Abstract
Infection of mice with murine gammaherpesvirus 68 (γHV68; also referred to as MHV68) provides a tractable small-animal model with which to address the requirements for the establishment and maintenance of gammaherpesvirus infection in vivo. The M2 gene of γHV68 is a latency-associated gene that encodes a protein lacking discernible homology to any known viral or cellular proteins. M2 gene transcripts have been detected in latently infected splenocytes (S. M. Husain, E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample, Proc. Natl. Acad. Sci. USA 96:7508-7513, 1999; H. W. Virgin IV, R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck, J. Virol. 73:2321-2332, 1999) and peritoneal exudate cells (H. W. Virgin IV, R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck, J. Virol. 73:2321-2332, 1999), as well as in a latently γHV68-infected B-lymphoma cell line (S. M. Husain, E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample, Proc. Natl. Acad. Sci. USA 96:7508-7513, 1999). Here we describe the generation of γHV68 mutants with disruptions in the M2 gene. Mutation of the M2 gene did not affect the ability of the virus to replicate in tissue culture, nor did it affect γHV68 virulence in B6.Rag1 deficient mice. However, we found that M2 was differentially required for acute replication in vivo. While mutation of M2 did not affect acute phase of virus replication in the lungs of mice following intranasal inoculation, acute-phase virus replication in the spleen was decreased compared to that of the wild-type and marker rescue viruses following intraperitoneal inoculation. Upon intranasal inoculation, M2 mutant viruses exhibited a significant decrease in the establishment of latency in the spleen on day 16 postinfection, as measured by the frequency of viral genome-positive cells. In addition, M2 mutant viral genome-positive cells reactivated from latency inefficiently compared to wild-type and marker rescue viruses. By day 42 after intranasal inoculation, the frequencies of M2 mutant and wild-type viral genome-positive cells were nearly equivalent and little reactivation was detected from either population. In sharp contrast to the results obtained following intranasal inoculation, after intraperitoneal inoculation, no significant defect was observed in the establishment or reactivation from latency with the M2 mutant viruses. These results indicate that the requirements for the establishment of latency are affected by the route of infection.
Gammaherpesviruses are characterized by their ability to establish latency in lymphocytes and by an association with tumors in immunosuppressed hosts. Murine gammaherpesvirus 68 (γHV68; also referred to as MHV68) is a gamma-2 herpesvirus that is closely related to the human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) (3, 4, 23). γHV68 infection of mice provides a genetically tractable small-animal model for the analysis of gammaherpesvirus pathogenesis (reviewed in references 2, 10-13, and 25). γHV68 infection of inbred mice results in an acute, productive infection of multiple organs, including the lung and spleen, and CD4+ T-cell-dependent splenomegaly (5, 14, 16, 18). While acute infection is largely cleared 2 to 3 weeks postinfection (16, 27), a latent infection is established that presumably persists for the life of the host. During latency, the γHV68 genome is maintained in cells in the absence of detectable preformed infectious virus (24, 26, 28, 29). γHV68 establishes latency in B cells and macrophages (6, 17, 29). Lung epithelial cells and dendritic cells have also been implicated as sites of viral latency (6, 15).
Sequence analysis of γHV68 revealed that the majority of its open reading frames (ORFs) are homologous to genes present in other gammaherpesviruses (23). However, γHV68, like the other sequenced gammaherpesviruses, encodes a limited number of unique ORFs. Virus-specific ORFs are located in similar regions of the γHV68, EBV, KSHV, and herpesvirus saimiri genomes (13, 23, 25). In EBV, herpesvirus saimiri, and KSHV, many of the unique genes appear to be involved in either latency or growth transformation (13, 23-25). Initially, on the basis of its genomic position, the γHV68 M2 ORF was identified as a candidate latency-associated gene (24). While M2 expression has been detected in latently infected splenocytes (7, 24)and peritoneal exudate cells (PECs) (24) in the absence of detectable lytic-gene expression, M2 transcripts have also been detected in the spleen and lung within the first month postinfection in the presence of a lytic-cycle transcript, ORF50 (19). The latter study raises the issue that M2 expression may be present during lytic viral replication, as well as during latency. In addition to analyses of in vivo expression, M2 has been shown to be expressed in latently infected murine B-cell lymphoma line S11 (7), which was derived from a persistently infected mouse (20).
M2 contains a classic H-2Kd epitope and is recognized by CD8+ T cells from infected mice (7). Adoptive-transfer studies have demonstrated a role for the CD8+ T-cell response to M2 in reducing the initial, but not long-term, load of latently infected cells (19). In addition, it has been shown that vaccination with M2 reduces the load of latently infected cells in the spleen at early, but not later, times postinfection (22). To evaluate the contribution of M2 to γHV68 pathogenesis, we have characterized acute and latent infection with viral recombinants containing targeted mutations in the M2 gene.
MATERIALS AND METHODS
Viruses and tissue culture.
γHV68 WUMS (ATCC VR1465) is the wild-type (wt) virus from which all of the mutants used in this study were derived. γHV68 was passaged on NIH 3T12 cells as previously described (26). NIH 3T12 cells and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine (complete DMEM). Cells were maintained in a 5% CO2 tissue culture incubator at 37°C. MEFs were obtained from BALB/c mouse embryos as previously described (26).
Generation of virus mutants.
All recombinant viruses were generated via homologous recombination following calcium phosphate or Superfect (Qiagen, Hilden, Germany) cotransfection of NIH 3T12 cells with infectious viral DNA and the appropriate gene-targeting plasmid as previously described (1). Briefly, recombinant viruses were purified from infected NIH 3T12 monolayers overlaid with methylcellulose and subsequently stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Viruses were purified to blue or white plaque homogeneity after selection of well-circumscribed plaques by further rounds of plaque purification. Homogeneity was confirmed via Southern blot analysis. Viral DNA used for cotransfection and Southern blot analysis was generated as previously described (23).
A γHV68 genomic fragment that contained the region from bp 2403 (NgoMI site) to bp 6262 (HindIII site) (WUMS sequence; 23) was subcloned into the Litmus-38 vector (Lit38-M2). This fragment contained the M2 ORF (bp 4031 to 4627) and 1.6 kb of both the 5" and 3" flanking genome sequences to facilitate homologous recombination. A vector-derived SpeI site was removed from the Lit38-M2 construct via digestion with SnaBI and SfiI, subsequent blunting of the SfiI 3" overhang with T4 DNA polymerase, and religation of the construct. This intermediate construct was designated Lit38-M2 SpeKill24. The γHV68M2.LacZ construct, in which bp 4314 to 4632 (the first 313 bp of the ORF, as well as 5 bp 5") were deleted by insertion of a β-galactosidase expression cassette, was generated as follows. Lit38-M2SpeKill24 was digested with SacII (T4 DNA polymerase blunted) and SpeI, followed by ligation of a human cytomegalovirus (HCMV) immediate-early promoter- and enhancer-driven β-galactosidase expression cassette (SpeI-SmaI fragment of pBlu-M1-LacZ, a gift from Paul Olivo and David Leib; see reference 1). The γHV68M2.LacZ recombinant virus was generated by calcium phosphate cotransfection of the M2.LacZ construct with wt γHV68 infectious DNA in NIH 3T12 cells as described in detail elsewhere (1). γHV68M2.LacZ virus was isolated after six rounds of plaque purification.
The M2 marker rescue virus (γHV68M2.MR) was generated by using Superfect (Qiagen) cotransfection of γHV68M2.LacZ viral DNA and the Lit38-M2 construct, followed by selection of white plaques. γHV68M2.MR was isolated after three rounds of plaque purification.
The γHV68 M2.Stop targeting construct was generated by the insertion of a 26-bp linker into the SacII site within the M2 ORF at bp 4314 (oligonucleotides Oligo1 [5"AAG CTT AGG CTA GTT AAC TAG CCA GC] and Oligo2 [5"TGG CTA GTT AAC TAG CCT AAG CTT GC]). The linker contained a diagnostic HindIII site. Oligo1 and Oligo2 were annealed and ligated into SacII-digested Lit38-M2. The addition of the oligomer resulted in a translational stop codon after residue 108 of the genomic M2 ORF. The γHV68M2.Stop construct was sequenced over the entire ORF with the Big Dye DNA sequencing kit (Applied Biosystems, Foster City, Calif.). A silent T-to-C mutation (that did not alter the predicted amino acid sequence but did result in the loss of a PstI site) at bp 4271 in the Lit38-M2.Stop construct was noted. This silent mutation was also present in γHV68M2.MR, as confirmed by the loss of the PstI site at bp 4275 via Southern blot analysis. The γHV68M2.Stop virus was generated via Superfect (Qiagen) cotransfection of the γHV68M2.Stop targeting construct with γHV68M2.LacZ viral DNA and selection of white plaques. γHV68M2.Stop was isolated after three rounds of plaque purification and verified by Southern blot analyses.
