Abstract
The mechanisms underlying cleavage of herpesvirus genomes from replicative concatemers are unknown. Evidence from herpes simplex virus type 1 suggests that cleavage occurs by a nonduplicative process; however, additional evidence suggests that terminal repeats may also be duplicated during the cleavage process. This issue has been difficult to resolve due to the variable numbers of reiterated terminal repeats that the herpes simplex virus type 1 genome can contain. Guinea pig cytomegalovirus is a herpesvirus with a simple terminal repeat arrangement that defines two genome types. Type II genomes have a single copy of a 1-kb terminal repeat at both their left and right termini, whereas type I genomes have only one copy at their left termini and lack the repeat at their right termini. In a previous study, we constructed a recombinant guinea pig cytomegalovirus in which certain cis elements were disrupted such that only type II genomes were produced. Here we show that double repeats that are formed by circularization of infecting genomes are rapidly converted to single repeats, such that the junctions between genomes within replicative concatemers formed late in infection almost exclusively contain single copies of the terminal repeat. Therefore, for the recombinant virus, each cleavage event begins with a single repeat within a concatemer yet produces two repeats, one at each of the resulting termini, demonstrating that terminal repeat duplication occurs in conjunction with cleavage. For wild-type guinea pig cytomegalovirus, the formation of type I genomes further suggests that cleavage can also occur by a nonduplicative process and that duplicative and nonduplicative cleavage can occur concurrently. Other herpesviruses having terminal repeats, such as the herpes simplex viruses and human cytomegalovirus, may also utilize repeat duplication and deletion; however, the biological importance of these events remains unknown.
The Herpesviridae family of viruses includes several significant human pathogens, including herpes simplex virus type 1 (HSV-1) and HSV-2, varicella-zoster virus, Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus, and human cytomegalovirus. Herpesviruses have large (130 to 235 kb), double-stranded linear DNA genomes that circularize shortly after infection (16, 22, 29). Viral DNA synthesis leads to the formation of concatemers of head-to-tail linked genomes that are packaged and cleaved to produce encapsidated unit-length genomes (2, 3, 18, 21, 22, 29, 30, 36). Concatemer cleavage and packaging are highly conserved among the herpesviruses, but little is known about the mechanisms or the machinery involved.
Cleavage occurs only at sites defined by specific cis-acting signals in the DNA. Two sequence elements, denoted pac1 and pac2, are conserved near genomic termini and are essential for cleavage or packaging of concatemeric DNA to produce infectious viral progeny (4, 5, 8, 10-12, 17, 23, 24, 28, 31, 33, 37). In HSV-1, pac1 and pac2 lie at opposite ends of a small terminal repeat called the a sequence. One copy of the a sequence is located at the short-arm end of the HSV-1 genome, and one to several copies are located both at the long-arm terminus and at an internal site where the long and short arms are joined (29).
Several mechanisms have been proposed for HSV-1 cleavage. In the staggered-cut mechanism proposed by Mocarski and Roizman (27), cleavage is a simple, single-base, staggered cut and does not involve duplication of terminal repeat sequences. This was suggested by comparing the sequences of cleavage sites to the sequences of genomic termini from HSV-1 (27). Sequences from several other herpesviruses with genomes that lack terminal repeats are also consistent with this model (5, 8, 17, 20, 23, 26, 32). However, in order to produce genomes with at least one a sequence at each end, cleavage by a staggered-cut mechanism must occur between two a sequences, and this has been difficult to reconcile with observations that approximately half of the cleavage sites within HSV-1 DNA contain only single a sequences (9, 19, 34). Moreover, single a sequences inserted into the thymidine kinase gene of HSV-1 (33) or within plasmids used to construct HSV-1 amplicons (11, 33) become amplified to form tandem arrays containing variable numbers of a sequences. These observations have led to proposals that the HSV-1 a sequence is duplicated during cleavage (11, 33). This hypothesis has been difficult to prove owing to the variable numbers of reiterated a sequences present both within cleavage sites and at the termini of HSV-1 genomes; however, analysis of ectopically engineered cleavage sites within the murine cytomegalovirus genome revealed that the 30-bp terminal repeat was only duplicated when the ectopic site was able to be cleaved, thus demonstrating a clear link between repeat duplication and cleavage (24). Even so, because rare duplications could have accumulated over successive replication cycles, it was not clear from either the HSV-1 or murine cytomegalovirus studies whether duplication occurred at each cleavage event or only very rarely.
