Abstract
Previous studies have shown that Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) is uniquely able to up-regulate the expression of the peptide transporters (referred to as TAP-1 and TAP-2) and major histocompatibility complex (MHC) class I in Burkitt's lymphoma (BL) cell lines. This up-regulation is often accompanied by a restoration of antigen-presenting function as measured by the ability of these cells to present endogenously expressed viral antigen to cytotoxic T lymphocytes. Here we show that the expression of LMP1 resulted in up-regulation and nuclear translocation of RelB that were coincident with increased expression of MHC class I in BL cells. Deletion of the C-terminal activator regions (CTARs) of LMP1 significantly impaired the abilities of LMP1 to translocate RelB into the nucleus and to up-regulate the expression of antigen-processing genes. Further analysis with single-point mutations within the CTARs confirmed that the residues critical for NF-κB activation directly contribute to antigen-processing function regulation in BL cells. This LMP1-mediated effect was blocked following expression of either dominant negative IκBα S32/36A, an NF-κB inhibitor, or antisense RelB. These observations indicate that upregulation of antigen-presenting function in B cells mediated by LMP1 is signaled through the NF-κB subunit RelB. The data provide a mechanism by which LMP1 modulates immunogenicity of Epstein-Barr virus-infected normal and malignant cells.
The interaction of cytotoxic T lymphocytes (CTLs) with the major histocompatibility complex (MHC)-peptide complex is a critical step toward the initiation and propagation of specific immune responses against viral infection. The specificity of this interaction is determined by two distinct components, namely, MHC-restricted presentation of a peptide epitope and a heterodimeric αβ cell surface protein called the T-cell receptor. It is well recognized that many viruses that establish persistent infections or are involved in malignant processes have evolved unique mechanisms by which to evade this potent antiviral CTL response in the immune-competent host. These escape mechanisms include limited gene expression during latent infection, virus replication in immune-privileged tissues, downregulation of MHC and adhesion molecules, and sequence variation affecting peptide binding to MHC or recognition by the T-cell receptor of CTLs (36).
The Epstein-Barr virus (EBV)-specific CTL response plays a crucial role in controlling the outgrowth of EBV-infected B cells in immunocompetent hosts (19, 22). Any relaxation of this immune control often results in massive expansion of EBV-infected B cells, as seen in immunocompromised transplant recipients. EBV-infected normal B cells (also referred to as lymphoblastoid cell lines [LCLs]) and some Burkitt's lymphoma (BL) cell lines (commonly referred to as group III BL lines), which express the complete array of latent antigens (referred to as EBNA1 to -6 and LMP1and−2) consistently express very high levels of peptide transporters (TAP-1 and −2) and surface HLA class I and show no resistance to CTL-mediated immune recognition. In contrast to EBV-infected normal B cells, EBV-positive tumor cells from BL patients (referred to as group I BL lines) display strong resistance to virus-specific CTL-mediated lysis (13). Contributory factors include down-regulated expression of either the immunodominant latent antigens EBNA2 to−6 and LMP1 and−2 or expression of antigen-processing genes, such as those that encode TAP-1 and TAP-2 and surface MHC class I antigens, thereby leading to impaired endogenous processing and presentation of CTL epitopes. Extensive analysis of individual EBV latent antigens indicated that the gene for LMP1 was sufficient to induce TAP and MHC class I antigen expression in group I BL cells. This immunomodulatory property is unique to LMP1, as none of the other EBV latent antigens display any regulatory effect on antigen-processing genes in B cells (23). However, the precise mechanism by which LMP1 modulates the antigen-processing function is unclear. Previous studies have shown that LMP1 acts as a constitutively active receptor-like molecule independent of the binding of a ligand (5, 11, 15, 38). The transmembrane domains mediate oligomerization of LMP1 molecules in the plasma membrane, a prerequisite for LMP1 function (9, 11). The C terminus of LMP1 initiates signaling through C-terminal activator regions (referred to as CTAR1 [amino acids 194 to 231] and CTAR2 [amino acids 351 to 386]) (16, 29). Both CTARs are involved in the induction of NF-κB, CTAR2 being the principal NF-κB activator site. Recent studies by our group have shown that the RelB subunit of the NF-κB family directly modulates the antigen-presenting function of B cells (32). Therefore, the present studies were undertaken to examine whether LMP1-mediated activation of NF-κB results in nuclear translocation of RelB, thereby influencing the antigen-presenting function of B cells. To address this issue, we used a series of deletions and point mutants to precisely map the domain(s) of LMP1 that influences antigen-processing gene expression and endogenous CTL epitope presentation in B cells. The data we obtained demonstrate that the expression of full-length LMP1 is coincident with strong induction and nuclear translocation of RelB and that this effect is mediated primarily through the CTARs. Furthermore, LMP1-mediated nuclear translocation of RelB is sufficient for antigen-presenting cell function in group I BL cells normally resistant to CTL lysis.
MATERIALS AND METHODS
Establishment and maintenance of LCL and BL cell lines.
LCLs from seropositive donors were established by exogenous virus transformation of peripheral B cells using EBV derived from type 1 (B95.8) cell lines (10). The mutant LCL × T-lymphoblastoid hybrid cell line 174 × CEM.T2 (referred to as the T2 cell line) does not express the gene for TAP1, TAP2, LMP2, or LMP7 (34). T2 cells were used as negative controls for TAP-1, TAP-2, LMP2, and LMP7 reverse transcription (RT)-PCR analysis. The TAP-deficient B-cell lymphoma cell lines BJAB and BL30 and derivatives of the BJAB cell line stably transfected with LMP1 (BJAB-MTLM6) or a control plasmid (BJAB-gpt2) were also used (39). All cell lines were routinely maintained in RPMI 1640 medium containing 2 mM glutamine, penicillin at 100 IU/ml, and streptomycin at 100 μg/ml plus 10 to 20% fetal calf serum (growth medium).