Stocks of mutant viruses were generated as previously described (1). Briefly, NIH 3T12 cells were infected with 0.05 PFU per cell and harvested 4 to 5 days postinfection. Samples were homogenized, clarified, and aliquoted for storage at −80°C. Titers of viral stocks were determined by averaging the values obtained from a minimum of three independent plaque assays.
Mice, infections, and organ harvests.
Female C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). C57BL/6J-Ragtm1Mom mice were bred at Washington University, St. Louis, Mo. All mice were housed in a specific-pathogen-free barrier facility at Washington University in accordance with federal and university guidelines. The mice used for experiments were females between 8 and 12 weeks of age. Mice were placed under metofane anesthesia and infected with 106 PFU of virus in 0.5 ml of complete DMEM by intraperitoneal injection or infected intranasally by administration of 4 × 105 PFU of virus in 40 μl of complete DMEM into the nostril. Upon sacrifice, organs were harvested into 1 ml of complete DMEM on ice and frozen at −80°C. Resident PECs were harvested by peritoneal lavage with 10 ml of DMEM supplemented with 1% fetal calf serum.
Plaque assay.
Plaque assays were performed as described previously with modifications (1). Briefly, 3 × 105 NIH 3T12 cells were plated in six-well plates 1 day prior to infection. Organs were thawed and subjected to mechanical disruption with 1.0-mm zirconia/silica beads (Biospec Products, Bartlesville, Okla.) in a Mini-Beadbeater-8 (Biospec Products) for two rounds of 1 min each. This organ disruption procedure resulted in viral titers comparable to those obtained previously through homogenization (1; M. A. Jacoby, L. F. Van Dyk, S. H. Speck, and H. W. Virgin IV, unpublished observations). Serial 10 fold dilutions of organ homogenate were added to NIH 3T12 monolayers in 200 μl and allowed to adsorb for 1 h at 37°C. Samples were overlaid with medium containing Noble agar immediately after infection and on day 3 and stained with a Noble agar-neutral red overlay on day 6. Plaques were scored on day 7. All titers were determined in parallel with a known laboratory standard titer. The limit of detection by this assay is 50 PFU per organ.
Limiting-dilution ex vivo reactivation assay.
The frequency of cells carrying virus capable of reactivating from latency was determined as previously described (26, 29). Briefly, PECs and splenocytes were harvested from mice either 16 to 18 days or 6 weeks postinfection and single-cell suspensions were generated. Cells were resuspended in complete DMEM and plated in a twofold dilution series (starting with 105 splenocytes or 4 × 10 4 PECs per well) onto MEF monolayers in 96-well tissue culture plates. Wells were scored microscopically for a cytopathic effect (CPE) 21 to 28 days postplating. In some cases, samples were replated onto fresh MEF monolayers to confirm the presence of infectious virus, particularly for wells containing large numbers of cells, where a CPE was difficult to discern. Twenty-four wells were plated per dilution, and 12 dilutions were plated per experimental sample. Preformed infectious virus was detected by plating parallel samples of mechanically disrupted cells onto MEF monolayers. Mechanically disrupted cells contained <1% live cells, and thus, the presence of preformed infectious virus could be discerned from virus reactivating from latently infected cells (26, 28, 29). The level of sensitivity of the limiting-dilution assay is 0.2 PFU (26).
Limiting-dilution nested-PCR detection of γHV68 genome-positive cells.
We determined the frequency of cells harboring the γHV68 genome by using a previously described, single-copy-sensitive, nested-PCR assay (28, 29) with modifications. Briefly, splenocytes and PECs harvested from mice either 16 to 18 days or 6 weeks postinfection were frozen in complete DMEM plus 10% dimethyl sulfoxide and stored at −80°C. Cells were thawed, counted, resuspended in isotonic buffer, and then plated in serial threefold dilutions in a background of 104 uninfected NIH 3T12 cells in 96-well plates (MWG Biotech, High Point, N.C.). Plates were covered with PCR foil (Eppendorf Scientific, Westbury, N.Y.). Cells were lysed with proteinase K at 56°C for 6 to 12 h., 10 μl of round 1 PCR mix was added, and the first round of PCR was performed on a Primus thermal cycler (MWG Biotech). Ten microliters of round 2 PCR mix was added to the plate, and the samples underwent a second round of PCR with nested primers. Products were analyzed by ethidium bromide staining of a 2% agarose gel. Twelve PCRs were performed for each cell dilution, and a total of six dilutions (starting at 104 cells) of each sample were analyzed. Control reactions (uninfected cells and 10 copies, 1 copy, and 0.1 copy of plasmid DNA in a background of 104 cells) were included in each experiment as previously described (28, 29). All of the assays reported here demonstrated approximately single-copy sensitivity with no false positives.
Statistical analysis.
All data were analyzed by using GraphPad Prism software (GraphPad Software, San Diego, Calif.). Frequencies of reactivation and viral genome-positive cells were obtained from the cell number at which 63% of the wells scored positive for reactivating virus or presence of the viral genome based on the Poisson distribution; data were subjected to nonlinear regression analysis to obtain the single-cell frequency for each limiting-dilution analysis.
RESULTS
Targeted disruption of the M2 gene.
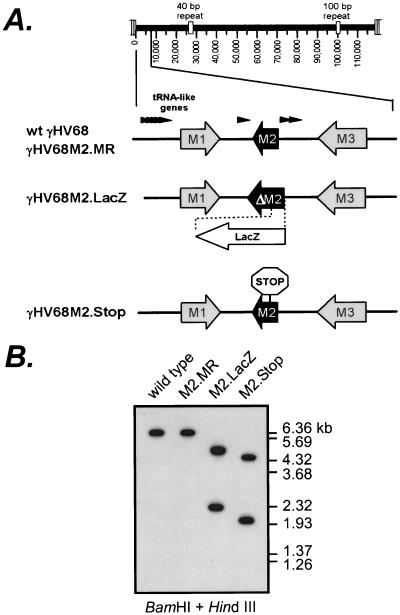
Previous studies have shown that the M2 transcript cloned from the B-lymphoma cell line S11 consists of a noncoding 110-nucleotide (nt) 5" exon and a 1,235-nt 3" exon that contains the M2 ORF and a predicted 656-nt 3" untranslated region (7). The M2 ORF in the cDNA is 7 amino acids shorter than the genomic ORF because of the splice acceptor site used (7), resulting in the predicted utilization of the in-frame translation initiation site at the ATG codon at bp 4606 of the virus genome (23). To study the contribution of the M2 gene to viral pathogenesis, we generated two recombinant viruses containing mutations in M2 (γHV68M2.LacZ and γHV68M2.Stop). γHV68M2.LacZ was generated by the insertion of a β-galactosidase expression cassette containing the β-galactosidase gene under the control of the HCMV immediate-early promoter and enhancer, as described in Materials and Methods. This resulted in the deletion of the first 313 bp of the genomic M2 ORF, as well as 5 bp 5" to the genomic ORF start site (bp 4314 to 4632) (Fig. 1A). This deletion does not disrupt any other known viral coding sequences. After six rounds of plaque purification, the structure of γHV68M2.LacZ was confirmed by Southern blot analysis of viral DNA doubly digested with BamHI and HindIII (Fig. 1B). The hybridization of a 32P-labeled M2 region probe (containing the viral sequence from bp 2815 to bp 5362) to 4.7- and 2.3-kb fragments in γHV68M2.LacZ, but to a 6.15-kb fragment in the wt virus, was consistent with the targeted insertion of the β-galactosidase expression cassette in γHV68M2.LacZ. (Fig 1B). Overexposure of the blot did not reveal any contamination of the γHV68M2.LacZ stock with wt virus (data not shown). Further Southern blot analysis using a HindIII and EcoRI double digest and the M2 region probe also confirmed that γHV68M2.LacZ had the expected structure (data not shown), and a probe containing lacZ gene sequences hybridized to a fragment with the expected size in γHV68M2.LacZ (data not shown).