To better understand the mechanisms of herpesvirus genome cleavage, we chose to study guinea pig cytomegalovirus (GPCMV), a herpesvirus with a simple genome structure that lacks reiterated terminal repeats, internal repeats, or invertible elements (15, 23). GPCMV has the feature of forming two genome types in approximately equimolar amounts. Type II genomes have one copy of a 1-kb terminal repeat at both left and right ends, whereas type I genomes have the repeat at the left end but lack the repeat at the right end (15) (Fig. 1A). In earlier studies (23), we constructed the two recombinant GPCMVs, RG5001 and RG5201, illustrated in Fig. 1A. In virus RG5001, O-terminal sequences including a presumed pac2 cis element were displaced by insertion of a 1-kb xanthine-guanine phosphoribosyltransferase (gpt) marker cassette between unique O sequences and the terminal repeat. Only type II genomes were produced by this virus (23). In the control virus RG5201, reintroduction of 64 bp of O-terminal sequences containing the presumed cis cleavage/packaging sequences between the terminal repeat and the gpt cassette restored a wild-type phenotype, that is, an approximately equimolar mixture of type I and type II genomes (23). We reasoned that, if cleavage occurs by a nonduplicative mechanism, then RG5001 genomes would have to be cleaved from concatemer junctions containing two repeats. Alternatively, if duplication occurs during cleavage, RG5001 genomes could be cleaved from single-repeat-containing junctions. Therefore, we sought to determine whether junctions within RG5001 concatemeric DNA contain predominantly double or single repeats.
FIG. 1.
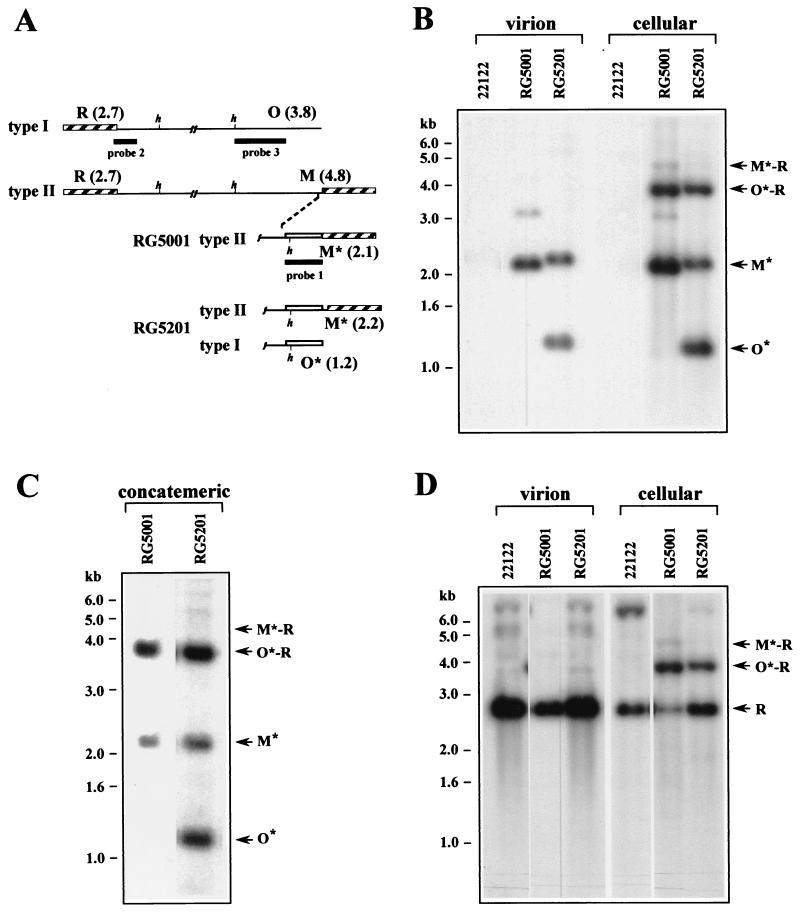
(A) Type I and type II genomes of wild-type GPCMV are illustrated with the terminal repeats shown as hatched boxes and the HindIII sites proximal to the termini indicated (h). Illustrated below are the right termini of RG5001 and RG5201 with inserted gpt sequences shown as open boxes. The locations and sizes (in kilobases) of the terminal HindIII restriction fragments R, O, M, O*, and M* are indicated. Thick lines indicate the regions contained in probes used for hybridizations. (B) GEFs were infected with wild-type GPCMV or with recombinant viruses RG5001 and RG5201 at an MOI of 3. Four days after infection, DNA was extracted from extracellular culture supernatants (virion) or from infected cells (cellular) and was digested with HindIII. The fragments were subjected to agarose electrophoreses, transferred to a nylon membrane, and hybridized with probe 1 (Fig. 1A) containing gpt sequences. (C) Cells were infected with viruses RG5001 and RG5201 at an MOI of 3, and 4 days after infection, total cell-associated DNA was separated by PFGE. Concatemeric DNA that failed to migrate was then removed from the PFGE gel and digested with HindIII. The fragments were separated and hybridized as described for panel B. (D) The nylon membrane shown in panel B was rehybridized with probe 2 (Fig. 1A) containing unique sequences from HindIII R. In panels B to D, the sizes (in kilobases) and locations of molecular-weight markers are indicated on the left and the positions of relevant fragments are indicted with arrows on the right.