DNA expression vectors.
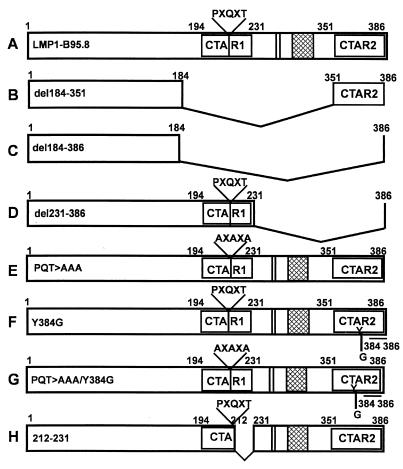
All full-length and deletion mutant LMP1-expressing constructs were based on the pSG5 or pHEBO eukaryotic expression vector, in which the inserted genes are placed downstream of a simian virus 40 promoter and a β-globin intron. The pSG5-LMP1 plasmid containing a cDNA of B95.8 has been described previously (16). In addition, a panel of deletion mutants expressing truncated forms of LMP1 protein was used (Fig. 1A to D). These expression vectors were designed to express an LMP1 protein in which CTAR1 and/or CTAR2 was deleted. To precisely map the site within LMP1 that is crucial for modulating the expression and function of antigen-processing genes in B cells, a panel of single-point mutant forms of the gene for LMP1 was used (Fig. 1E to H). Previous studies have shown that these point mutations within CTAR1 and/or CTAR2 of LMP1 block TRAFF/TRADD binding and NF-κB activation (12, 25, 27). Detailed descriptions of these deletion mutant and single-point mutant expression vectors have been published elsewhere (10, 26). In addition, expression vectors encoding dominant negative IκBα S32/36A (serines 32 and 36 mutated to alanine; a kind gift from Francis Shanon, ANU, Canberra, Australia) and antisense RelB were also used in this study (33, 37). When transfected into LCLs, antisense RelB blocked RelB but not p50 or p65 nuclear activity (data not shown).
FIG. 1.
Schematic representation of the LMP1 constructs used in this study. Expression constructs encoding either full-length LMP1 or a series of truncated forms of LMP1 were used (A to D and H). The deletion mutant forms were designed to express a LMP1 protein in which CTAR1 and/or CTAR2 were deleted. In addition, a panel of single-point mutant forms of the LMP1-encoding gene were also used (E to G). The point mutations within the CTARs of LMP1 (PQT>AAA, Y384G, and PQT>AAA/Y384G) block TRAFF/TRADD binding and NF-κB activation. Further details of these constructs can be found in references 12, 25, and 27.
Cellular staining for RelB in LMP1-transfected BJAB cells.
RelB was detected by immunocytochemical staining. Briefly, 105 cells were cytospun, fixed in paraformaldehyde, stained with anti-RelB (Santa Cruz Biotechnology, Santa Cruz, Calif.), followed by swine anti-rabbit immunoglobulin (Ig; DAKO, Carpinteria, Calif.), and revealed with diaminobenzidine as previously described (32, 33). Either full-length B95.8 LMP1 or a mutant form, i.e., the 184-to-386 deletion mutant or the PQT>AAA/Y384G point mutant form, was cotransfected with green fluorescent protein (GFP) into 5 × 106 BJAB cells and used for immunocytochemical detection of RelB as described above.
Transfection of lymphoid cell lines.
For electroporation of B-cell lymphoma cell lines, 5 × 106 cells in growth medium and DNA were placed in electroporation cuvettes with 0.4 mm of space between the electrodes. For cells intended for fluorescence-activated cell sorter (FACS) and RT-PCR analysis, 2 μg of an expression vector encoding enhanced GFP (EGFP) (pEGFP-N1) was added to the cell suspension, together with 4 μg of the pSG5 vector or an LMP1 expression vector. The cells were then pulsed with a Bio-Rad Gene Pulser at 960 μF and either 240 (BJAB) or 320 (BL30) mV and subsequently transferred to 15 ml of RPMI 1640 growth medium in a T25 tissue culture flask. After 48 h, the LMP1 expression in these cells was analyzed by immunoblotting using an LMP1-specific antibody (CS1-4) and RT-PCR using LMP1 sequence-specific primers.
For transfection of RelB inhibitors, 10 μg of cDNA encoding full-length LMP1-B95.8 was cotransfected with either antisense RelB or IκBα S32/36A into 5 × 106 BJAB cells. At 24 to 48 h after transfection, these cells were used as targets in CTL assays or analyzed by FACScan for surface HLA class I expression.
CTL clones and peptide epitopes.
The EBV-specific CTL clones used in this study were LC13 (EBNA3 specific, HLA B8 restricted), NB37 (EBNA3 specific, HLA B35 restricted), and NB26 (EBNA3 specific, HLA B35 restricted). The specificity of these clones has been defined at the peptide epitope level: LC13 recognizes the minimal epitope sequence FLRGRAYGL (3), while NB37 and NB26 recognize the minimal epitope sequence YPLHEQHGM (2, 24). These clones were maintained in growth medium containing highly purified recombinant human interleukin-2 from Escherichia coli.
Vaccinia virus recombinants.