FIG. 1.
Generation and confirmation of γHV68M2.LacZ, γHV68M2.MR, and γHV68M2.Stop. (A) Genomic structure of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, and γHV68M2.Stop in the region containing the M2 gene. Genomic coordinates of the wt γHV68 ORFs in the M2 region are as follows: M1, bp 2023 to 3282; M2, bp 4031 to 4627; M3, bp 6060 to 7277. All genomic coordinates are based on the γHV68 WUMS sequence (23). γHV68M2.LacZ was generated by excision of bp 4314 to 4632 using SacII and SpeI and subsequent insertion of the LacZ expression cassette (see Materials and Methods). γHV68M2.MR was generated by restoration of the wt sequences to γHV68M2.LacZ. γHV68M2.Stop was constructed by insertion of an oligomer containing a translational stop codon and a diagnostic HindIII site as described in detail in Materials and Methods. (B) Southern blot analysis of the wt γHV68, γHV68M2.LacZ, γHV68M2.MR, and γHV68M2 Stop genomes. Viral DNA was purified from viral stocks and subsequently digested with BamHI and HindIII, electrophoresed, blotted, and hybridized with a probe spanning the M2 region (bp 2815 to 5362). Expected sizes are as follows: wt γHV68 and γHV68M2.MR, 6.1 kb; γHV68M2.LacZ, 4.7 and 2.3 kb; γHV68M2.Stop, 4.2 and 1.9 kb. 32P-labeled molecular size standards (Lambda DNA-BstEII Digest; New England Biolabs, Beverly, Mass.) were included in each Southern blot analysis.
To rule out the possibility that phenotypic alterations in γHV68M2.LacZ were caused by spurious distal mutations rather than the loss of M2, we generated a marker rescue virus (γHV68M2.MR) in which the wt M2 sequences were reconstituted in the γHV68M2.LacZ virus (see Materials and Methods). In parallel, we rescued the γHV68M2.LacZ mutant with a targeting construct containing a translational stop codon after residue 108 of the genomic M2 ORF (γHV68M2.Stop; see Materials and Methods) to rule out the possibility that the β-galactosidase expression cassette itself contributed to phenotypic changes, as has been previously observed by our lab (1; S. Kapadia, S. H. Speck, and H. W. Virgin IV, unpublished observations). The translation stop codon was introduced by inserting a 26-bp oligomer containing a diagnostic HindIII site into the M2 gene via the SacII site at bp 4314 of the viral genome (see Materials and Methods). γHV68M2.MR and γHV68M2.Stop were each isolated after three rounds of plaque purification, and the viral DNA was subjected to Southern blot analysis to confirm the structure of the genomes. After BamHI and HindIII double digestion, the M2 region probe hybridized to 1.9- and 4.2-kb fragments of γHV68M2.Stop (consistent with the addition of the diagnostic HindIII site) and a 6.15-kb fragment of γHV68M2.MR (consistent with restoration of wt M2 sequences) (Fig. 1B). Further Southern blot analysis using HindIII and EcoRI also confirmed that the γHV68M2.Stop and γHV68M2.MR genomes had the expected structures (data not shown). In addition, a probe containing the lacZ gene failed to hybridize to either γHV68M2.Stop or γHV68M2.MR, demonstrating that the β-galactosidase cassette had been removed from these viral genomes and that these viral stocks were not contaminated with γHV68M2.LacZ virus (data not shown).
The predicted molecular mass of the M2 protein is 22 kDa, although immunoblot analysis of whole-cell lysates prepared from Cos-1 cells either transiently or stably transfected with an M2 gene expression vector, using a polyclonal rabbit antiserum generated against purified M2 protein, demonstrated that the M2 protein migrates on sodium dodecyl sulfate-polyacrylamide gels with an apparent molecular mass of ca. 30 kDa (M. A. Jacoby, H. W. Virgin IV, and S. H. Speck, unpublished data). We were unable to detect M2 protein in cell lysates generated from murine fibroblasts productively infected with wt γHV68. The latter finding is consistent with our previous observation that M2 transcripts are not readily detectable in lytically infected murine fibroblasts (24). As a result of our inability to detect M2 protein in productively infected fibroblasts, we were not able to determine whether truncated forms of the M2 protein are expressed from the M2 mutant viruses.
M2 is not required for in vitro replication or virulence in B6.Rag1 deficient mice.
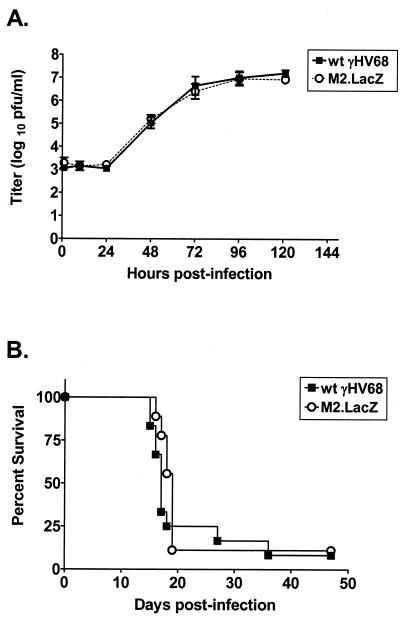
(24) The observation that M2 transcripts are not readily detectable by Northern blot analysis (24), together with the relative ease with which we were able to isolate the γHV68M2.LacZ mutant, led to our expectation that M2 would be dispensable for lytic replication in vitro. We compared γHV68M2.LacZ and the wt virus for growth in vitro in a multistep growth assay with NIH 3T12 cells (Fig. 2A). We did not observe any growth deficit of the γHV68M2.LacZ mutant compared to the wt virus.
FIG. 2.
M2 is not required for in vitro replication or virulence in B6.Rag1 deficient mice. (A) γHV68M2.LacZ replicates comparably to wt γHV68 in vitro. NIH 3T12 monolayers were infected with 0.05 PFU of wt γHV68 or γHV68M2.LacZ per cell and harvested at the indicated times. Samples were freeze-thawed three times, and titers were subsequently determined by plaque assay on NIH 3T12 monolayers. Data were compiled from three independent experiments. (B) γHV68M2.LacZ kills B6.Rag1 deficient mice with the same kinetics as wt γHV68. B6.Rag1 deficient mice were infected intraperitoneally with 10 PFU of wt γHV68 or γHV68M2.LacZ, and mortality over a 47-day period was recorded. The data shown are compilations of two (γHV68M2.LacZ) or three (wt γHV68) independent experiments with three to five mice each.
To begin our assessment of γHV68M2.LacZ viral replication in vivo, we compared the kinetics of lethality in B6.Rag1 deficient mice, which lack mature B and T cells (9), after infection with M2.LacZ or wt γHV68. We infected B6.Rag1 deficient mice intraperitoneally with 10 PFU of wt or γHV68M2.LacZ. Notably, γHV68M2.LacZ killed B6.Rag1 deficient mice with kinetics comparable to those of the wt virus (Fig. 2B), indicating that in the absence of the specific immune system, acute virus replication does not require M2.
M2 is differentially required for acute virus replication in vivo.
