduction of genetically distinct pleural quasispecies in the blood due to pleural TB could contribute to HIV-1 heterogeneity in the blood. Prior to the removal, there was a significant increase in pleural over blood quasispecies heterogeneity in three of six patients. After the removal of these migrants, five of six patients showed an even greater difference in the heterogeneity of HIV-1 quasispecies isolated from the pleural space compared to the blood. The significant difference between pleural and blood HIV-1 heterogeneity was absent in two patients prior to the removal of these obvious migrants. Therefore, it appears that pleura-derived HIV-1 quasispecies are migrating from the pleural space to the blood, causing the blood population to become more heterogeneous.
DISCUSSION
We have previously shown that HIV-1 patients with pulmonary TB harbor a more heterogeneous HIV-1 population in the blood than individuals with HIV-1 alone (5). Quasispecies from HIV-1-infected participants tended to have phylogenies with few phylogenetic clusters and relatively short branch lengths. In contrast, quasispecies from HIV/TB individuals had phylogenies with longer intrapatient branches and more genetically divergent populations (5). Since TB also mediates increased HIV-1 heterogeneity in the lung (24), we hypothesized that a TB-associated, site-specific increase in HIV-1 replication and heterogeneity in the lung (24) may migrate to the blood to increase systemic HIV-1 heterogeneity.
To test this hypothesis, we used pleural TB as a model of the effects of TB on intrapatient HIV-1 evolution. Pleural TB is found in 28 to 38% of HIV-1-infected patients with TB (21, 37) and is diagnosed by culturing M. tuberculosis from the pleural fluid and/or histological examination of pleural biopsies. We performed an extensive, cross-sectional phylogenetic analysis on HIV-1 env quasispecies from the pleural space and blood of HIV-1-infected patients with pleural TB. Despite increased viral loads in the pleural space, there was only a slight trend for increased heterogeneity in the pleural space. However, phylogenetic separation of the blood quasispecies from the pleural quasispecies in several patients was suggestive of compartmentalization and divergent evolution in pleural effusions. Also, there were substantial migration events between compartments in all of the patients. Upon removing migrants from the heterogeneity analysis, the pleural compartments contained a more heterogeneous HIV-1 population than that found in the blood. In addition, the majority of these migrants tended to be viral particles that relocated from the pleural space to the blood. These results support our initial hypothesis that an increase in systemic heterogeneity associated with pulmonary TB may be a result of divergent viral particles migrating from the site of TB infection to the blood.
In several patients, there was an anatomical compartmentalization of HIV-1 quasispecies between the blood and the pleural space. Previous studies suggest a compartmentalization of HIV-1 quasispecies in the brain (13, 20, 58), cerebrospinal fluid (45, 53), spleen (58), lymph node (13, 58), lung, and semen (6, 7). Compartmentalization in the pleural space is likely partly due to a physical separation of fluids between the blood and pleural space. An initial infiltration of the pleural space with leukocytes may lead to secretion of interleukin-8 and other cytokines and chemokines and the subsequent recruitment of lymphocytes and monocytes to the pleural space (2, 27). A subset of CD4+ T lymphocytes and monocytes infiltrating the M. tuberculosis-infected pleural space may be infected with HIV-1 and harbor integrated provirus. TB-mediated cytokine and chemokine expression (e.g., tumor necrosis factor alpha and MCP-1) may then cause in situ activation of HIV-1 replication. TB may also enhance the susceptibility of newly recruited monocytes to HIV-1 infection at sites of M. tuberculosis infection (9, 50). Thus, the microenvironment generated by M. tuberculosis in the pleural space is likely responsible for the observed increase in pleural viral load compared to the plasma and for further intracompartmental HIV-1 evolution.