Recombinant vaccinia virus constructs encoding EBNA3 of EBV and a vaccinia virus construct made by insertion of the pSC11 vector alone and negative for thymidine kinase (Vacc.TK−) have been previously described (21). BL cells were infected with recombinant vaccinia virus at a multiplicity of infection of 10:1 for 1 h at 37°C as described earlier (14). After overnight infection, cells were washed with growth medium and processed for CTL assays.
Cytotoxicity assay.
LMP1-transfected BL cells infected with recombinant vaccinia virus (Vacc.EBNA3 or Vacc.TK−) were incubated with 51Cr for 90 min and used as targets in a standard 5-h 51Cr release assay (31).
Analysis of MHC expression on BL cells.
MHC expression on BL cells was analyzed by surface immunofluorescence labeling, followed by flow cytofluorometry (FACS). Cells were initially incubated with the HLA allele-specific monoclonal antibodies, washed, and further incubated with polyclonal, affinity-purified, phycoerythrin-conjugated goat anti-mouse or -rat Ig (Silenus). The monoclonal antibodies used were HB54 (ATCC, Manassas, Va.) specifying HLA A2, A11.1 M (ATCC) specifying HLA A11, SFR6-Bw6 (ATCC) specifying HLA Bw6, and 0289HA (One Lambda Inc.) specifying HLA A1. Labeled cells were analyzed with the FACScan (Becton Dickinson & Co., Mountain View, Calif.), and results are expressed as mean fluorescence intensity.
Analysis of antigen-processing genes in BL cells.
Expression of peptide transporters (TAP-1 and TAP-2) and/or proteasome components (LMP2 and LMP7) in LCLs and LMP-transfected BL cells was analyzed by RT-PCR. Total RNA from BL cells, LCLs, and TAP- and LMP-negative T2 cells was isolated by using a single-step RNA isolation reagent (Advanced Biotechnologies Ltd.) and reverse transcribed with oligo-p(dT) primer. Semiquantitative PCR analysis of relative amounts of TAP-1, TAP-2, LMP2, LMP7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts was performed with the following sequence-specific primers: TAP-2 FOR, 5" ATGTCTCGAATCAACTTGCGGA 3"; TAP-2 REV, 5" CGCCAATACAGCTGCCGACATT 3"; TAP-1 FOR, 5" CTGCACTGGGGAAGTCACCCTA 3"; TAP-1 REV, 5" GAGCATGATCCCCAAGAGACATA 3"; LMP2 FOR, 5" ATGGGATAGAACTGGAGGAACC 3"; LMP2 REV, 5" ACCATAGATAAAGGTGCTGCCG 3"; LMP7 FOR, 5" CAGATTGAGATGGCCCATGGCA 3"; LMP7 REV, 5" TGCACATCATGTTGGACAGCAGC 3"; GAPDH FOR, 5" CTCAGACACCATGGGGAAGGTG 3"; GAPDH REV, 5" CAGGGGTGCTAAGCAGTTGGTG 3". Amplifications were performed in 20-μl reaction volumes using 10 μl of cDNA, 10 pmol of each primer, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 1 U of Taq polymerase. Amplified products were separated on 2% agarose gels and quantitated densitometrically using ImageQuant software version 3.3 (Molecular Dynamics Inc., Sunnyvale, Calif.). The integrity of these primers was tested in LCLs and T2 cells. LCLs consistently showed strong expression of the genes for TAP and LMP2/7, while expression of none of these genes was detected in T2 cells.
RESULTS
Regulation of RelB expression by LMP1 and its effect on antigen-presenting function.
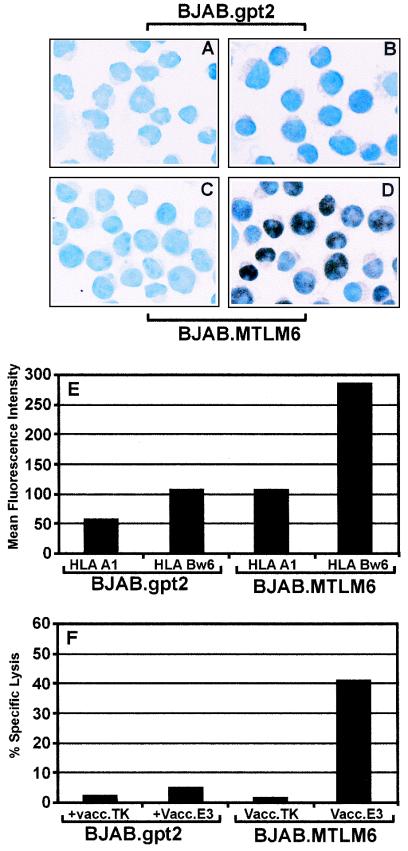
We have previously shown that, unlike EBV-transformed LCLs, the BL cell lines display defects in antigen presentation due to down-regulated expression of essential components of the antigen-processing machinery (e.g., TAP-1 and−2 and MHC class I). However, this defect can be completely reversed following the expression of EBV-encoded LMP1. To delineate the mechanism of LMP1-mediated up-regulation of the antigen-processing function in BL cells, we initially explored the possibility that the LMP1-mediated immunomodulatory effect might be mediated through the RelB subunit of the NF-κB transcription factor family. This hypothesis was based on our recent finding that RelB transcriptional activity can directly affect the antigen-presenting function of B cells (32). To examine the effect of LMP1 on RelB expression, stable LMP1 transfectants of the BJAB cell line (BJAB.MTLM6) and control cells (BJAB.gpt2) were stained with RelB-specific antibody and detected by immunocytochemical staining. Representative data from one such analysis are presented in Fig. 2. More than 90% of the BJAB cells stably transfected with the LMP1 expression vector (BJAB.MTLM6) showed strong expression of RelB, which was localized primarily within the nucleus. On the other hand, LMP1-negative cells (BJAB.gpt2) showed very weak staining with a predominantly cytoplasmic localization of RelB. Furthermore, analysis of surface HLA class I expression indicated that BJAB.MTLM6 cells expressed two- to threefold higher HLA A1 and Bw6 levels than did BJAB.gpt2 cells (Fig. 2E). This increased surface HLA expression correlated with efficient presentation of endogenously processed CTL epitopes through the class I pathway (Fig. 2F). Taken together, the data indicate that MHC class I expression, antigen-presenting function, and translocation of RelB to the nucleus are correlated in BJAB.MTLM6 cells that express LMP1.
