Abstract
To date, the insect nodavirus flock house virus (FHV) is the only virus of a higher eukaryote that has been shown to undergo a full replicative cycle and produce infectious progeny in the yeast Saccharomyces cerevisiae. The genome of FHV is composed of two positive-sense RNA segments: RNA1, encoding the RNA replicase, and RNA2, encoding the capsid protein precursor. When yeast cells expressing FHV RNA replicase were transfected with a chimeric RNA composed of a selectable gene flanked by the termini of RNA2, the chimeric RNA was replicated and transmitted to daughter cells indefinitely. In the work reported here, we developed a system in which a selectable chimeric RNA replicon was transcribed from an inducible RNA polymerase II (polII) promoter in vivo in yeast. To render marker gene expression absolutely dependent on RNA replication, the primary polII transcript was made negative in sense and contained an intron that blocked the translation of cryptic transcripts from the opposite DNA strand. The RNA products of DNA-templated transcription, processing, and RNA replication were characterized by Northern blot hybridization and primer extension analysis. Marker gene expression and colony growth under selective conditions depended strictly on FHV RNA replication, with background colonies arising at a frequency of fewer than 1 in 108 plated cells. The utility of the system was demonstrated by introducing a second chimeric replicon and showing that at least two different selectable markers could be simultaneously expressed by means of RNA replication. This is the first example of FHV RNA1-dependent selectable marker expression initiated in vivo and will greatly facilitate the identification and characterization of the requirements and inhibitors of RNA replication.
Nodaviruses are positive-strand RNA animal viruses with bipartite genomes that are among the smallest RNA virus genomes known (5). This genetic simplicity has facilitated research and the development of the key tools required for the dissection of these model positive-sense RNA viruses (5). Flock house virus (FHV) is the best-studied member of the family Nodaviridae. Stocks of FHV, which was originally isolated from the grass grub Costelytra zealandica (11), are typically prepared in Drosophila cells (14), although the virus is able to replicate in many different cells (4, 26, 28). The larger segment of the FHV genome (RNA1) is an autonomous replicon that encodes all of the viral activities required for the viral RNA-dependent RNA replicase (RdRp) and also serves as a replication template (18). The smaller genome segment (RNA2) relies on RNA1 for its replication and encodes only the viral capsid protein precursor (19), which is required for virion formation and cell-to-cell spread. FHV RNA2 is one of a very few positive-sense viral RNAs discovered to have the ability to initiate its replication from the antigenomic RNA (3).
The yeast Saccharomyces cerevisiae is perhaps the premier eukaryotic laboratory organism, with a long history of molecular and genetic analyses (22). We showed previously that this yeast, when transfected with FHV RNA1 and RNA2, supports FHV replication and produces infectious virus particles at levels similar to those obtained from infected Drosophila cells (26). As the foundation for genetic analyses, yeast cells constitutively expressing RNA1 from DNA plasmids supported the replication of transfected chimeric RNA2 replicons that carried selectable markers. At cell division, these RNA replicons were transmitted to daughter cells, giving rise to selectable colonies. In addition, we have developed a system for inducible expression of positive-sense FHV RNA1 (25), which was amplified to levels approaching those of rRNA. This system was used to characterize FHV RNA1 replication and to express green fluorescent protein.
To facilitate the analysis and manipulation of FHV RNA2 replication, we synthesized selectable chimeric RNA2-based replicons in vivo by inducing primary transcription from DNA plasmids. Colonies were obtained following the induction of either positive- or negative-sense primary transcripts. In the latter case, expression of the selectable marker was absolutely dependent on FHV RNA replication, with a background colony frequency of less than 10−8. This improved method of introducing FHV RNA2 replicons into yeast cells will facilitate the use of S. cerevisiae for the characterization of nodavirus RNA replication requirements and expedite the identification and development of inhibitors of viral RNA replication.
MATERIALS AND METHODS
Cells, transformation, growth, and induction.