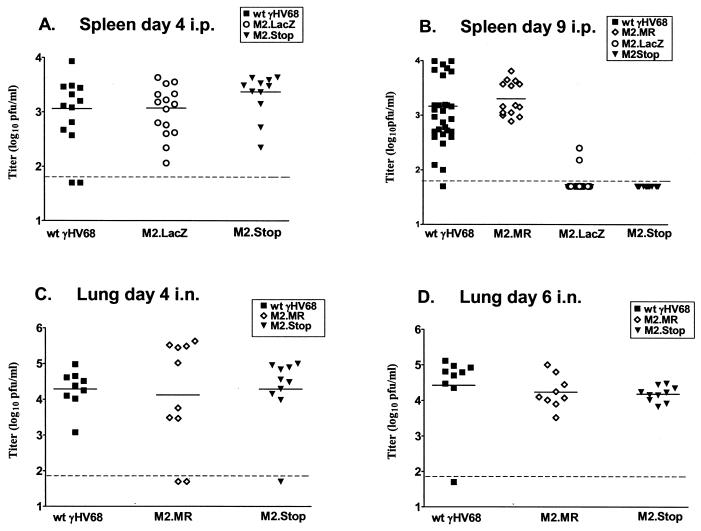
To assess acute virus replication in vivo, we infected C57BL/6J mice either intraperitoneally with 106 PFU or intranasally with 4 × 105 PFU and quantified viral titers by plaque assay at various times postinfection in the spleen (intraperitoneal inoculation, days 4 and 9) and lung (intranasal inoculation, days 4, 6, and 9). After intraperitoneal inoculation, splenic virus titers 4 days postinfection with γHV68M2.LacZ and γHV68M2.Stop were comparable to that of the wt virus (Fig. 3A). In contrast, 9 days after intraperitoneal infection, splenic titers of γHV68M2.LacZ had decreased to the limit of detection of the plaque assay (50 PFU/organ), while the wt and marker rescue (γHV68M2.MR, which contained wt M2 sequences restored to γHV68M2.LacZ) virus titers remained comparable to those measured on day 4 (Fig. 3B). This finding indicated that the M2 locus (the M2 gene and/or flanking sequences) is required for splenic viral replication on day 9 after intraperitoneal infection. To confirm that this phenotype was due to the mutation of the M2 gene rather than an untoward effect resulting from the presence of the β-galactosidase expression cassette (perhaps due to disruption and/or altered transcription of surrounding genes), splenic virus titers were assessed after intraperitoneal infection with γHV68M2.Stop. As seen with γHV68M2.LacZ, splenic virus titers were decreased 9 days, but not 4 days, after intraperitoneal infection with γHV68M2.Stop (Fig. 3A and B). Therefore, the M2 gene contributes to splenic replication 9 days, but not 4 days, after intraperitoneal inoculation. We were unable to determine whether the M2 mutants were impaired in acute replication in other organs, since the virus titers in the lung and liver were at or below the limit of detection of the plaque assay by day 9 after intraperitoneal infection with γHV68M2.LacZ, γHV68M2.Stop, or the wt virus (data not shown).
FIG. 3.
M2 is differentially required for acute replication in vivo. (A) C57BL/6 mice were infected intraperitoneally with 106 PFU of wt γHV68, γHV68M2.LacZ, or γHV68M2.Stop, and spleens were harvested 4 days postinfection. The data shown are compilations of four (wt γHV68 and γHV68M2.LacZ) or three (γHV68M2.Stop) independent experiments with three to five mice each. (B) C57BL/6 mice were infected intraperitoneally (i.p.) with 106 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and spleens were harvested 9 days postinfection. The data shown are compilations of seven (wt γHV68 and γHV68M2.LacZ) or three (γHV68M2.MR and γHV68M2.Stop) independent experiments with three to five mice each. (C) C57BL/6 mice were inoculated intranasally (i.n.) with 4 × 105 PFU of wt γHV68, γHV68M2.MR, or γHV68M2.Stop, and the right lung was harvested 4 days postinfection. The data shown were compiled from two independent experiments with five mice each. (D) C57BL/6 mice were inoculated intranasally with 4 × 105 PFU of wt γHV68, γHV68M2.MR, or γHV68M2.Stop, and the right lung was harvested 6 days postinfection. The data shown were compiled from two independent experiments with five mice each. Virus titers in organs were determined by plaque assay on NIH 3T12 monolayers. Each point represents the virus titer from an individual mouse. The dashed line indicates the level of detection of the plaque assay (50 PFU), and the solid line indicates the mean virus titer of each group.
We investigated whether the requirement for M2 in ongoing acute replication was organ specific. To simplify this analysis, we used γHV68M2.Stop, which contained the more subtle mutation. We observed that after intranasal inoculation, γHV68M2.Stop, γHV68M2.MR, and wt virus titers in the lung were comparable at days 4 and 6 postinfection (Fig. 3C and D). These results are consistent with findings from previous studies showing that neither adoptive transfer of M2-specific CD8+ T cells (19) nor immunization with M2 (22) had an effect on acute lung infection. By day 9, no virus was detectable in the lung after intranasal inoculation with γHV68M2.Stop, γHV68M2.MR, or the wt virus (data not shown). These data indicate that M2 is not necessary for acute replication in the lung after intranasal inoculation. Notably, no virus was detectable by plaque assay in the spleen after intranasal infection with γHV68M2.Stop, γHV68M2.MR, or the wt virus at day 4, 6, or 9 postinfection (data not shown).
M2 is important for both establishment of γHV68 and reactivation from latency in the spleen after intranasal inoculation.
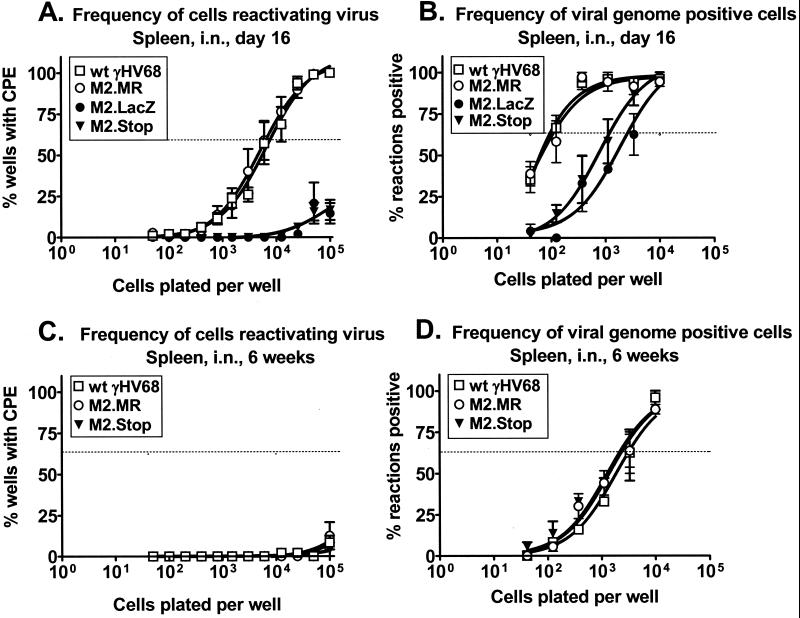
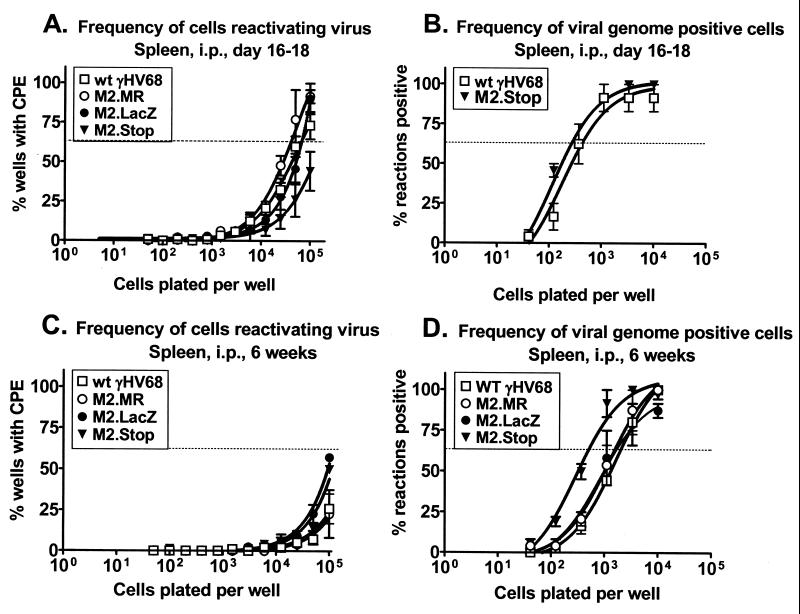
As discussed above, M2 is a candidate latency-associated gene. We investigated the requirement for M2 during latency after intranasal infection. By day 16 postinfection, wt γHV68 establishes a latent infection with no preformed infectious virus present in wt (C57BL/6) mice after either intraperitoneal or intranasal inoculation (Fig. 4,5, and 6; see also references 26, 28, and 29) and both PECs and splenocytes harbor latent virus (28). We investigated the ability of the M2 mutant viruses to reactivate from latency in splenocytes by using an ex vivo, limiting-dilution reactivation assay (see Materials and Methods and references 26, 28, and 29). To ensure that the virus CPE was due to reactivating virus and not preformed infectious (lytic) virus, mechanically disrupted cells were plated in parallel. In this study, no preformed infectious virus of any of the strains tested was detected at either 16 to 18 days or 6 weeks postinfection (Fig. 4,5, and 6).