FIG. 2.
Regulation of RelB expression by LMP1 and its effect on antigen-presenting function. (A to D) immunohistochemical analysis of RelB expression in LMP1-negative (BJAB.gpt2) and LMP1-positive (BJAB.MTM6) cell lines. Cells (105) were cytospun, fixed in paraformaldehyde, and stained with anti-RelB, followed by swine anti-rabbit Ig, and revealed with diaminobenzidine. These cells were counterstained with hematoxylin. (E) Surface HLA A1 and Bw6 expression in BLAB.gpt2 and BJAB.MTLM6 cells. BLAB.gpt2 and BJAB.MTLM6 cells were incubated with HLA allele-specific antibody, followed by fluorescein isothiocyanate-labeled anti-mouse Ig. Mean fluorescence intensity was determined by FACScalibur. (F) Endogenous processing of HLA B35-restricted EBV CTL epitopes by BJAB.gpt2 and BJAB.MTLM6 cells. BJAB.gpt2 and BJAB.MTLM6 cells were infected with either a control vaccinia virus (Vacc.TK) or a recombinant vaccinia virus encoding EBNA3. Following overnight infection, these cells were exposed to EBNA3-specific CTL clone NB26. Percent specific lysis from one of three different experiments is shown.
CTARs of LMP1 are critical for regulating the expression of antigen-processing genes and nuclear translocation of RelB.
To further delineate the mechanism by which LMP1 regulates antigen processing and RelB translocation in BL cells, we tested a panel of LMP1 constructs to assess their effects on the expression of MHC class I, peptide transporters (TAP-1 and −2), proteasome (LMP2 and LMP7)-encoding genes, and RelB. These constructs were designed to encode either the full-length LMP1 protein or truncated LMP1 containing specific deletions within CTAR-1 and/or CTAR-2 (Fig. 1A to D). Two B-cell lymphoma lines, BJAB and BL30, which were previously shown to have reduced expression of surface MHC class I and peptide transporters were used (20, 23).
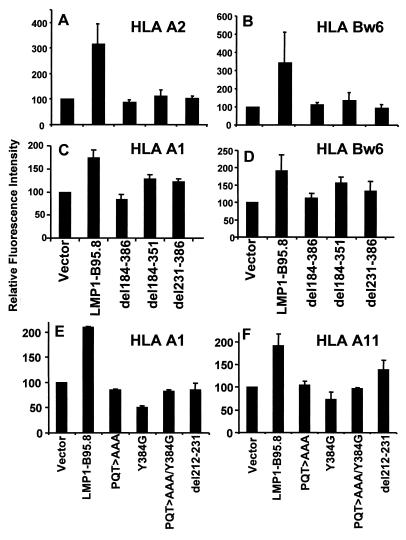
In the first set of experiments, BJAB and BL30 cells were cotransfected with the pEGFP-N1 expression vector and the LMP1 expression construct LMP1-B95.8, del184-386, del184-351, or del231-386. Expression of the various LMP1 constructs following transfection was confirmed by RT-PCR and immunoblotting. LMP1 expression was consistently detected by RT-PCR and immunoblotting after transfection with each construct (data not shown). At 48 h posttransfection, surface MHC class I expression was analyzed on cells based on green fluorescence. Data from the BJAB (panels A and B) and BL30 (panels C and D) cell lines are shown in Fig. 3. Expression of the full-length LMP1-B95.8 sequence was coincident with the strong induction of surface HLA A2 and Bw6 in BJAB cells and HLA A1 and Bw6 in BL30 cells. Deletion of the complete C-terminal domain (del184-386) or the CTARs (del231-386 and del184-351) significantly reduced the magnitude of cell surface HLA class I induction compared with that obtained with full-length LMP1 (Fig. 3).
FIG. 3.
Role of CTARs in regulating LMP1-mediated up-regulation of surface HLA class I expression on BL cells. BJAB (A and B) and BL30 (C and D) cells were transfected with either full-length LMP1 or deletion mutant forms (del184-386, del184-351, and del231-386). At 48 h after transfection, these cells were analyzed for surface HLA expression (HLA A1, A2, or Bw6) using allele-specific antibodies as described in Materials and Methods. To precisely map the motif within the CTAR, BL30 cells were transfected with single-point mutant forms (E and F) and surface HLA expression was analyzed as described above. Data are presented as the mean ± the standard error of three different experiments.