Plasmid DNAs were introduced into synthetic deletion strain BY4733 (MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0) (7) by using the Frozen-EZ yeast transformation kit (Zymo Research) and following the manufacturer's protocol. Yeast cells were grown at 30°C in medium selective for desired plasmids, with glucose or galactose as the carbon source as indicated. Throughout, figures and legends have been simplified to reflect only the presence or absence of uracil or histidine, but all of the synthetic media used also lacked leucine and tryptophan. The GAL1 promoter was induced as described in the figure legends or Results.
Plasmid constructions.
Where PCR was employed, cloned and sequenced fragments were used to construct desired vectors. Laboratory names of plasmids are indicated in brackets.
(i) pF2U and pU2F [LpGDUR and LpG(DU)R].
pF2U contains the EcoRI/PstI GAL1 promoter fragment of pF1 (25), the PstI/MluI 5" FHV RNA2 fragment of p2B10SP (10), the MluI/MluI URA3 open reading frame (ORF) fragment amplified from YEp24 (6) with primers 5"ACGCGTTTCGAAAGCTACATATAAG and 5"ACGCGTTTAGTTTTGCTGGCCGCA, the MluI/MscI fragment of the FHV DI634 cDNA (32), and the MscI/SacI 3" FHV RNA2/hepatitis delta virus (HDV) ribozyme (Rz) fragment of pFHV2[0,0] (2) cloned between the EcoRI and SacI sites of the multiple cloning sites of yeast 2μm high-copy plasmid YEp351 (16).
pU2F contains an EcoRI/MscI fragment containing a fusion (described below) of the GAL1 promoter to the 3" end of FHV RNA2, the MscI/SacI fragment of pF2U, and the SacI/SacI 5" FHV RNA2/HDV Rz fragment of p2VHF[1,0] (3) cloned between the EcoRI/SacI sites of the multiple cloning sites of YEp351 (16). The GAL1-3"FHV RNA2 fusion was obtained by blunt-end ligation of the SnaBI blunt-cut, pB3MI1-derived GAL1 promoter (17) to the XbaI-linearized, mung bean nuclease-blunted 3" end of p2B10SP-derived FHV RNA2 (10).
(ii) piU2F and piH2F [LpG(DUi-)R and LpG(DHi-)R].
A previously characterized yeast artificial intron (9, 12, 31) was reconstructed by PCR and cloned. A unique EagI restriction site was introduced into a multiple cloning site within the intron by XmaI digestion, filling in of the overhang, and ligation. piU2F was generated by antisense ligation of a blunt PCR product, amplified from the intron clone, into the unique StuI site of pU2F. The same blunt intron PCR product was ligated antisense into the HIS3 ORF MscI site within piH2F, which contains the MluI/MluI HIS3 ORF fragment amplified from pRS313 (29) with primers 5"ACGCGTGATGACAGAGCAGAAAGCCC and 5"ACGCGTTCACTACATAAGAACACCTT cloned into the MluI/MluI vector fragment of piU2F.
(iii) pT3F2U and pT3F2H [pDU and pDH].
pT3F2U and pT3F2H, vectors for the in vitro transcription of F2U and F2H, were constructed by ligation of the ORF-containing MluI fragments into the cDNA clone of FHV RNA2 DI634 (32). Incubation of XbaI-linearized pT3F2U or pT3F2H with T3 RNA polymerase produced RNAs starting at the FHV RNA2 5" end and containing up to four nonviral nucleotides 3" of the FHV RNA2 3" end.
RNA and DNA analysis.
Hot phenol extraction of total yeast RNA, formaldehyde denaturation, Northern blotting to Nytran nylon membranes (Schleicher & Schuell), and hybridization were performed as described previously (21, 23). Strand-specific, 32P-labeled in vitro transcripts were generated as previously described (20). The probes for negative- and positive-strand URA3 corresponded to or were complementary to the entire URA3 ORF, respectively. Analysis was done with a Molecular Dynamics PhosphorImager digital radioactivity imaging system.
RESULTS
Expression of reporter genes from RNA replicons transcribed from plasmids in yeast.