FIG. 4.
M2 is critical for both establishment of γHV68 and its reactivation from splenic latency at early times after intranasal (i.n.) inoculation. (A and B) C57BL/6 mice were inoculated intranasally with 4 × 105 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and spleens were harvested 16 days postinfection. (A) The frequency of splenocytes reactivating virus ex vivo 16 days postinfection was analyzed in six (γHV68M2.MR), four (wt γHV68 and γHV68M2.Stop), and two (γHV68M2.LacZ) independent experiments with four or five mice each. (B) In parallel, the frequency of viral genome-positive splenocytes was analyzed in four (wt γHV68 and γHV68M2.Stop), three (γHV68M2.MR), or two (γHV68M2.LacZ) of the above samples by limiting-dilution, nested PCR. (C and D) C57BL/6 mice were inoculated intranasally with 4 × 105 PFU of wt γHV68, γHV68M2.MR, or γHV68M2.Stop, and spleens were harvested 6 weeks postinfection. (C) The frequency of splenocytes reactivating virus ex vivo 6 weeks postinfection was analyzed in two independent experiments with four or five mice each. (D) The frequency of viral genome-positive splenocytes was analyzed by limiting-dilution, nested PCR for the above samples and one additional experiment, for a total of three independent experiments with four or five mice each. For the ex vivo reactivation assay (A and C), intact (live) cells were serially diluted onto a MEF indicator monolayer as described in Materials and Methods. In parallel, mechanically disrupted cells were plated to detect the presence of preformed infectious virus. In this study, no preformed infectious virus of any of the strains assayed was detected and these data are not presented in the figures for clarity. For each cell dilution, 24 wells were scored for CPE; data are shown as the mean percentage of wells positive for a CPE ± the standard error of the mean. In the limiting-dilution nested-PCR assay (B and D), 12 PCRs were performed per cell dilution for each experiment with the inclusion of PCR specificity and sensitivity controls as described in Materials and Methods. Data are expressed as the mean percentage of positive PCRs ± the standard error of the mean. For both assays, curve fit lines were derived from nonlinear regression analysis and the dashed line represents 62.5%, which was used to calculate the frequency of reactivation or the frequency of viral genome-positive cells by the Poisson distribution.
FIG. 5.
M2 is not required for establishment of γHV68 or its reactivation from splenic latency after intraperitoneal (i.p.) infection. (A and B) C57BL/6 mice were inoculated intraperitoneally with 106 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and spleens were harvested 16 to 18 days postinfection. (A) The frequency of splenocytes reactivating virus ex vivo at 16 to 18 days postinfection was analyzed in five (wt γHV68), four (γHV68M2.LacZ), three (γHV68M2.Stop), or two (γHV68M2.MR) independent experiments with four or five mice each. (B) In parallel, the frequency of viral genome-positive splenocytes was analyzed for wt γHV68 and γHV68M2.Stop in two of the above samples by limiting-dilution, nested PCR. (C and D) C57BL/6 mice were inoculated intraperitoneally with 106 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and spleens were harvested 6 weeks postinfection. (C) The frequency of splenocytes reactivating virus ex vivo at 6 weeks postinfection was analyzed in four (wt γHV68 and γHV68M2.LacZ), three (γHV68M2.Stop), or two (γHV68M2.MR) independent experiments with four or five mice each. (D) In parallel, the frequency of viral genome-positive splenocytes was analyzed for three (wt γHV68 and γHV68M2.Stop) or two (γHV68M2.MR and γHV68M2.LacZ) of the above samples by limiting-dilution, nested PCR. The ex vivo reactivation assay and the limiting-dilution nested-PCR assay are described in the legend to Fig. 4, as well as in Materials and Methods. Data are shown as the mean percentage of wells positive for a CPE ± the standard error of the mean (ex vivo reactivation assay) or as the mean percentage of positive PCRs ± the standard error of the mean. For both assays, curve fit lines were derived from nonlinear regression analysis and the dashed line represents 62.5%, which was used to calculate the frequency of reactivation or frequency of viral genome-positive cells by the Poisson distribution.
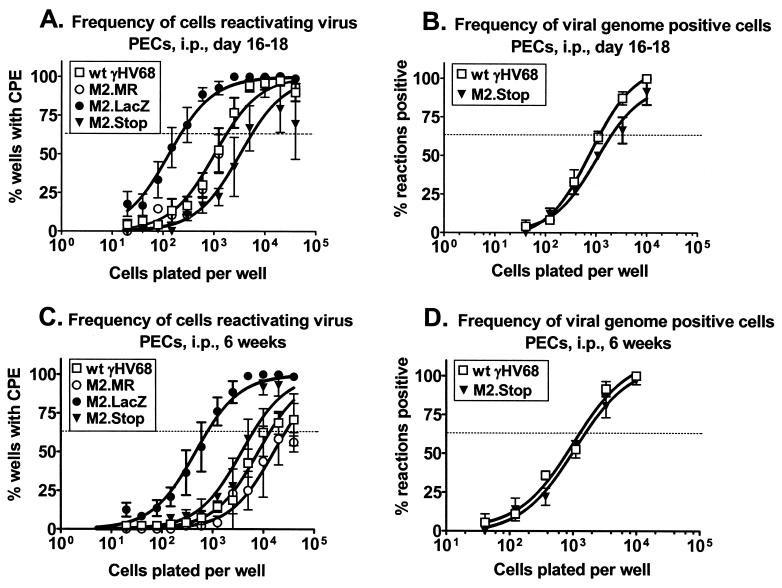
FIG. 6.
M2 is not required for establishment or reactivation from γHV68 latency in PECs after intraperitoneal (i.p.) infection. (A and B) C57BL/6 mice were inoculated intraperitoneally with 106 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and PECs were harvested 16 to 18 days postinfection. (A) The frequency of PECs reactivating virus ex vivo at 16 to 18 days postinfection was analyzed in five (wt γHV68), four (γHV68M2.LacZ), three (γHV68M2.Stop), or two (γHV68M2.MR) independent experiments with four or five mice each. (B) In parallel, the frequency of viral genome-positive PECs was analyzed for wt γHV68 and γHV68M2.Stop in two of the above samples by limiting-dilution, nested PCR. (C and D) C57BL/6 mice were inoculated intraperitoneally with 106 PFU of wt γHV68, γHV68M2.MR, γHV68M2.LacZ, or γHV68M2.Stop, and PECs were harvested 6 weeks postinfection. (C) The frequency of PECs reactivating virus ex vivo at 6 weeks postinfection was analyzed in four (wt γHV68 and γHV68M2.LacZ), three (γHV68M2.Stop), or two (γHV68M2.MR) independent experiments with four or five mice each. (D) In parallel, the frequency of viral genome-positive PECs was analyzed for wt γHV68 and γHV68M2.Stop in three of the above samples by limiting-dilution, nested PCR. The ex vivo reactivation assay and the limiting-dilution nested-PCR assay are described in the legend to Fig. 4, as well as in Materials and Methods. The data shown are the mean percentages of wells positive for a CPE ± the standard error of the mean (ex vivo reactivation assay) or the mean percentages of positive PCRs ± the standard error of the mean. For both assays, curve fit lines were derived from nonlinear regression analysis and the dashed line represents 62.5%, which was used to calculate the frequency of reactivation or the frequency of viral genome-positive cells by the Poisson distribution.