To precisely map the potential regulatory elements within the CTARs, we tested a series of single-point mutant forms of LMP1 (Fig. 1E to H). Previous studies have shown that these point mutations within CTAR1 and/or CTAR2 of LMP1 block TRAFF/TRADD binding and NF-κB activation (12, 25, 27). Considering the importance of the NF-κB pathway in the transcriptional regulation of genes within the MHC locus, we hypothesized that specific residues or motifs within the CTARs of LMP1 could control MHC class I expression. To address this hypothesis, BL30 cells were cotransfected with EGFP-N1 and LMP1 constructs, LMP1-B95.8, PQT>AAA, Y384G, PQT>AAA/Y384G, and del212-231. At 48 h after transfection, GFP-positive cells were sorted and then analyzed for surface HLA class I expression by FACS as described above. Significant loss of LMP1-mediated up-regulation of surface expression of HLA A1 and Bw6 alleles was noted when BL30 cells were transfected with the PQT>AAA, Y384G, or PQT>AAA/Y384G expression vector (Fig. 3E and F). These results demonstrate the contribution of the CTARs of LMP1 to the regulation of surface HLA class I expression in BL cells.
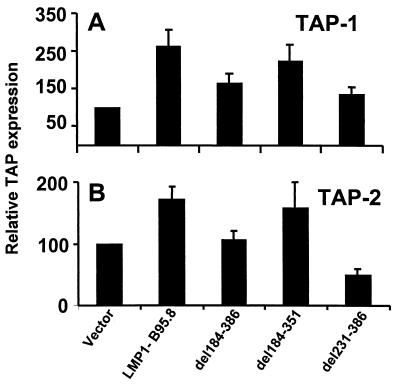
To determine whether the CTARs of LMP1 might have similar effects on other genes controlling the antigen-processing function of B cells, BL30 cells were cotransfected with EGFP-N1 and LMP1 constructs and then GFP-positive cells were sorted flow cytometrically. The expression of peptide transporters (TAP-1 and−2) and proteasomes (LMP2 and LMP7) in these GFP-positive cells was analyzed by RT-PCR. The mRNA expression of each antigen-processing component was quantitated densitometrically by using ImageQuant software. The analysis of the expression of TAP-1 and−2 and LMP-2 and−7 was standardized by using the housekeeping gene for GAPDH as an internal control, and the ratio of TAP to GAPDH was calculated. A 1.5- to 2.5-fold increase in the expression of both TAP-1 and TAP-2 was observed in BL30 cells following transfection of full-length LMP1 (Fig. 4). This up-regulation was impaired when the complete LMP1 C-terminal domain (del184-386) was deleted. Since the transfection of an LMP1 expression vector lacking CTAR1 (del184-351) induced TAP-1 and TAP-2 expression almost as efficiently as did full-length LMP1, the data suggest that up-regulation of TAP expression in BL 30 cells was mediated primarily through CTAR2 (Fig. 4). Further analysis with single-point mutant proteins indicated that the Y384G and PQT>AAA/Y384G mutant forms impaired this effect on TAP-1 and TAP-2 (data not shown). LMP1-transfected BL30 cells were also analyzed in the same way for the expression of inducible proteasome components LMP2 and LMP7. In contrast to its effect on TAP and MHC class I, LMP1 had little effect on LMP2 and LMP7 expression (data not shown). These data together indicate that LMP1 induces both MHC class I and TAP expression through its CTARs. The effect of deletion of CTAR1 and CTAR2 on RelB nuclear translocation was therefore examined.
FIG. 4.
Effects of CTARs on regulation of TAP-1 (A) and TAP-2 (B) expression in BL cells. BL30 cells were transfected with either full-length LMP1 or deletion mutant forms (del184-386, del184-351, and del231-386). At 48 h after transfection, these cells were assessed for TAP-1 and TAP-2 expression by RT-PCR using sequence-specific primers. A detailed description of the RT-PCR method used is given in Materials and Methods. Data are presented as the mean ± the standard error of five different experiments.
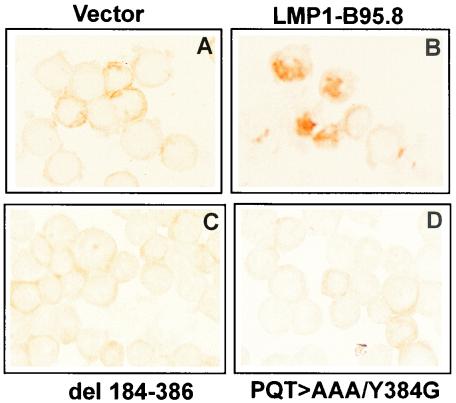
BJAB cells were transfected with either a full-length LMP1, del184-386, or PQT>AAA/Y384G expression vector. At 48 h after transfection, these cells were assessed for RelB expression by immunocytochemical staining with a RelB-specific antibody (Fig. 5). BJAB cells transfected with full-length LMP1 (panel B) showed strong nuclear expression of RelB, while BJAB cells transfected with a control vector (panel A), del184-386 (panel C), or PQT>AAA/Y384G (panel D) lacked nuclear expression of RelB, despite the presence of RelB in the cytoplasmic compartment.
FIG. 5.
Immunohistochemical analysis of RelB expression in BJAB cells following transfection of either a control vector (A), full-length LMP1-B95.8 (B), del184-386 (C), or PQT>AAA/Y384G (D). Cells (105) were cytospun, fixed in paraformaldehyde, and stained with anti-RelB, followed by swine anti-rabbit Ig (DAKO), and revealed with diaminobenzidine. No counterstain was used in this analysis.
Cytotoxic T-cell recognition of BL cells transfected with LMP1 expression vectors is RelB dependent.