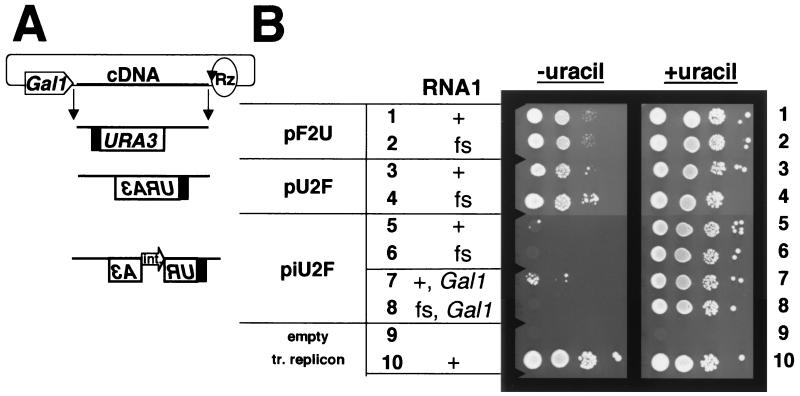
To facilitate the manipulation and analysis of FHV RNA replication in yeast, we sought to express RNA replicons from plasmids in vivo. The structures of plasmids designed to transcribe URA3-containing replicons based on FHV RNA2 are shown schematically in Fig. 1A. To drive primary transcription, they all used the inducible GAL1 polII promoter, which is actively repressed when cells are grown on dextrose but strongly induced when cells are grown on galactose (24). The 3" end of the RNA replicon was generated by the self-cleaving Rz of HDV, positioned to cleave precisely at the 3" end of the FHV RNA2 sequence (2). In each experiment shown in Fig. 1B, transcription of the replicon was transiently induced by growing BY4733 cells in galactose-containing liquid medium. Serial 50-fold dilutions of the cultures were spotted onto plates containing dextrose, with or without uracil to select for URA3 expression. As a positive control, we used cells in which the F2U RNA replicon was derived from a transcript made in vitro and replicated by functional FHV RdRp (Fig. 1B, row 10). Cells that contained no plasmids or replicons failed to grow under either set of conditions (Fig. 1B, row 9).
FIG. 1.
FHV dependence of the Ura+ phenotype of yeast cells that contain replicon expression plasmids. (A) Schematic of RNA2-based replicons transcribed from plasmids. The GAL1 promoter (pentagon) is fused to the 5" end of the positive- or negative-sense FHV RNA2 sequences as shown (see Materials and Methods for details). The circle and arrow denote a cDNA of the HDV self-cleaving Rz positioned to cleave at the exact 3" end of the FHV RNA2 positive or negative strand, as indicated. The line and filled box indicate FHV RNA2-derived sequences. The open box denotes the yeast URA3 ORF. The polarity of the replicon cDNA is indicated by the orientation of the URA3 lettering. The open arrow indicates an artificial yeast intron (int.) (31), whose polarity is indicated by the orientation of the arrow and lettering. A detailed schematic of the expected steps following induction from piU2F is presented in Fig. 2. (B) Replicon induction. Yeast cells in rows 1 to 6 contain the indicated plasmid for FHV RNA2-based replicon expression, with plasmids for low-level constitutive expression of wild-type or frameshifted (fs) FHV RNA1. Cells in rows 7 and 8 contain the indicated replicon plasmid and plasmids for high-level GAL1-induced expression of wild-type or frameshifted RNA1. Cells in row 9 are the parent yeast strain BY4733 with no plasmids or replicons (empty), which was used as a negative control. Cells in row 10 are a positive control containing the transfected (tr.) replicon and the plasmid for constitutive expression of FHV RNA1. Dextrose-grown cells in rows 1 to 8 were induced in medium containing galactose and uracil and cultured for 2 days before spotting of 5 μl of serial 50-fold dilutions (from left to right) onto synthetic dextrose medium lacking or containing uracil, as indicated. Cells in row 9 were cultured in rich dextrose medium, and cells in row 10 were cultured in synthetic dextrose medium lacking uracil prior to dilution and spotting. For all rows, the spot in the first column contains approximately 2.5 × 105 cells. Plates were incubated for 4 days at 30°C and then photographed.