Guinea pig embryonic fibroblasts (GEFs) were propagated as previously described (23) and were infected at an multiplicity of infection (MOI) of 3 with either RG5001, RG5201, or wild-type GPCMV (ATCC 22122). After 4 days, virion and total cellular DNAs were prepared as previously described (23). DNAs were digested with HindIII, separated electrophoretically, transferred to a nylon membrane, and hybridized with a gpt sequence probe that had been 32P labeled by the random hexamer method as previously described (22). A 2.1-kb terminal fragment, designated M*, was predicted from type II genomes, and a 1.2-kb terminal fragment, designated O*, was predicted from type I genomes (Fig. 1A). Single-repeat-containing O*-R junctions were predicted to be 3.9 kb (1.2 + 2.7), and double-repeat-containing M*-R junctions were predicted to be 4.8 kb (2.1 + 2.7).
Consistent with results of our previous studies (23), virion DNA from RG5001 contained M*- but lacked O*-terminal fragments, indicating that only type II genomes are formed by this virus, whereas virion DNA from RG5201 contained both O*- and M*-terminal fragments, indicating that formation of both type I and type II genomes was restored in this virus (Fig. 1B). Similar terminal fragments were observed in infected-cell DNA; however, the presence of concatemeric DNA resulted in additional fragments corresponding to junctions between genomes. Single-repeat-containing O*-R junction fragments were abundant for both viruses, whereas only a faint signal was observed for double-repeat-containing M*-R junction fragments (Fig. 1B). Therefore, junctions within concatemeric DNA from both RG5001 and RG5201 almost exclusively contained single repeats.
To confirm these results, concatemeric and genomic DNAs from RG5001- and RG5201-infected cells were separated by pulsed-field gel electrophoresis (PFGE) as previously described (22). Concatemeric DNAs were cut from the PFGE gel and were digested with HindIII (25). The DNA fragments were separated by agarose electrophoresis and were then transferred to a nylon membrane and hybridized with gpt sequences. Consistent with the previous experiment in which total infected-cell DNA was used, isolated concatemeric DNA from both viruses contained only O*-R junction fragments (Fig. 1C). That PFGE separation was effective in removing genomic DNA was confirmed in a parallel experiment by failure of a HindIII R-specific probe to detect R-terminal fragments, which were previously shown to be absent from GPCMV concatemeric DNA (25).
One alternative to duplication of terminal repeats has been termed the “theft” model, in which every other genome in the concatemer is packaged and the intervening genomes, which lack terminal repeats, are discarded and presumably degraded (10, 33). This model predicts that termini lacking repeats should be formed, if only transiently. Figure 1B clearly shows that O* termini, which lack a repeat, are not detectable in RG5001-infected cells. To determine if R termini are formed lacking a repeat, the membrane from Fig. 1B was rehybridized with a probe containing sequences from the unique region of the terminal HindIII R fragment (described in reference 23). For both virion and infected-cell DNAs, only the 2.7-kb R-terminal fragment was observed; a 1.7-kb fragment corresponding to R termini which lack a repeat was not detected, indicating that all left termini contained a repeat. As before, single-repeat-containing O*-R junctions were predominantly observed for both recombinant viruses (Fig. 1D).