Although HIV-1 compartmentalization is evident in the pleural space, there is considerable flow of HIV-1 quasispecies between the blood and pleural space in each patient (Fig. 3 and 5). Higher viral loads in the pleural space support the findings that the majority of quasispecies are migrating from the pleural space to the blood (Table 1 and Fig. 5) and that pleural quasispecies are contributing to the increase in systemic HIV-1 heterogeneity. Seeding of the systemic HIV-1 population with quasispecies from other compartments has been observed in patients on antiretroviral therapy (30). Poor drug penetration of various tissues (e.g., central nervous system) often leads to tissue-specific selection of drug-resistant quasispecies, which can continue to seed the systemic population and lead to emergence of HIV-1 variants resistant to several antiretroviral drugs (55). Migrations between the blood and semen have been documented by Delwart et al. (7), where some patients had clear communication between the compartments, and other patients had more defined compartments. This variability may be due to the intermittent shedding of HIV-1 from the blood to the semen (12).
Our data suggest that TB may be associated with increased HIV-1 heterogeneity at the pleural site of M. tuberculosis infection. There is a slight trend for enhanced pleural heterogeneity before removal of migrant HIV-1 quasispecies (Table 2). Upon removal of the migrants, there is a further increase in genetic diversity in the pleural space compared to the blood (Fig. 5C). An increase in HIV-1 quasispecies heterogeneity has also been demonstrated in the lung and has been attributed to enhanced virus replication (24). A localized immune response (e.g., in the pleural space) may lead to an HIV-1-specific immune response at the site of coinfection.
An analysis of ds/dn ratios in the pleural space and blood revealed limited selection of HIV-1 env sequences in either compartment. Our previous study did show greater selection for amino acid changes (ds/dn < 1) in env quasispecies in HIV/TB patients compared to HIV-1-infected individuals (5). More selective HIV-1 evolution in patients with pulmonary TB compared to patients with pleural TB may be due to the establishment of HIV-1 infection in the lung prior to active TB versus the more acute nature of M. tuberculosis infection in the pleural space, associated with a rapid and intense recruitment of leukocytes. In addition, an HIV-1-specific immune response may be more active in the lung than in the pleural space.
As mentioned earlier, we used pleural TB as a model for pulmonary TB and its effects on HIV-1 heterogeneity and quasispecies distribution. We must acknowledge that differences in HIV-1 replication within the lung and pleural space may affect HIV-1 heterogeneity and quasispecies distribution. HIV-1 replicates within the lung in the absence of active TB (39, 46). Thus, HIV-1 evolves continuously within the lung and prior to the development of active TB. Recent studies suggest that the lung may also be a continual HIV-1 reservoir and harbor a very diverse population of HIV-1 quasispecies, i.e., ancestral quasispecies of the blood (J. Mullins, personal communication). As quasispecies migrate from the lung, the systemic population would become more diverse. Stimulation of lung quasispecies as a result of pulmonary TB or other coinfections would lead to a greater diversity in the lung and a corresponding increase in systemic HIV-1 heterogeneity.
In conclusion, we have found that the increase in systemic HIV-1 heterogeneity in patients coinfected with TB may be due to a site-specific activation of HIV-1. We have also observed compartmentalization of HIV-1 quasispecies in the blood and pleural space. A unidirectional migration of quasispecies from the pleural space to the blood suggests a contribution of pleural isolates to the HIV-1 genetic diversity in the blood.
Acknowledgments
We thank the National Tuberculosis Treatment Centre, Mulago Hospital, and the Uganda-Case Western Reserve University Research Collaboration for administrative support and use of their research facilities and clinics. We also thank the members of the Z. Toossi and E. J. Arts groups' laboratories for their invaluable support and suggestions.
M.E.Q.-M. was supported by an NHLB1/NIH research grant (K01-HL67610-01). Z.T. and E.J.A. were supported by an NHLBI/NIH grant (HL51636-06). Additional support was provided by a CWRU Center for AIDS Research grant (AI36219) and a Tuberculosis Research Unit grant (A1-45244) from NIAID/NIH.