To introduce a replicable RNA2 derivative into the cells, we first constructed a plasmid, pF2U, for the transcription of a positive-sense replicon (Fig. 1A). Promoter and Rz positioning was based on previous expression of FHV RNA1 in yeast (25) and FHV RNA2 in mammalian cells (2). Because the primary transcript from this plasmid served as an mRNA for the expression of the URA3 gene, we hoped to be able to transcribe the replicon transiently in the presence of galactose (i.e., under inducing conditions) and then select for RNA replication and maintenance of the replicon during growth on dextrose, where primary transcription would be repressed. But, as shown in Fig. 1B, rows 1 and 2, cells containing pF2U grew equally well under selection for Ura+, irrespective of whether the FHV RNA1 plasmid expressed wild-type RNA replicase (pF1ΔP) or a frame-shifted inactive mutant enzyme [pF1fsΔP, described previously as p1(fs)R (26)]. Both of these RNA1 expression plasmids lack an explicit polII promoter (hence ΔP), but pF1ΔP expresses sufficient levels of RNA1 to maintain FHV RNA2 replicons without cytotoxicity (25). Evidently, the basal level of transcription from pF2U was sufficient for cell growth even in the absence of RNA replication.
We tried to eliminate this background by reversing the orientation of the replicon with respect to the GAL1 promoter and Rz, generating plasmid pU2F (Fig. 1A). Previous work has shown that the FHV RdRp can replicate negative-sense primary transcripts of FHV RNA2 (3). The 5"-most nucleotide of negative-sense RNA2 cDNA was positioned at one of the previously observed GAL1 transcription start sites (25), and the Rz was positioned to cleave at the 3" end as before (3). We reasoned that galactose-induced transcription from pU2F should produce a negative-sense RNA that would require replication to produce translatable mRNA. However, cells containing pU2F grew equally well under Ura+ selection in the presence or absence of functional FHV RNA replicase (Fig. 1B, rows 3 and 4). We attribute this growth to the generation of positive-sense transcripts from a cryptic promoter on the opposite DNA strand, which then served as mRNAs for URA3 expression. This interpretation was supported by Northern blot analysis (see Fig. 3B).
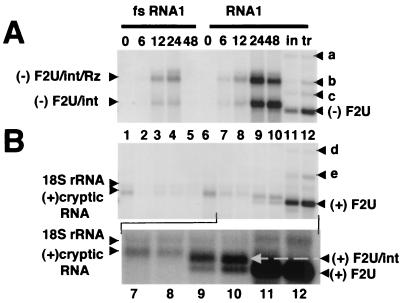
FIG. 3.
Northern blot analyses of the RNA products of polII transcription and FHV-mediated amplification of RNA replicons in yeast. For the induction time course, total RNA was isolated from cells carrying piU2F and plasmids for GAL1-driven expression of FHV RNA1 encoding frameshifted (fs) or wild-type FHV protein A. These cells were grown in medium containing glucose, pelleted, and resuspended in medium containing galactose to induce the GAL1 promoter of the plasmids, as described previously (25). Cells were harvested for RNA extraction at 0, 6, 12, 24, and 48 h postinduction (as indicated). For comparison, total RNA was isolated from Ura+ cells containing pF1ΔP and transfected (tr) or induced (in) replicon F2U. (A) Northern blot analysis of negative-strand RNA accumulation. A 1.5-μg sample of total RNA per lane was denatured in 50% formamide-6% formaldehyde at 65°C, electrophoresed on a 1% agarose-formaldehyde gel, transferred to a nylon membrane, hybridized to a 32P-labeled in vitro transcript probe complementary to negative-strand URA3, and visualized by PhosphorImager analysis. The origin of the RNA is indicated above each lane. The positions of the intron-containing/Rz-containing transcripts [(−)F2U/int/Rz] and the cleaved, intron-containing RNAs [(−)F2U/int] are indicated at the left. Products of replication, including cleaved, spliced negative-sense F2U [(−)F2U], are indicated at the right. The fainter bands above the replicon (designated a, b, and c) are discussed more fully in Results. (B) Northern blot analysis of positive-strand replicon accumulation performed as described for panel A except that the blot was hybridized to a 32P-labeled in vitro transcript probe complementary to positive-strand URA3 and printed at a fourfold lower intensity level. The positions of the 18S rRNA (nonspecific detection) and a positive-sense transcript from an unidentified promoter [(+)cryptic RNA] are indicated at the left. Products of replication following selection, including positive-sense F2U [(+)F2U], are indicated at the right. The fainter bands above the replicon (designated d and e) are discussed in Results. Lanes 7 to 12 are magnified and printed at a darker intensity to clearly distinguish the cryptic RNA from positive-sense unspliced F2U [(+)F2U/int], indicated at the right.