The capacity of splenocytes to reactivate virus from latency ex vivo after intranasal inoculation 16 days postinfection was severely compromised by the loss of M2 (Fig. 4A). The frequency of splenocyte reactivation from latency was approximately 1 in 9,000 in wt virus- and γHV68M2.MR-infected animals. In contrast, the frequency of splenocyte reactivation of virus from γHV68M2.LacZ-infected mice was too low to be accurately determined but was ca. 100-fold less than that of the wt (Fig. 4A). These data indicate that the M2 locus (the M2 gene and flanking sequences) is required for virus reactivation from latently infected splenocytes. To confirm that this phenotype was due to mutation of the M2 gene rather than the presence of the β-galactosidase expression cassette, the frequency of splenocyte reactivation from latency was assessed after infection with γHV68M2.Stop. As observed with γHV68M2.LacZ, the frequency of splenocyte reactivation from latency after infection with γHV68M2.Stop was too low to be accurately determined but was at least 100-fold less than that of the wt (Fig. 4A). Thus, the M2 gene is important for splenic latency, as assessed by reactivation 16 days after intranasal infection.
This decreased frequency of reactivation from latency after the loss of M2 could be due to a decreased efficiency of reactivation, a decrease in the frequency of latently infected cells (establishment of latency), or both. To determine the contribution of M2 to the establishment of latency in the spleen, we determined the frequency of viral genome-positive cells present in the spleen by using a previously described limiting-dilution PCR assay capable of detecting a single copy of the viral genome in a background of 104 uninfected cells (see Materials and Methods and references 28 and 29). Loss of M2 resulted in a defect in the ability of γHV68 to establish latency in the spleen on day 16 postinfection (Fig. 4B). The frequency of splenocytes harboring the viral genome after infection with γHV68M2.LacZ was approximately 1 in 2,500, compared with approximately 1 in 100 in wt virus- and γHV68M2.MR-infected animals (Fig. 4B). To confirm that the defect in establishment of latency was due to loss of the M2 gene rather than the presence of the β-galactosidase expression cassette, we assessed the frequency of viral genome-positive splenocytes after infection with γHV68M2.Stop. As observed with γHV68M2.LacZ, the frequency of viral genome-positive splenocytes was decreased compared to that obtained with the wt virus (approximately 1 in 1,000) (Fig. 4B). Thus, loss of the M2 gene results in decreased establishment of splenic latency 16 days after intranasal infection.
By comparing the frequency of viral genome-positive cells (1 in 100) to the frequency of splenocytes that reactivate virus (1 in 9,000) after intranasal inoculation, we determined that approximately 1 in 100 viral genome-positive splenocytes from wt virus- and γHV68M2.MR-infected animals reactivates ex vivo 16 days postinfection. If the efficiency of reactivation were the same in mice infected with γHV68M2.LacZ and γHV68M2.Stop, we would expect approximately 1 in 105 splenocytes to reactivate the virus (i.e., 1% of the viral genome-positive cells); however, the frequency of cells reactivating is much lower than 1 in 105 (Fig. 4A). Therefore, these data show that mutation of M2 results in a defect in both establishment and reactivation from latency after intranasal inoculation at early times postinfection.
We investigated the requirement for M2 in latency at later times postinfection. To simplify this analysis, we examined latency after infection with γHV68M2.Stop, the M2 mutant virus that contained the more subtle mutation. Six weeks postinfection, the frequency of splenocytes reactivating virus from wt virus-, γHV68M2.MR-, and γHV68M2.Stop-infected animals was extremely low (Fig. 4C). Therefore, it was not possible to compare the reactivation frequency of γHV68M2.Stop to that of the wt virus at 6 weeks postinfection. Notably, the frequency of viral genome-positive splenocytes in wt virus- and γHV68M2.MR-infected animals decreased from approximately 1 in 100 at 16 days postinfection to approximately 1 in 3,000 at 6 weeks postinfection (Fig. 4B and D). However, the frequency of viral genome-positive cells in γHV68M2.Stop-infected mice remained relatively constant from 16 days to 6 weeks postinfection (1 in 1,000 versus 1 in 3,000, respectively). Thus, by 6 weeks postinfection, the frequency of virus-positive splenocytes from γHV68M2.Stop infected mice was comparable to that of wt virus- and γHV68M2.MR-infected mice (Fig. 4D). These data indicate that M2 is not necessary for maintenance of latency, as measured by the frequency of viral genome-positive cells 6 weeks postinfection and that the decrease in viral genome-positive cells from 16 days to 6 weeks postinfection is less in γHV68M2.Stop infected animals than in wt virus-infected animals. Notably, it has been shown that vaccination with M2 causes a significant decrease in latency at 14 days postinfection but does not reduce the long-term level of latency in the spleen (22).
M2 is not required for establishment or reactivation of γHV68 from latency after inoculation via the intraperitoneal route.
Previously, we have demonstrated that intraperitoneal, but not intranasal, inoculation of B-cell-deficient mice leads to efficient establishment of latency in the spleen (28). We considered the possibility that the role of M2 in latency might be dependent on the route of infection. Thus, we examined latency at both early (16 to 18 days) and later (6 weeks) times postinfection in both splenocytes and PECs after intraperitoneal inoculation.
At early times in latency (days 16 to 18), we determined that reactivation frequencies from splenocytes after infection with γHV68M2.LacZ, γHV68M2.MR, and the wt virus were comparable (between approximately 1 in 40,000 and 1 in 60,000) (Fig. 5A). The reaction frequency after infection with γHV68M2.Stop appeared to be mildly decreased (approximately 1 in 2 × 105), although this is a minor difference (Fig. 5A). Notably, the frequency of splenocyte reactivation from latency on ex vivo day 16 after intranasal infection with the wt virus and γHV68M2.MR was higher than after intraperitoneal infection (compare Fig. 4A with Fig. 5A). By day 42 postinfection, there was a decrease in the frequency of splenocyte reactivation from latency compared to that on day 16 postinfection; however, consistent with results from day 16, no defect in the ability of the M2 mutant viruses to reactivate from splenocytes compared to those of the wt virus and γHV68M2.MR was observed (all reactivation frequencies were less than 1 in 105; Fig. 5B). In addition, we determined that the frequency of viral genome-positive splenocytes detected in γHV68M2.Stop-infected animals on days 16 to 18 postinfection was comparable to that in wt virus-infected animals (1 in 300 and 1 in 450, respectively; Fig. 5C). By day 42, the frequency of viral genome-positive splenocytes was actually slightly higher in γHV68M2.Stop-infected animals (1 in 500) than in wt virus-infected animals (1 in 1,500) (Fig. 5D), although this difference was minimal and its biological significance remains to be determined. Taken together, these data indicate that M2 is dispensable for the establishment of latency in the spleen after intraperitoneal inoculation. In addition, these results indicate that the requirements for the establishment of latency are affected by the route of infection (compare Fig. 4 and 5).
We investigated whether M2 contributed to latency in PECs after intraperitoneal infection. The frequency of PEC reactivation from latency was approximately 1 in 2,000 in wt virus- and γHV68M2.MR-infected mice 16 to 18 days postinfection (Fig. 6A). In contrast, the frequency of PEC reactivation of virus from γHV68M2.LacZ-infected mice was enhanced (approximately 1 in 250) (Fig. 6A). To determine if this phenotype was due to the presence of the β-galactosidase expression cassette, we measured the frequency of PEC reactivation from latency after infection with γHV68M2.Stop. Notably, the frequency of PEC reactivation from γHV68M2.Stop-infected mice (1 in 6,000) was not enhanced but rather appeared to be slightly less than that of wt virus- and γHV68M2.MR-infected mice at days 16 to 18 postinfection (1 in 2,000) (Fig. 6A). Therefore, the enhanced reactivation from latency observed after infection with γHV68M2.LacZ may be due to altered expression of genes closely linked to the M2 gene by the presence of the β-galactosidase expression cassette (see Discussion). To determine whether establishment of latency was altered in PECs due to the loss of M2, we measured the frequency of viral genome-positive PECs. The frequency of genome-positive PECs detected in γHV68M2.Stop-infected animals on days 16 to 18 postinfection was comparable to that in wt virus-infected animals (1 in 2,000 and 1 in 1,200, respectively; Fig. 6B). Thus, M2 is not required for the establishment of latency in PECs at early times postinfection.