REFERENCES
- 1.Ait-Khaled, M., J. E. McLaughlin, M. A. Johnson, and V. C. Emery. 1995. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS 9:675-683. [DOI] [PubMed] [Google Scholar]
- 2.Antony, V. B., S. W. Godbey, S. L. Kunkel, J. W. Hott, D. L. Hartman, M. D. Burdick, and R. M. Strieter. 1993. Recruitment of inflammatory cells to the pleural space. Chemotactic cytokines, IL-8, and monocyte chemotactic peptide-1 in human pleural fluids. J. Immunol. 151:7216-7223. [PubMed] [Google Scholar]
- 3.Beck, J. M. 1998. Pleural disease in patients with acquired immune deficiency syndrome. Clin. Chest Med. 19:341-349. [DOI] [PubMed] [Google Scholar]
- 4.Boyum, A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of mononuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 97:77-89. [PubMed] [Google Scholar]
- 5.Collins, K. R., H. Mayanja-Kizza, B. A. Sullivan, M. E. Quinones-Mateu, Z. Toossi, and E. J. Arts. 2000. Greater diversity of HIV-1 quasispecies in HIV-infected individuals with active tuberculosis. J. Acquir. Immune Defic. Syndr. 24:408-417. [DOI] [PubMed] [Google Scholar]
- 6.Coombs, R. W., C. E. Speck, J. P. Hughes, W. Lee, R. Sampoleo, S. O. Ross, J. Dragavon, G. Peterson, T. M. Hooton, A. C. Collier, L. Corey, L. Koutsky, and J. N. Krieger. 1998. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. J. Infect. Dis. 177:320-330. [DOI] [PubMed] [Google Scholar]
- 7.Delwart, E. L., J. I. Mullins, P. Gupta, G. H. Learn, Jr., M. Holodniy, D. Katzenstein, B. D. Walker, and M. K. Singh. 1998. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 72:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrait, V., J. Cadranel, H. Esvant, I. Herry, P. Morinet, C. Mayaud, and D. Israel-Biet. 1997. Tuberculosis generates a microenvironment enhancing the productive infection of local lymphocytes by HIV. J. Immunol. 159:2824-2830. [PubMed] [Google Scholar]
- 10.Gojobori, T., Y. Yamaguchi, K. Ikeo, and M. Mizokami. 1994. Evolution of pathogenic viruses with special reference to the rates of synonymous and nonsynonymous substitutions. Jpn. J. Genet. 69:481-488. [DOI] [PubMed] [Google Scholar]
- 11.Goletti, D., D. Weissman, R. W. Jackson, N. M. Graham, D. Vlahov, R. S. Klein, S. S. Munsiff, L. Ortona, R. Cauda, and A. S. Fauci. 1996. Effect of Mycobacterium tuberculosis on HIV replication. Role of immune activation. J. Immunol. 157:1271-1278. [PubMed] [Google Scholar]
- 12.Gupta, P., C. Leroux, B. K. Patterson, L. Kingsley, C. Rinaldo, M. Ding, Y. Chen, K. Kulka, W. Buchanan, B. McKeon, and R. Montelaro. 2000. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral quasispecies between blood and semen. J. Infect. Dis. 182:79-87. [DOI] [PubMed] [Google Scholar]
- 13.Haggerty, S., and M. Stevenson. 1991. Predominance of distinct viral genotypes in brain and lymph node compartments of HIV-1-infected individuals. Viral Immunol. 4:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch, C. S., Z. Toossi, J. L. Johnson, H. Luzze, L. Ntambi, P. Peters, M. McHugh, A. Okwera, M. Joloba, P. Mugyenyi, R. D. Mugerwa, P. Terebuh, and J. J. Ellner. 2001. Augmentation of apoptosis and interferon-gamma production at sites of active Mycobacterium tuberculosis infection in human tuberculosis. J. Infect. Dis. 183:779-788. [DOI] [PubMed] [Google Scholar]
- 15.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 16.Itescu, S., P. F. Simonelli, R. J. Winchester, and H. S. Ginsberg. 1994. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc. Natl. Acad. Sci. USA 91:11378-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiessling, A. A., L. M. Fitzgerald, D. Zhang, H. Chhay, D. Brettler, R. C. Eyre, J. Steinberg, K. McGowan, and R. A. Byrn. 1998. Human immunodeficiency virus in semen arises from a genetically distinct virus reservoir. AIDS Res. Hum. Retrovir. 14(Suppl. 1):S33-S41. [PubMed] [Google Scholar]