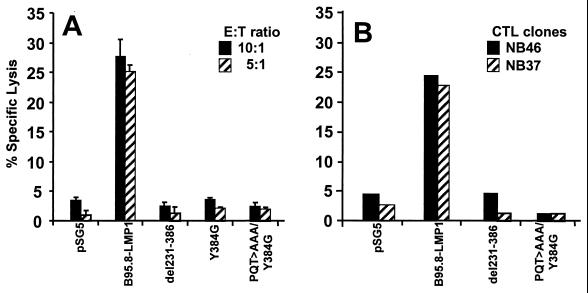
Since normal expression of surface MHC class I antigens and antigen-processing components is essential for the endogenous processing of CTL epitopes and CTARs of LMP1 are involved in the up-regulation of antigen-processing genes, the effects of LMP1 and its deletion mutant forms on the CTL-mediated immune recognition of EBV epitopes were tested. TAP-deficient BL30 and BJAB cells were transiently transfected with each of the LMP1 expression constructs (B95.8-LMP1, del231-386, and PQT>AAA/Y384G) or a control vector (pSG5). At 24 h after transfection, these cells were infected with a recombinant vaccinia virus encoding EBNA3 for 18 h and then exposed to EBNA3-specific CTL clones in a standard 51Cr release assay. A summary of three different sets of experiments is presented in Fig. 6. Only BL30 and BJAB cells transfected with the full-length LMP1 sequence from the B95.8 isolate were efficiently recognized by the EBNA3-specific CTL clones. In contrast, cells transfected with either del231-386 or point mutants Y384G and PQT>AAA/Y384G were poorly recognized by these CTLs (Fig. 6A and B).
FIG. 6.
Endogenous processing function in BL cells transfected with LMP1 expression vectors is dependent on the presence of the CTARs. BJAB (A) and BL30 (B) cells were transfected with the pSG5, LMP1-B95.8, del231-386, Y384G, or PQT>AAA/Y384G expression vector. At 24 h after transfection, these cells were infected with a recombinant vaccinia virus expression vector encoding EBNA3 for 14 to 16 h. After infection, these cells were used as targets in a CTL assay. For BL30 cells, an HLA B8-restricted, EBNA3-specific LC13 CTL clone was used as an effector, while for BJAB cells, HLA B35-restricted, EBNA3-specific CTL clones NB26 and NB37 were used as effector cells. Results are expressed as percent specific lysis. E:T ratio, effector-to-target cell ratio.
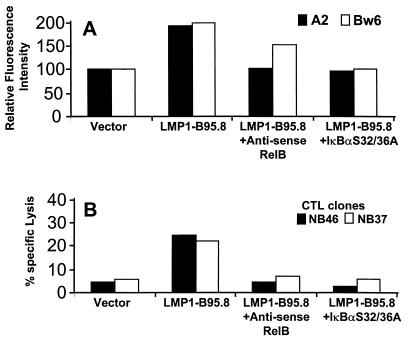
To determine whether the augmentation of antigen-presenting function conferred by LMP1 was dependent on RelB, BJAB cells transiently transfected with full-length LMP1 were cotransfected with expression vectors encoding either antisense RelB or dominant negative IκBα S32/36A (Fig. 7). The increased surface HLA A2 and Bw6 expression on BJAB cells following transfection of full-length LMP1 was impaired in the presence of an antisense RelB or IκBα S32/36A expression vector (Fig. 7A). Similarly, endogenous processing of class I-restricted CTL epitopes by BJAB cells was blocked when full-length LMP1 was coexpressed with either antisense RelB or IκBα (Fig. 7B). The data indicate that the enhancement of antigen-presenting function conferred by LMP1 is directly related to the expression of NF-κB, specifically, the RelB subunit.
FIG. 7.
Effects of antisense RelB and IκBα S32/36A on the LMP1-mediated modulation of surface HLA class I expression and antigen-presenting cell function. BJAB cells were cotransfected with full-length LMP1 and antisense RelB or IκBα S32/36A, and 24 h after transfection, these cells were analyzed for surface HLA class I expression (A) and antigen-presenting cell function (B). BJAB cells cotransfected with an expression vector encoding full-length LMP1 and a control vector were used as positive controls.
DISCUSSION
Group I BL cell lines retaining the original BL tumor cell phenotype are impaired in the ability to present endogenously expressed antigens to HLA class I-restricted cytotoxic T cells. Compared with the group III BL lines, features of the BL phenotype, including very low levels of TAP-1/TAP-2 and HLA class I expression, contribute to this effect (20). Expression of the EBV-encoded oncogene for LMP1 can independently up-regulate the expression of TAP and MHC class I expression in group I BL cells (23). To delineate the pathway of this immunomodulation, we used a series of deletion and point mutant forms of LMP1 to study the effects of individual domains and residues within this protein on the regulation of TAP and MHC expression. Since previous studies have shown that the amino terminus and transmembrane domains primarily contribute to the structural integrity of LMP1 rather than the LMP1-mediated effects on B cells, we retained these domains in all of the mutant constructs that were used in this study. We focused primarily on the LMP1 carboxyl terminus, which includes CTAR1 and CTAR2, to study its effect on the antigen-presenting cell function of B cells.
CTAR1 and CTAR2 of LMP1 are involved in the activation of NF-κB, which plays a crucial role in EBV-mediated cellular phenotypic changes during the transformation process (16, 17). NF-κB proteins are also important players in the immune control of various pathogens. Studies with various transgenic mouse models have indicated that individual Rel family members (RelA, c-Rel, p50, p52, and RelB) contribute specifically to the generation of immune responses. In particular, a recent report from our group has shown that RelB can directly affect the antigen-presenting cell function of B cells. This suggested the possibility that LMP1 might specifically regulate the function of RelB and thus directly influence the antigen-processing function of B cells. Previous studies have shown that nuclear translocation of RelB confers a qualitative effect on antigen-presenting cell function (32). Similar to these observations, LMP1-expressing BJAB cells expressed two- to threefold higher levels of surface HLA class I molecules and displayed an efficient antigen-presenting function in conjunction with RelB nuclear translocation.