Our previous work suggested that circularization of the wild-type GPCMV genome occurs by end-to-end ligation, resulting in a nearly equimolar ratio of junctions containing double and single repeats within circular DNA (23). Within concatemeric DNA, however, the ratio observed was approximately 1:4 (23), and the data in Fig. 1C indicate a substantially lower prevalence of double-repeat-containing junctions. That these experiments were done using different host cell lines (104C1 cells in the previous study; GEFs in this study) suggested the possibility that host factors might influence the M-R/O-R ratio, but in either case, the discrepancy in ratios between circular DNA and concatemeric DNA suggested that, at some point in the replication cycle, double repeats are converted to single repeats. To determine the stage at which this occurs, PFGE-isolated circular, concatemeric, and 230-kb DNAs from wild-type GPCMV-infected 104C1 (ATCC CRL-1405) and GEFs were analyzed for terminal and junction fragments at different times after infection. At early time points, cycloheximide (Sigma) was added to block concatemer synthesis but to permit circularization, resulting in DNA that fails to migrate upon PFGE that contains predominantly circularized genomes (22, 23). At 4 h postinfection (hpi), nearly equimolar amounts of M-R and O-R junction fragments were detected in both cell types. These fragments were absent from the inoculum DNA used to infect the cells and therefore represent circularization of input genomes (Fig. 2). Although the presence of terminal fragments indicates some contamination of the circular DNA by linear input genomes, the substantial amount of linear DNA in the 230-kb sample indicates that PFGE was effective in removing the vast majority of linear genomes (Fig. 2).
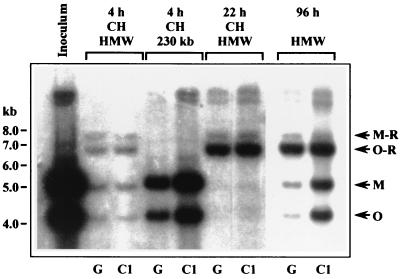
FIG. 2.
GEFs (G) or 104C1 cells (C1) were infected at an MOI of 5 with wild-type GPCMV in the presence or absence of 50 μg of cycloheximide (CH)/ml. Infected-cell DNA was harvested at the times postinfection indicated and separated by PFGE. DNA which failed to enter the PFGE gel (HMW), consisting of either circular or concatemeric DNA (23), and 230-kb DNA were excised from the PFGE gel, digested with HindIII, separated by electrophoresis, and hybridized using probe 3 (Fig. 1A) to specifically detect junction fragments as well as M- and O-terminal fragments. Inoculum DNA was prepared directly from the stocks used to infect the cells as previously described (23). The sizes (in kilobases) and locations of molecular-weight markers are indicated on the left, and the positions of relevant fragments are indicted with arrows on the right.
A marked shift in the M-R/O-R ratio was observed at 22 hpi, indicating conversion of double-repeat-containing M-R junctions to single-repeat-containing O-R junctions. That this conversion occurred in the presence of cycloheximide indicates that loss of one repeat, presumably by homologous recombination, does not require de novo synthesis of either viral or host factors. The same M-R/O-R ratio was observed in concatemeric DNA formed in the absence of cycloheximide both at early times in infection (22 hpi; data not shown) and at late times (96 hpi; Fig. 2). Thus, the shift in repeat number appears to occur after genome circularization but prior to the onset of concatemer synthesis. That both cell lines exhibited similar ratios within concatemeric DNA further demonstrated that the discrepancy between the ratios observed within concatemers in the previous report (23) and in this report is not due to differences in the host cell factors but presumably results from variations in cell culture or infection conditions.
These results reveal that, for recombinant virus RG5001, concatemers in which genomes are separated by single copies of the repeat are cleaved to form genomes containing one repeat at each terminus. As double-repeat-containing fragments are rare in concatemeric DNA, duplication is required for virtually all cleavage events in RG5001. For both RG5201 and wild-type GPCMV, the proportion of type II genomes is approximately 50%, yet in concatemeric DNA, M-R junctions are consistently underrepresented relative to O-R junctions (23; this study). Thus, a significant proportion of type II genomes in RG5201 and in wild-type GPCMV are likely formed by duplicative cleavage at O-R sites. But wild-type GPCMV also produces type I genomes, and as these lack a terminal repeat at their right end, it is probable that they are formed by a simple, nonduplicative cleavage between unique sequences and the terminal repeat. Thus, it appears that that both duplicative and nonduplicative cleavage can occur concurrently within a single virus, although a more complicated process in which duplication occurs followed by excision of the repeat from the right terminus cannot be excluded.