- 18.Korber, B. 2000. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 19.Korber, B. T., B. T. Foley, C. L. Kuiken, S. K. Pillai, and J. G. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG, p. 102-111. In G. Myers, B. Korber, B. H. Hahn, K.-T. Jeang, J. W. Mellors, F. E. McCutchan, L. E. Henderson, and G. N. Pavlakis (ed.), Human retroviruses and AIDS. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 20.Korber, B. T., K. J. Kunstman, B. K. Patterson, M. Furtado, M. M. McEvilly, R. Levy, and S. M. Wolinsky. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467-7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, F., T. Modilevsky, A. R. Waliany, J. M. Leedom, and P. F. Barnes. 1990. Delayed diagnosis of tuberculosis in patients with human immunodeficiency virus infection. Am. J. Med. 89:451-456. [DOI] [PubMed] [Google Scholar]
- 22.Lukashov, V. V., and J. Goudsmit. 1998. HIV heterogeneity and disease progression in AIDS: a model of continuous virus adaptation. AIDS 12:S43-S52. [PubMed] [Google Scholar]
- 23.Maddison, W. P., and D. R. Maddison. 1989. Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol. 53:190-202. [DOI] [PubMed] [Google Scholar]
- 24.Nakata, K., W. N. Rom, Y. Honda, R. Condos, S. Kanegasaki, Y. Cao, and M. Weiden. 1997. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication in the lung. Am. J. Respir. Crit. Care Med. 155:996-1003. [DOI] [PubMed] [Google Scholar]
- 25.Nakata, K., M. Weiden, T. Harkin, D. Ho, and W. N. Rom. 1995. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-1-infected patients: evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol. Med. 1:744-757. [PMC free article] [PubMed] [Google Scholar]
- 26.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 27.Pace, E., M. Gjomarkaj, M. Melis, M. Profita, M. Spatafora, A. M. Vignola, G. Bonsignore, and C. H. Mody. 1999. Interleukin-8 induces lymphocyte chemotaxis into the pleural space. Role of pleural macrophages. Am. J. Respir. Crit. Care Med. 159:1592-1599. [DOI] [PubMed] [Google Scholar]
- 28.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 29.Petito, C. K., H. Chen, A. R. Mastri, J. Torres-Munoz, B. Roberts, and C. Wood. 1999. HIV infection of choroid plexus in AIDS and asymptomatic HIV-infected patients suggests that the choroid plexus may be a reservoir of productive infection. J. Neurovirol. 5:670-677. [DOI] [PubMed] [Google Scholar]
- 30.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 31.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. de Vange Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinones-Mateu, M. E., J. Dopazo, J. A. Este, T. R. Rota, and E. Domingo. 1995. Molecular characterization of human immunodeficiency virus type 1 isolates from Venezuela. AIDS Res. Hum. Retrovir. 11:605-616. [DOI] [PubMed] [Google Scholar]
- 34.Quinones-Mateu, M. E., V. Soriano, E. Domingo, and L. Menendez-Arias. 1997. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology 236:364-373. [DOI] [PubMed] [Google Scholar]
- 35.Retief, J. D. 2000. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132:243-258. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigo, A. G., and J. I. Mullins. 1996. Human immunodeficiency virus type 1 molecular evolution and the measure of selection. AIDS Res. Hum. Retrovir. 12:1681-1685. [DOI] [PubMed] [Google Scholar]
- 37.Saks, A. M., and R. Posner. 1992. Tuberculosis in HIV positive patients in South Africa: a comparative radiological study with HIV negative patients. Clin. Radiol. 46:387-390. [DOI] [PubMed] [Google Scholar]
- 38.Sanders-Buell, E., M. Salminen, and F. McCutchan. 1995. Sequencing primers for HIV-1, p. 15-21. In G. Myers, B. Korber, B. H. Hahn, K.-T. Jeang, J. W. Mellors, F. E. McCutchan, L. E. Henderson, and G. N. Pavlakis (ed.), Human retroviruses and AIDS. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 39.Sei, S., D. E. Kleiner, J. B. Kopp, R. Chandra, P. E. Klotman, R. Yarchoan, P. A. Pizzo, and H. Mitsuya. 1994. Quantitative analysis of viral burden in tissues from adults and children with symptomatic human immunodeficiency virus type 1 infection assessed by polymerase chain reaction. J. Infect. Dis. 170:325-333. [DOI] [PubMed] [Google Scholar]