By analysis of deletion and point mutant forms of LMP1, the regions involved in the regulation of antigen-presenting function were identified. Deletion or mutation of CTAR-1 and/or CTAR-2 significantly impaired the LMP1-mediated up-regulation of surface HLA class I, TAP-1, and TAP-2 expression; antigen presentation; and RelB nuclear translocation. Point mutant forms of LMP1 demonstrated that inhibition of the TRAFF/TRADD binding and NF-κB activation regions also blocked the LMP1-induced immunomodulatory effect on antigen presentation. The reduction of both MHC class I and TAP by the Y384G and PQT>AAA mutant forms was not unexpected since downstream pathways regulating the expression of antigen-processing components are controlled by common regulatory elements (11). In this regard, nuclear localization of RelB might lead to induction of interferon-regulatory factors (e.g., IRF-4 and IRF-7) that are known to regulate lymphocyte activation and expression of antigen-processing genes (14, 40). Furthermore, nuclear translocation of RelB is completely blocked when either the CTARs are deleted or single-point mutations are introduced at the sites that are critical for NF-κB activation. Previously, mutations within CTAR1 and CTAR2 have been shown to reduce the ability of LMP1 to activate NF-κB, whose activation is probably initiated by two independent effector pathways (16). Indeed, the PQT>AAA and Y384G mutant forms disrupt the binding of tumor necrosis factor receptor-associated factors such as TRAF1, −2, −3, and −5 or TRADD, respectively, to the CTARs, which in turn blocks the activation of NF-κB (1, 6, 7).
Previous studies have shown that addition of increasing amounts of IκBα can inhibit p50/RelB and p52/RelB reporter induction (8). In contrast, antisense RelB specifically blocked RelB but not p50 or p65 nuclear activity. Based on these observations, we overexpressed dominant negative IκBα S32/36A or antisense RelB in the presence of full-length LMP1. Each blocked LMP1-induced up-regulation of surface HLA class I and endogenous processing of CTL epitopes. These observations indicate that LMP1-mediated nuclear translocation of NF-κB plays an important role in restoring endogenous processing function in BL cells. Furthermore, they strongly suggest that RelB is the predominant regulatory NF-κB subunit. The observed nuclear translocation of RelB may be initiated by a signaling cascade activated through LMP1, which induces degradation of IκBα (18). The constitutive nuclear expression of RelB in BJAB.MTLM6 cells that stably express LMP1 supports this mechanism. In contrast BJAB cells stably expressing RelB constitutively regulate RelB, so that it is located predominantly in the cytoplasm but translocated to the nucleus in response to CD40 ligation (32).
Overall, the present study has enhanced our understanding of the mechanism by which LMP1 modulates the antigen-processing and presentation function of EBV-infected B cells. The data presented here provide convincing evidence that a motif(s) within the CTARs initiates downstream activation of the NF-κB pathway, and one of the components of this pathway, namely, RelB, appears to be one of the most critical players in the regulation of the antigen-presenting function of B cells. These observations also have important implications for our understanding of how an immunogenic virus like EBV is able to persist for life in an immunocompetent host by balancing its own need for survival without jeopardizing the survival of the host. Previous studies suggest that this is achieved by a long-lived, virus-carrying B-cell pool in which EBV gene expression is restricted to LMP2 and EBNA1 but not LMP1 (35). Similarly, EBV-positive BL cell lines, which are believed to be transformed counterparts of germinal center centroblasts, express only EBNA1 but not LMP1 (4, 28). Thus, long-term persistence could be facilitated by the inability of these cells to process and present antigens, as was observed in our earlier studies using BL cells (23). Moreover, these resting EBV-infected B cells express very low levels of costimulatory molecules (CD80 and CD23) and thus provide additional mechanisms by which to escape immunosurveillance (30). On the contrary, it is likely that when EBV-infected B cells express a full spectrum of latent proteins, LMP1 up-regulates antigen-processing function in virus-infected cells, resulting in the activation of a strong CTL response. This CTL response allows the host to maintain a constant check in the overall pool of virus-infected B cells. Any alteration of this immune regulation, as seen in immunocompromised individuals, often results in the outgrowth of EBV-infected B cells. Thus, LMP1 could, in effect, act as an expression switch that, depending on the stage of EBV infection, could maneuver a pathway that modulates the immune system either toward or against its survival.