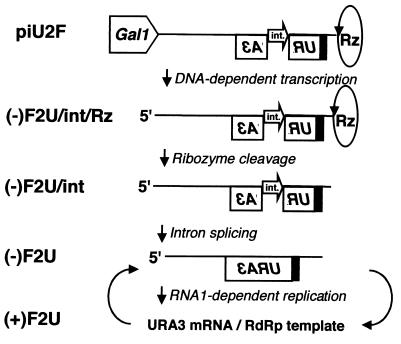
To prevent the cryptic positive-sense RNA from serving as an mRNA, an artificial intron (9, 12, 31) was incorporated into pU2F within, and antisense to, the URA3 ORF, generating piU2F (Fig. 1A). The intent was that the intron could be spliced out of the negative-sense GAL1 promoter-driven transcripts but not the cryptic positive-sense transcripts. The expected path from piU2F to replication-dependent URA3 expression is shown in Fig. 2. Galactose-induced cells containing piU2F and pF1fsΔP exhibited no growth on plates lacking uracil (Fig. 1B, row 6). In contrast, colonies were obtained from induced cells that contained piU2F and functional RdRp expressed from pF1ΔP (Fig. 1B, row 5). In this representative experiment, two colonies were recovered from approximately 2.5 × 105 cells (a frequency of 8 × 10−6). This frequency could be increased to 2 × 10−3 by inducing these cells directly on galactose-containing solid media (data not shown).
FIG. 2.
Schematic illustration of piU2F and the events leading to RNA-dependent RNA replication and URA3 expression. The line and filled box indicate FHV RNA2-derived sequences. The open box denotes the yeast URA3 ORF, whose polarity is indicated by lettering orientation. The flanking GAL1 promoter and self-cleaving HDV Rz are indicated. For junction details, see Materials and Methods. The open arrow indicates an artificial intron (int.) inserted antisense to the URA3 ORF. Initial RNA synthesis is by DNA-dependent RNA polII transcription. This RNA is cleaved by the HDV Rz to generate the correct 3" end and processed by the cellular splicing machinery to remove the intron. The resulting negative-sense replicon serves as the template for the synthesis of a positive-strand RNA by the FHV RNA-dependent RNA polymerase. This positive-sense RNA serves as mRNA for the URA3 protein and as the template for the production of new negative-strand RNA.
Since the overall level of RNA1 replication initiated by pF1ΔP was only about 2% of the level supported by an FHV RNA1 expression plasmid with an explicit, high-level GAL1 promoter, pF1 (25), we tested whether increasing the level of RNA1 replication would increase the frequency of colony formation. Cells containing piU2F and the GAL1-driven pF1 (or pF1fs encoding frameshifted RNA1 as a negative control) were transiently induced and plated (Fig. 1B, rows 7 and 8). Cells containing pF1 (Fig. 1B, row 7) formed approximately 50-fold more colonies (a frequency of 4 × 10−4) than did cells containing pF1ΔP (Fig. 1B, row 5), and those containing pF1fs still exhibited no background growth (Fig. 1B, row 8).
To determine the level of background colony formation from piU2F in the absence of RNA replication, cells containing pF1fsΔP and piU2F were induced and plated on medium lacking uracil. When this experiment was performed with a parental yeast strain from which the URA3 ORF was precisely deleted (BY4733 [7]), no RNA1-independent colonies were observed among 108 plated cells (data not shown). However, when the same experiment was repeated with a parental yeast strain in which the URA3 gene was inactivated by the insertion of a Ty retrotransposon (YPH500, carrying the ura3-52 allele [27]), RNA1-independent colonies were formed with a low frequency (data not shown). We attribute this background to the restoration of a functional URA3 ORF by homologous DNA recombination between the plasmid-borne gene and the chromosomal locus. Analogous events have been previously documented at similar frequencies (8). For this reason, strain BY4733 was used throughout the work presented in this paper.