We also measured latency in PECs at 6 weeks postinfection. As observed at earlier times, PECs from γHV68M2.LacZ-infected mice displayed enhanced reactivation from latency (1 in 800) compared to wt virus-infected mice (1 in 12,000) and γHV68M2.MR-infected mice (1 in 25,000) at 6 weeks postinfection (Fig. 6C). However, as observed at 16 days postinfection, the frequency of PECs reactivating from latency after infection with γHV68M2.Stop was 1 in 6,000, approximately that seen after infection with the wt (Fig. 6C). In addition, we determined that the frequency of viral genome-positive PECs detected in γHV68M2.Stop-infected animals 6 weeks postinfection was comparable to that in wt-infected animals (approximately 1 in 1,000; Fig. 6D). Taken together, these data indicate that M2 is not crucial for establishment or reactivation from latency in PECs after intraperitoneal infection. In addition, we conclude that the enhanced reactivation from PEC latency observed after infection with γHV68M2.LacZ is likely not a result of specific disruption of the M2 gene but rather an effect resulting from the presence of the β-galactosidase expression cassette (see Discussion).
DISCUSSION
Assignment of phenotypes to the M2 gene.
Based on positional homology to latency- and transformation-associated genes in other characterized gammaherpesviruses, as well as its expression in latently infected tissues (7, 19, 24), M2 was identified as a candidate γHV68 latency-associated gene (24). In this study, we demonstrated, through targeted mutagenesis of the γHV68 genome, that the M2 gene is a critical determinant of γHV68 pathogenesis during both the acute and latent phases of infection. We constructed three recombinant viruses to evaluate the role of M2 in γHV68 pathogenesis. (i) γHV68M2.LacZ had approximately half of the M2 ORF deleted via the insertion of a β-galactosidase expression cassette. (ii) A marker rescue virus (γHV68M2.MR) was generated from γHV68M2.LacZ in which M2 wt sequences were restored to γHV68M2.LacZ. (iii) γHV68M2.Stop (generated in parallel with the generation of γHV68M2.MR), which contained a translational stop codon after residue 108 of the genomic M2 ORF, was generated to further refine the mutation in M2 by eliminating the bulky β-galactosidase expression cassette and the strong transcriptional activating sequences present in the HCMV immediate-early promoter and enhancer. To ensure that the loss of the M2 gene product was responsible for the phenotypes observed after infection with γHV68M2.LacZ, all experiments in which γHV68M2.LacZ exhibited phenotypic differences from the wt virus were repeated with γHV68M2.Stop (to evaluate the possibility that the phenotypic changes were due to the presence of the β-galactosidase expression cassette), as well as γHV68M2.MR (to rule out the effects of spurious distal mutations). We mapped phenotypic changes to the M2 gene only if differences from the wt virus observed upon infection with γHV68M2.LacZ were recapitulated upon infection with γHV68M2.Stop and were rescued to the wt with γHV68M2.MR. By this criterion, we found that M2 is differentially required for acute virus replication in vivo and is a critical determinant of establishment and efficient reactivation from splenic latency at early times (16 days) after intranasal, but not intraperitoneal, inoculation (see below).
Distinguishing between M1 and M2 mutant phenotypes.
Previously, we have observed that the β-galactosidase expression cassette contributed to phenotypic changes noted after infection with γHV68 viral mutants (1; S. Kapadia, S. H. Speck, and H. W. Virgin IV, unpublished observations). In this study, we observed that the enhanced reactivation from latency in PECs after intraperitoneal infection with γHV68M2.LacZ was not recapitulated upon infection with γHV68M2.Stop, although the wt reactivation frequency was restored after infection with γHV68M2.MR (Fig. 6; Table 1). It is formally possible that a truncated form of the M2 protein is expressed from the γHV68M2.Stop virus, leading to wt levels of reactivation from latency in PECs. However, mutation of the adjacent M1 ORF leads to enhanced efficiency of reactivation in PECs (1) (Table 1), whether generated via targeted insertion of a β-galactosidase expression cassette into the M1 ORF (γHV68M1.LacZ) or deletion of the identical genomic sequences without a β-galactosidase expression cassette (γHV68M1.Δ511). Thus, a phenotype exhibited by γHV68M2.LacZ that was not recapitulated with γHV68M2.Stop was observed after infection with both γHV68M1.LacZ and γHV68M1.Δ511, raising the possibility that the enhanced reactivation from PECs actually maps to the adjacent M1 ORF. Furthermore, decreased acute-phase virus replication in the spleen was observed after infection with γHV68M1.LacZ but was not recapitulated after infection with γHV68M1.Δ511 (1) (Table 1). As discussed above, a decrease in acute-phase splenic replication was also noted after both γHV68M2.LacZ and γHV68M2.Stop infections (Fig. 3 and Table 1). Thus, the acute-phase replication defect exhibited by γHV68M1.LacZ which was not recapitulated with the γHV68 M1.Δ511 mutant may map to the M2 gene. In summary, the γHV68M1.LacZ and γHV68M2.LacZ mutants exhibit similar acute-phase replication and latency phenotypes (Table 1). However, distinct phenotypes mapping to either the M1 or M2 gene emerge upon the analysis of more subtle mutants lacking the β-galactosidase expression cassette. Taken together, these data argue strongly that use of the β-galactosidase expression cassette in this locus leads to complex, overlapping phenotypes of mutants (perhaps due to disruption and/or altered transcription of adjacent genes) and that these phenotypes can be teased out by using alternative mutagenesis strategies. Importantly, none of the above phenotypes were observed upon infection with a recombinant virus containing a mutation that ablates expression of the M3 protein, encoded by the M3 gene located immediately upstream of the M2 gene (V. van Berkel, S. H. Speck, and H. W. Virgin IV, unpublished observations).
TABLE 1.
Summary of M1 and M2 mutant phenotypesa
| Virus | Mean virus titer (PFU) in spleen, day 9 | Latencyb in spleen
|
Latency in PECs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 16 (i.n.)
|
Day 16 (i.p.)
|
Day 42 (i.n.)
|
Day 42 (i.p.)
|
Day 42 (i.p.)
|
|||||||||
| RF | GF | RF | GF | RF | GF | RF | GF | RF | GF | ||||
| wt γHV68 | 1,466 | 1/9,000 | 1/100 | 1/60,000 | 1/450 | <<1/105 | 1/3,000 | <1/105 | 1/1,500 | 1/10,000c-1/12,000 | 1/600c-1/1,000 | ||
| M2.LacZ | <50 | <1/105 | 1/2,500 | 1/60,000 | ND | ND | ND | <1/105 | 1/1,500 | 1/800 | ND | ||
| M2.Stop | <50 | <1/105 | 1/1,000 | ≈1/105 | 1/300 | <<1/105 | 1/3,000 | <1/105 | 1/500 | 1/6,000 | 1/1,000 | ||
| M1.LacZc | <50c | ND | ND | ND | ND | ND | ND | <1/105 | ND | 1/700c | 1/300c | ||
| M1.Δ511c | 2,070c | ND | ND | ND | ND | ND | ND | <1/105 | ND | 1/1,000c | ND | ||
Abbreviations: i.n., intranasal inoculation; i.p., intraperitoneal inoculation; RF, frequency of cells reactivating latent γHV68; GF, frequency of cells harboring the γHV68 genome; ND, not determined.
Frequency of cells harboring and reactivating virus.
Data are from reference 1.
M2 is differentially required for acute viral replication in vivo.
Work of several groups has shown that M2 is expressed both in vivo in latently infected tissues (7, 19, 24) and in the latently infected B-lymphoma cell line S11 (7). M2 transcripts are not readily detectable via Northern blot analysis in lytically infected fibroblasts (7, 24). Therefore, it was somewhat surprising that the effects of M2 mutagenesis were not restricted to latency. While some studies have shown M2 expression in latently infected splenocytes (7, 24) and PECs (24) in the absence of detectable lytic-cycle transcripts, another study (19) demonstrated M2 expression in the spleen and lung within the first month postinfection in the presence of the ORF 50 transcript (the γHV68 homolog of the EBV BRLF1 gene [23]), which is an immediate-early gene (8, 30) that has been shown to be sufficient to drive reactivation from latency in S11 cells (30). Although it is not clear that M2 and ORF 50 transcripts were present in the same cells, the latter study raises the possibility that M2 expression is present during lytic viral replication (de novo or reactivating from latency), as well during latency. Furthermore, we found that M2 is differentially required for acute replication in vivo. Splenic virus titers were unaffected at day 4 but were significantly decreased at day 9 postinfection compared to wt virus titers as a result of the loss of M2. This finding may indicate that in the absence of M2, γHV68 is cleared faster from the spleen. Alternatively, a cellular reservoir that contributes to the virus titer at day 9 postinfection in the spleen may not be infected, perhaps due to a trafficking defect, or may not support viral replication in the absence of M2. Another possibility is that the virus produced by cells reactivating from latency contributes to the virus titer measured at day 9 and that a defect in the ability of the M2 mutant virus to reactivate results in a decreased virus titer. Notably, mutagenesis of M2 does not result in a generalized in vivo acute replication defect, as the M2 mutant γHV68M2.LacZ kills B6.Rag1 deficient mice with kinetics identical to those of the wt virus and lung virus titers after intranasal infection with γHV68M2.Stop and the wt virus were comparable.