REFERENCES
- 1.Brodeur, S. R., G. Cheng, D. Baltimore, and D. A. Thorley-Lawson. 1997. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J. Biol. Chem. 272:19777-19784. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, S. R., J. Gardner, R. Khanna, T. Steward, D. J. Moss, S. Rodda, and A. Suhrbier. 1994. Five new cytotoxic T cell epitopes identified within Epstein-Barr virus nuclear antigen 3. J. Gen. Virol. 75:2489-2493. [DOI] [PubMed] [Google Scholar]
- 3.Burrows, S. R., S. J. Rodda, A. Suhrbier, H. M. Geysen, and D. J. Moss. 1992. The specificity of recognition of a cytotoxic T lymphocyte epitope. Eur. J. Immunol. 22:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., J. Z. Zou, L. di Renzo, G. Winberg, L. F. Hu, E. Klein, G. Klein, and I. Ernberg. 1995. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J. Virol. 69:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherney, B. W., C. Sgadari, C. Kanegane, F. Wang, and G. Tosato. 1998. Expression of the Epstein-Barr virus protein LMP1 mediates tumor regression in vivo. Blood 91:2491-2500. [PubMed] [Google Scholar]
- 6.Devergne, O., M. Hummel, H. Koeppen, M. M. Lebeau, E. C. Nathanson, E. Kieff, and M. Birkenbach. 1996. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr Virus infection in B lymphocytes. J. Virol. 70:1143-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devergne, O., E. C. McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrzanski, P., R. P. Ryseck, and R. Bravo. 1995. Specific inhibition of RelB/p52 transcriptional activity by the C-terminal domain of p100. Oncogene 10:1003-1007. [PubMed] [Google Scholar]
- 9.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C terminus activation region 2 (CTAR2) maps to the far C terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 10.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gires, O., M., Ueffing, and W. Hammerschmidt. 2001. Chimeric and mutated variants of LMP1. A helpful tool to analyze the structure-function relationship of a pseudoreceptor. Methods Mol. Biol. 174:313-323. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, C. D., R. J. Murray, C. F. Edwards, and A. B. Rickinson. 1988. Downregulation of cell adhesion molecules LFA-3 and ICAM-1 in Epstein-Barr virus-positive Burkitt's lymphoma underlies tumor cell escape from virus-specific T-cell surveillance. J. Exp. Med. 167:1811-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grumont, R. J., and S. Gerondakis. 2000. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor κB. J. Exp. Med. 191:1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, L. F., E. R. Zabarovsky, F. Chen, S. L. Cao, I. Ernberg, G. Klein, and G. Winberg. 1991. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J. Gen. Virol. 72:2399-2409. [DOI] [PubMed] [Google Scholar]
- 16.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 17.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karin, M. 1999. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 19.Khanna, R., and S. R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19-48. [DOI] [PubMed] [Google Scholar]
- 20.Khanna, R., S. R. Burrows, V. Argaet, and D. J. Moss. 1994. Endoplasmic reticulum signal sequence facilitated transport of peptide epitopes restores immunogenicity of an antigen processing defective tumour cell line. Int. Immunol. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 21.Khanna, R., S. R. Burrows, M. G. Kurilla, C. A. Jacob, I. S. Misko, T. B. Sculley, E. Kieff, and D. J. Moss. 1992. Localization of Epstein-Barr virus cytotoxic T-cell epitopes using recombinant vaccinia: implications for vaccine development. J. Exp. Med. 176:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khanna, R., D. J. Moss, and S. R. Burrows. 1999. Vaccine strategies against Epstein-Barr virus-associated diseases: lessons from studies on cytotoxic T-cell mediated immune regulation. Immunol. Rev. 170:49-64. [DOI] [PubMed] [Google Scholar]
- 23.Khanna, R., M. Rowe, C. A. Jacob, V. Argaet, A. Kelly, S. Powis, M. Belich, D. Croom Carter, S. Lee, S. R. Burrows, D. J. Moss, and A. B. Rickinson. 1995. Restoration of endogenous antigen processing in Burkitt's lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur. J. Immunol. 25:1374-1384. [DOI] [PubMed] [Google Scholar]
- 24.Khanna, R., S. L. Silins, Z. Weng, D. Gatchell, S. R. Burrows, and, L. Cooper. 1999. Cytotoxic T cell recognition of allelic variants of HLA B35 bound to an Epstein-Barr virus epitope: influence of peptide conformation and TCR-peptide interaction. Eur. J. Immunol. 29:1587-1597. [DOI] [PubMed] [Google Scholar]
- 25.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Raven Press, Philadelphia. Pa.
- 26.Kieser, A., C. Kaiser, and W. Hammerschmidt. 1999. LMP1 signal transduction differs substantially from TNF receptor 1 signaling in the molecular functions of TRADD and TRAF2. EMBO J. 18:2511-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 16:6478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLennan, I. C., S. Oldfield, Y. J. Liu, and P. J. Lane. 1989. Regulation of B-cell populations. Curr. Top. Pathol. 79:37-57. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss, D. J., I. S. Misko, S. R. Burrows, K. Burman, R. McCarthy, and T. B. Sculley. 1988. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature 331:719-721. [DOI] [PubMed] [Google Scholar]
- 32.O'Sullivan, B. J., K. P. MacDonald, A. R. Pettit, and R. Thomas. 2000. RelB nuclear translocation regulates B cell MHC molecule, CD40 expression, and antigen-presenting cell function. Proc. Natl. Acad. Sci. USA 97:11421-11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettit, A. R., K. P. MacDonald, B. O'Sullivan, and R. Thomas. 2000. Differentiated dendritic cells expressing nuclear RelB are predominantly located in rheumatoid synovial tissue perivascular mononuclear cell aggregates. Arthritis Rheum. 43:791-800. [DOI] [PubMed] [Google Scholar]
- 34.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant T × B cell hybrid. EMBO J. 5:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tierney, R. J., N. Steven, L. S. Young, and A. B. Rickinson. 1994. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier. J. Virol. 68:7374-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 37.Traenckner, E. B., H. L. Pahl, T. Henkel, K. N. Schmidt, S. Wilk, and P. A. Baeuerle. 1995. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 14:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida, J., T. Yasui, Y. Takaoka-Shichijo, M. Muraoka, W. Kulwichit, N. Raab-Traub, and H. Kikutani. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300-303. [DOI] [PubMed] [Google Scholar]
- 39.Wang, J. H., Y. W. Yan, T. P. Garrett, J. H. Liu, D. W. Rodgers, R. L. Garlick, G. E. Tarr, Y. Husain, E. L. Reinherz, and S. C. Harrison. 1990. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature 348:411-418. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J.Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]