- 40.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 41.Singh, A., G. Besson, A. Mobasher, and R. G. Collman. 1999. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J. Virol. 73:6680-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slatkin, M., and W. P. Maddison. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123:603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slatkin, M., and W. P. Maddison. 1990. Detecting isolation by distance using phylogenies of genes. Genetics 126:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smouse, P. E., J. C. Long, and R. R. Sokal. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35:627-632. [Google Scholar]
- 45.Staprans, S., N. Marlowe, D. Glidden, T. Novakovic-Agopian, R. M. Grant, M. Heyes, F. Aweeka, S. Deeks, and R. W. Price. 1999. Time course of cerebrospinal fluid responses to antiretroviral therapy: evidence for variable compartmentalization of infection. AIDS 13:1051-1061. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, M., and H. E. Gendelman. 1994. Cellular and viral determinants that regulate HIV-1 infection in macrophages. J. Leukoc. Biol. 56:278-288. [DOI] [PubMed] [Google Scholar]
- 47.Swofford, D. L., and W. P. Maddison. 1987. Reconstructing ancestral character states under Wagner parsimony. Math. Biosci. 87:199-229. [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toossi, Z., H. Mayanja-Kizza, C. S. Hirsch, K. L. Edmonds, T. Spahlinger, D. L. Hom, H. Aung, P. Mugyenyi, J. J. Ellner, and C. W. Whalen. 2001. Impact of tuberculosis (TB) on HIV-1 activity in dually infected patients. Clin. Exp. Immunol. 123:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toossi, Z., J. G. Sierra-Madero, R. A. Blinkhorn, M. A. Mettler, and E. A. Rich. 1993. Enhanced susceptibility of blood monocytes from patients with pulmonary tuberculosis to productive infection with human immunodeficiency virus type 1. J. Exp. Med. 177:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toossi, Z., L. Xia, M. Wu, and A. Salvekar. 1999. Transcriptional activation of HIV by Mycobacterium tuberculosis in human monocytes. Clin. Exp. Immunol. 117:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanham, G., K. Edmonds, L. Qing, D. Hom, Z. Toossi, B. Jones, C. L. Daley, B. Huebner, L. Kestens, P. Gigase, and J. J. Ellner. 1996. Generalized immune activation in pulmonary tuberculosis: coactivation with HIV infection. Clin. Exp. Immunol. 103:30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venturi, G., M. Catucci, L. Romano, P. Corsi, F. Leoncini, P. E. Valensin, and M. Zazzi. 2000. Antiretroviral resistance mutations in human immunodeficiency virus type 1 reverse transcriptase and protease from paired cerebrospinal fluid and plasma samples. J. Infect. Dis. 181:740-745. [DOI] [PubMed] [Google Scholar]
- 54.Waddle, D. M. 1994. Matrix correlation tests support a single origin for modern humans. Nature 368:452-454. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Y. M., W. B. Dyer, C. Workman, B. Wang, J. S. Sullivan, and N. K. Saksena. 2000. Molecular evidence for drug-induced compartmentalization of HIV-1 quasispecies in a patient with periodic changes to HAART. AIDS 14:2265-2272. [DOI] [PubMed] [Google Scholar]
- 56.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 57.Whalen, C., C. R. Horsburgh, D. Hom, C. Lahart, M. Simberkoff, and J. Ellner. 1995. Accelerated course of human immunodeficiency virus infection after tuberculosis. Am. J. Respir. Crit. Care Med. 151:129-135. [DOI] [PubMed] [Google Scholar]
- 58.Wong, J. K., C. C. Ignacio, F. Torriani, D. Havlir, N. J. Fitch, and D. D. Richman. 1997. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of Pol sequences from autopsy tissues. J. Virol. 71:2059-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., K. Nakata, M. Weiden, and W. N. Rom. 1995. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication by transcriptional activation at the long terminal repeat. J. Clin. Investig. 95:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]