The mechanism of duplicative cleavage in herpesviruses is unknown; however, duplication of terminal repeats during cleavage is not unprecedented. The genomes of several bacteriophages, including T3 and T7, have terminal repeats that are derived from single copies within concatemeric DNA (13, 14). Duplication has been proposed to occur via priming of DNA synthesis at nicks formed near the repeat, transit of the replication fork through the repeat, cleavage at opposite ends of the duplicated repeats, and resolution of the branch generated by the replication fork (6). In support of this model, the T7 DNA polymerase associates with the cleavage/packaging enzyme complex (1) and the absence of T7 DNA polymerase in in vitro packaging reactions results in genomes that lack terminal repeats at one end (35). A similar model could certainly be applied to herpesviruses. Evidence that branch resolution is important for the stability of HSV-1 genome packaging (21) is consistent with a bacteriophage-like mechanism. However, using HSV-1, Church et al. found that cleavage and packaging are not impaired by acyclovir, a specific inhibitor of the viral DNA polymerase. Thus, in contrast to bacteriophage, viral DNA polymerase activity does not appear to be required for HSV-1 cleavage and packaging (7). These results, however, do not exclude the possibility that a host DNA polymerase might complement the lack of viral DNA polymerase activity under these conditions. Similar experiments using human cytomegalovirus confirm that inhibitors of the viral DNA polymerase do not significantly impair DNA cleavage and packaging but that compounds such as aphidicolin and 9-(2-phosphonylmethoxyethyl)guanine, which inhibit both viral and host DNA polymerases, profoundly inhibit cleavage and packaging (M. Davis and M. A. McVoy, unpublished data). Thus, the possibility remains that duplication of herpesvirus terminal repeats may require a DNA polymerase activity. The less interesting theft model (10, 33), in which every other genome is packaged, while a formal possibility, would be highly inefficient and should result in at least transient formation of termini which lack repeats. Both we (this study) and others (11, 27) have failed to detect such termini; however, the possibility that rapid degradation of such termini prevents their detection cannot be excluded.
Our data further indicate that repeat sequences are lost from circular viral genomes prior to concatemer synthesis. This most probably occurs by intramolecular homologous recombination between the two direct repeats. As this process is not inhibited by cycloheximide, it does not require synthesis of viral or host proteins following infection. The biological advantages of duplicating and deleting repeats remain unknown, but it is remarkable that these advantages appear to be common to both bacteriophage and herpesviruses.
Our findings for GPCMV have interesting implications for how we consider cleavage in other herpesviruses. Many herpesviruses lack terminal repeats and presumably must use nonduplicative cleavage exclusively. Like GPCMV, both human and murine cytomegaloviruses form a significant proportion of type I genomes that lack a terminal repeat at the right end (20, 32). It is reasonable to assume that these viruses can also utilize both duplicative and nonduplicative cleavage. HSV-1, which can have either single or multiple a sequences at its cleavage sites, may likewise have the ability to utilize either mechanism as circumstances dictate. For example, at tandem a sequences, either a simple or a duplicative cleavage might occur, whereas upon encountering a single a sequence, duplicative cleavage would be obligatory. For each of these viruses, the arrangement in which the cleavage machinery encounters the cis cleavage/packaging elements (pac1 and pac2 at opposite ends of the terminal repeat versus flanking the point of cleavage) or the presence of additional cis elements may play a significant role in determining the mechanism of cleavage that is utilized; however, the roles that these and perhaps other cis elements play in orchestrating these events remain to be elucidated.
Acknowledgments
This work was supported by Public Health Service grants R21AI43527, RO1AI46668, and KO8AI01435 (to D.E.N.) from the National Institutes of Health, by grant IN-105V from the American Cancer Society, and by funds from the A. D. Williams Fund of the Medical College of Virginia, Virginia Commonwealth University.