RNA replication was initiated from negative-sense primary transcripts.
We identified the primary transcripts responsible for the Ura+ phenotype by both genetic and biochemical methods. The pathway shown in Fig. 2 predicted that colony formation would depend on removal of the intron from transcripts of piU2F by RNA splicing. To test this prediction, several mutations were introduced into the intron of piU2F.
The genome of S. cerevisiae contains 228 known introns, and the splice donor, branch point, and acceptor sites of each have been cataloged (30). The nucleotide at the fourth position of the splice donor site is somewhat variable, being a U, C, A, or G in 196, 27, 5, and 0 of the naturally occurring introns, respectively. When the U at this position in piU2F was mutated to C or G, colony formation was reduced to 0.40% ± 0.17% and 0.10% ± 0.11%, respectively, of the levels conferred by wild-type piU2F. The intron branch acceptor is an A residue that is absolutely conserved in all known yeast introns. When this A residue in piU2F was mutated to G, colony formation was abolished (<0.001% of the wild-type level). In contrast to the deleterious effects of these point mutations, removal of 39 nucleotides from the nonconserved region within the artificial intron did not decrease the frequency of Ura+ colonies (123% ± 23%). These genetic experiments established that the RNA replication responsible for the Ura+ phenotype was indeed dependent on splicing of negative-sense primary transcripts.
Biochemical characterization of URA3 RNAs.
We next examined the transcribed, processed, and replicated RNAs by using biochemical methods. Cells containing piU2F and either pF1 or the frameshifted version, pF1fs, were induced to transcribe high levels of the replicon and wild-type or frameshifted FHV RNA1 from their GAL1 promoters. No negative-sense RNAs were detected by Northern blot hybridization using a URA3 riboprobe in uninduced BY4733 cells (Fig. 3A, lanes 1 and 6), but 6 to 12 h after induction, two major negative-sense RNA species began to accumulate (Fig. 3A, lanes 2 and 3 and 7 and 8). These RNAs, which are designated (−)F2U/int/Rz and (−)F2U/int in Fig. 3A, persisted as long as 24 h after induction in the absence of RNA replication (Fig. 3A, lanes 3 and 4) and through at least 48 h postinduction in the presence of the RdRp (Fig. 3A, lanes 8 to 10).
Several lines of evidence indicated that the larger band [(−)F2U/int/Rz] was a transcript that initiated at the GAL1 promoter, extended through the replicon and HDV Rz, and terminated at an undefined cryptic polyadenylation site within the plasmid (data not shown). The smaller negative-sense RNA [(−)F2U/int in lanes 2 to 4 and 7 to 10] was likely the upstream product of Rz-mediated cleavage of the larger species (data not shown). Surprisingly, both of these negative-sense RNAs contained the intron (hybridization data not shown).
Following Ura+ selection of induced cells carrying piU2F and pF1ΔP, the URA3 riboprobe detected a negative-sense RNA corresponding in size to authentic intronless negative-sense F2U [band designated (−)F2U in Fig. 3A, lanes 11 and 12], and Northern blot hybridization confirmed that this RNA did not contain the intron (data not shown). A minor RNA species of the same size was also detected 24 and 48 h postinduction in cells containing pF1(Fig. 3A, lanes 9 and 10) but not in cells containing pF1fs (Fig. 3A, lanes 4 and 5). In addition to (−)F2U, three other minor RNA species were detected by the probe for negative-sense URA3 following induction and selection (Fig. 3A, lanes 11 and 12; bands labeled a, b, and c). We interpret these RNAs as being the F2U analogues of minor products of authentic FHV RNA2 replication which we have characterized in some detail in other work (1). According to this interpretation, the largest of these RNAs (band a) corresponded to a covalent homodimer consisting of the F2U replicon and the smallest (band c) corresponded to a covalent heterodimer consisting of F2U and a subgenomic RNA, FHV RNA3. The identity of band b remains undetermined.