M2 is important for both establishment of γHV68 and reactivation of γHV68 from splenic latency after intranasal, but not intraperitoneal, inoculation.
M2 plays a role in establishment and reactivation from latency in the spleen at early times after intranasal inoculation. In contrast, M2 is dispensable for splenic latency after intraperitoneal infection. These data imply that the requirements for latency, at least at early times postinfection, are route dependent. Notably, there have been no published reports investigating the natural route of γHV68 infection. Thus, the biological significance of the observed differences between intranasal and intraperitoneal inoculations are unclear. The route dependence of the M2 phenotype suggests that the M2 protein plays a role in trafficking of γHV68 to the spleen after intranasal inoculation. Since splenocytes are capable of reactivating virus ex vivo in the absence of M2 upon intraperitoneal inoculation, M2 is not required for the γHV68 reactivation program per se. However, interpretation of these results is complicated by the fact that, in the spleen, multiple cell types are latently infected (6). Therefore, further analyses of the impact of M2 mutation on establishment and reactivation from specific cell types is required. After intranasal infection, M2 may be required to seed a splenic latency reservoir capable of reactivation; intraperitoneal infection may bypass this requirement, either by directly seeding this reservoir independently of M2 gene expression or by seeding an alternative reservoir that does not require M2 expression for reactivation.
Previously, it has been demonstrated that intraperitoneal, but not intranasal, inoculation of B-cell-deficient mice with wt γHV68 led to efficient establishment and reactivation from latency in the spleen. In addition, after intraperitoneal infection of B-cell-deficient mice with wt γHV68, no virus replication in the spleen was detected (21, 26, 28). The similarity of the splenic phenotype of wt γHV68 infection in B-cell-deficient mice to that of wt mice infected with the M2 mutant viruses raises the intriguing possibility that M2 is necessary for some aspect of infection and/or latency in B cells. Further studies are necessary to elucidate the mechanism of the latency defect that occurs after the loss of M2.
While the frequency of viral genome-positive cells after intranasal infection with γHV68M2.Stop was lower than after infection with the wt at 16 days postinfection, the frequencies were comparable by 6 weeks postinfection. Although the loss of M2 impacts establishment of latency after intranasal infection, these data indicate that expression of M2 is not necessary for maintenance of the γHV68 genome 6 weeks postinfection. Notably, the frequency of viral genome-positive cells from γHV68M2.Stop-infected animals remained relatively constant from 16 days to 6 weeks postinfection, while the frequency of wt γHV68-infected cells decreased more than 10-fold. These data raise the possibility that clearance of viral genome-positive cells by the immune system differs after mutation of M2, a phenomenon which could occur for a number of reasons. For example, mutation of M2 could affect the γHV68 latency program, leading to a change in or lack of expression of viral gene products (including M2 itself) presented to the immune system. Previous studies have shown that M2 contains a classic H-2Kd epitope recognized by CD8+ T cells from infected mice (7) and have demonstrated a role of the CD8+ T-cell response to M2 in reducing the initial, but the not long-term, load of latently infected cells (19). Recently, it has been shown that immunization with M2 significantly reduces the load of latently infected cells in the spleen at early, but not later, times postinfection (22). However, these studies used BALB/c mice (H-2d), while our studies used C57BL/6 mice (H-2b), and therefore it is unclear whether an M2 epitope is presented to the immune system in this context.
Alternatively, the observed difference between wt γHV68 and M2 mutant virus in the decrease in viral genome-positive splenocytes over time may reflect differences in seeding of cellular latency reservoirs. Since the frequency of splenocyte reactivation of virus from latency is extremely low by 6 weeks postinfection, even from wt-infected animals at this time, it is not possible to compare reactivation of the M2 mutant virus and that of the wt virus at late time points. It is interesting to consider the kinetics of M2 expression from the studies discussed above in light of this data. It has been shown that M2 expression in latently infected splenocytes from B-cell-deficient mice on day 42 (24) and days 14 and 28 postinfection (7). In another study, M2 expression was detected in the spleen and lung within the first month postinfection but undetectable by 10 months (19). Thus, it is possible that M2 expression is more important during the initial establishment of latency. Further studies are required to define the role of M2 in long-term latency.
Conclusions.
In summary, we conclude that (i) use of an HCMV immediate-early promoter-driven β-galactosidase expression cassette to generate γHV68 mutants in the M1-M2 locus leads to complex, overlapping phenotypes; (ii) M2 is differentially required for acute-phase virus replication in vivo; (iii) M2 is important for both establishment and reactivation at early times from splenic latency after intranasal, but not intraperitoneal, inoculation; and (iv) M2 is not required for the maintenance of γHV68 genome-positive cells as determined 6 weeks after intranasal inoculation.
Acknowledgments
S. H. Speck was supported by NIH grants CA43143, CA52004, CA58524, and CA74730. H. W. Virgin IV was supported by NIH grants AI39616, CA74730, and HL60090.
We acknowledge helpful discussions with members of the Speck and Virgin labs, as well as discussions during joint lab meetings with members of the labs of David Leib and Lynda Morrison.
REFERENCES
- 1.Clambey, E. T., H. W. Virgin IV, and S. H. Speck. 2000. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J. Virol. 74:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty, P. C., R. A. Tripp, A. M. Hamilton-Easton, R. D. Cardin, D. L. Woodland, and M. A. Blackman. 1997. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr. Opin. Immunol. 9:477-483. [DOI] [PubMed] [Google Scholar]
- 3.Efstathiou, S., Y. M. Ho, S. Hall, C. J. Styles, S. D. Scott, and U. A. Gompels. 1990. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J. Gen. Virol. 71:1365-1372. [DOI] [PubMed] [Google Scholar]
- 4.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 5.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 7.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, S., I. V. Pavlova, H. W. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mombaerts, P., J. Iacomini, R. S. Johnson, K. Herrup, S. Tonegawa, and V. E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869-877. [DOI] [PubMed] [Google Scholar]
- 10.Nash, A. A., and N. P. Sunil-Chandra. 1994. Interactions of the murine gammaherpesvirus with the immune system. Curr. Opin. Immunol. 6:560-563. [DOI] [PubMed] [Google Scholar]
- 11.Nash, A. A., E. J. Usherwood, and J. P. Stewart. 2001. Immunological features of murine gammaherpesvirus infection. Semin. Virol. 7:125-130. [Google Scholar]
- 12.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 13.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson, P. G., and P. C. Doherty. 1999. Non-antigen-specific B-cell activation following murine gammaherpesvirus infection is CD4 independent in vitro but CD4 dependent in vivo. J. Virol. 73:1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 17.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 18.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 19.Usherwood, E. J., D. J. Roy, K. Ward, S. L. Surman, B. M. Dutia, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2000. Control of gammaherpesvirus latency by latent antigen-specific CD8+ T cells. J. Exp. Med. 192:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usherwood, E. J., J. P. Stewart, K. Robertson, D. J. Allen, and A. A. Nash. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 77:2819-2825. [DOI] [PubMed] [Google Scholar]
- 22.Usherwood, E. J., K. Ward, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2001. Latent antigen vaccination in a model gammaherpesvirus infection. J. Virol. 75:8283-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virgin, H. W., IV, R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 26.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin IV. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weck, K. E., A. J. Dal Canto, J. D. Gould, A. K. O'Guin, K. A. Roth, J. E. Saffitz, S. H. Speck, and H. W. Virgin. 1997. Murine gamma-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat. Med. 3:1346-1353. [DOI] [PubMed] [Google Scholar]
- 28.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]