REFERENCES
- 1.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. [DOI] [PubMed] [Google Scholar]
- 2.Bataille, D., and A. Epstein. 1994. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology 203:384-388. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Porat, T. 1983. Replication of herpesvirus DNA, p. 81-86. In B. Roizman (ed.), The herpesviruses. Plenum Press, New York, N.Y.
- 4.Broll, H., H. J. Buhk, W. Zimmermann, and M. Goltz. 1999. Structure and function of the prDNA and the genomic termini of the γ2-herpesvirus bovine herpesvirus type 4. J. Gen. Virol. 80:979-986. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury, S. I., H. J. Buhk, H. Ludwig, and W. Hammerschmidt. 1990. Genomic termini of equine herpesvirus 1. J. Virol. 64:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, Y. B., C. Nardone, and D. C. Hinkle. 1990. Bacteriophage T7 DNA packaging. III. A “hairpin” end formed on T7 concatemers may be an intermediate in the processing reaction. J. Mol. Biol. 216:939-948. [DOI] [PubMed] [Google Scholar]
- 7.Church, G. A., A. Dasgupta, and D. W. Wilson. 1998. Herpes simplex virus DNA packaging without measurable DNA synthesis. J. Virol. 72:2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J. 1984. Structure of the genome termini of varicella-zoster virus. J. Gen. Virol. 65:1969-1977. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., and N. M. Wilkie. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55:315-331. [DOI] [PubMed] [Google Scholar]
- 10.Deiss, L. P., J. Chou, and N. Frenkel. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deiss, L. P., and N. Frenkel. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57:933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., and S. Dewhurst. 1998. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J. Virol. 72:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa, H., and K. Sugimoto. 1983. On the terminally redundant sequences of bacteriophage T3 DNA. Virology 124:251-258. [DOI] [PubMed] [Google Scholar]
- 15.Gao, M., and H. C. Isom. 1984. Characterization of the guinea pig cytomegalovirus genome by molecular cloning and physical mapping. J. Virol. 52:436-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber, D. A., S. M. Beverley, and D. M. Coen. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt, W., H. Ludwig, and H. J. Buhk. 1988. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J. Virol. 62:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob, R. J., L. S. Morse, and B. Roizman. 1979. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 29:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locker, H., and N. Frenkel. 1979. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J. Virol. 32:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks, J. R., and D. H. Spector. 1988. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology 162:98-107. [DOI] [PubMed] [Google Scholar]
- 21.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McVoy, M. A., D. E. Nixon, and S. P. Adler. 1997. Circularization and cleavage of guinea pig cytomegalovirus genomes. J. Virol. 71:4209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McVoy, M. A., D. E. Nixon, S. P. Adler, and E. S. Mocarski. 1998. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 72:48-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McVoy, M. A., D. E. Nixon, J. K. Hur, and S. P. Adler. 2000. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 74:1587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mocarski, E. S., A. C. Liu, and R. R. Spaete. 1987. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain). J. Gen. Virol. 68:2223-2230. [DOI] [PubMed] [Google Scholar]
- 27.Mocarski, E. S., and B. Roizman. 1982. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell 31:89-97. [DOI] [PubMed] [Google Scholar]
- 28.Nasseri, M., and E. S. Mocarski. 1988. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology 167:25-30. [DOI] [PubMed] [Google Scholar]
- 29.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1048-1066. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fundamental virology. Raven Press, New York, N.Y.
- 30.Severini, A., A. R. Morgan, D. R. Tovell, and D. L. Tyrrell. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200:428-435. [DOI] [PubMed] [Google Scholar]
- 31.Tamashiro, J. C., D. Filpula, T. Friedmann, and D. H. Spector. 1984. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J. Virol. 52:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamashiro, J. C., and D. H. Spector. 1986. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J. Virol. 59:591-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varmuza, S. L., and J. R. Smiley. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793-802. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, M. J., and W. C. Summers. 1978. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J. Virol. 27:374-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, J. H., and C. C. Richardson. 1987. Processing of concatemers of bacteriophage T7 DNA in vitro. J. Biol. Chem. 262:8851-8860. [PubMed] [Google Scholar]
- 36.Zhang, X., S. Efstathiou, and A. Simmons. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202:530-539. [DOI] [PubMed] [Google Scholar]
- 37.Zimmermann, J., and W. Hammerschmidt. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 69:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]