Northern blot analysis also identified positive-sense RNAs containing URA3 sequences, including the replication products of the primary transcripts. A URA3 probe detected a single positive-sense RNA in uninduced cells containing piU2F and pF1fs or pF1, and this RNA diminished in abundance 6 and 12 h postinduction (Fig. 3B, lanes 1 to 3 and 6 to 8, respectively). This RNA likely resulted from transcription from an undefined cryptic promoter on the opposite DNA strand, as suspected from the phenotype of cells carrying pU2F (Fig. 1B, rows 3 and 4). At 24 and 48 h postinduction, two new positive-sense RNA species were detected, but only in cells expressing functional RNA replicase (Fig. 3B, compare lanes 4 and 5 with lanes 9 and 10). The smaller of these RNAs comigrated with the predominant species found after Ura+ selection and with authentic positive-sense F2U (Fig. 3B, lanes 11 and 12, respectively). Lanes 7 to 12 are presented again in Fig. 3B as a magnified longer exposure of the blot to clarify the migration of these positive-sense RNAs. The larger positive-sense RNA [(+)F2U/int] and the cryptic promoter-driven transcript [(+)cryptic RNA] contained the intron, whereas the RNA comigrating with authentic F2U [(+)F2U] did not (hybridization data not shown). In confirmation of the Northern blot, reverse transcription-PCR analysis identified an intronless positive-sense URA3 RNA, but not DNA, in cells containing pF1ΔP and piU2F following induction and selection (data not shown).
In addition to positive-sense F2U RNA, two other minor species were detected in Ura+-selected cells following induction or transfection (bands labeled d and e in Fig. 3B, lanes 11 and 12). The larger of these bands (d) likely represented a covalent F2U dimer, as identified previously for other FHV replicons (1). The identity of the smaller band (e) is unclear, although it was probably the complement of negative-sense band b (Fig. 3A). A low level of cross-hybridization to 18S rRNA is visible in all lanes.
Primer extension was used to map the GAL1 promoter start sites and to examine the 5" termini of negative- and positive-sense RNAs following selection of Ura+ colonies. As expected, a primer extension product corresponding to the authentic 5" end of the negative strand was detected in samples containing functional FHV RNA1 harvested at 48 h postinduction (data not shown). This product was also present, but at lower levels, in samples harvested from cells that contained frameshifted RNA replicase (data not shown). Following induction and selection in the presence of the replicase, primer extension products corresponding to the 5" end of the positive-sense F2U replicon were detected as expected (data not shown). In summary, the biochemical characterization of the products of RNA transcription, processing, and replication reinforced the genetic data and confirmed that the Ura+ phenotype was due to induction and replication of an RNA replicon from DNA.
Simultaneous expression of two selectable marker genes via RNA replication.
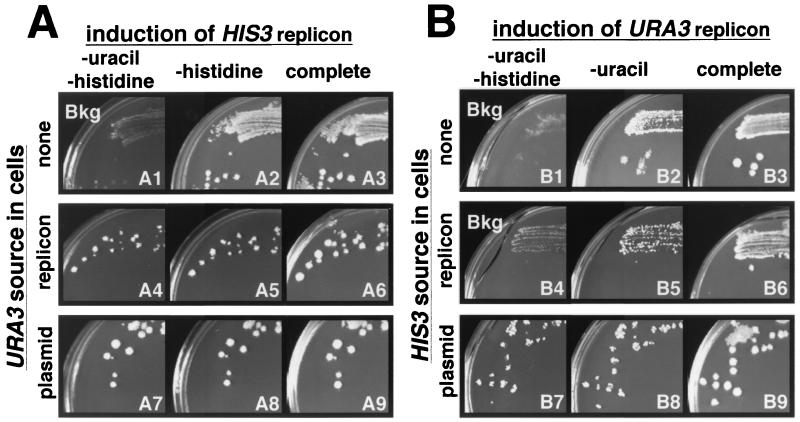
To determine the general applicability of this system for the expression of other replicons, we constructed piH2F. This plasmid was similar to piU2F but expressed the negative strand of F2H (for FHV RNA 2 plus HIS3), a replicon carrying the yeast HIS3 ORF in the same context as the URA3 ORF in F2U. F2H was a functional, FHV RNA1-dependent replicon since transfection of in vitro transcribed F2H complemented the HIS3 defect (his3Δ200, a deletion larger than the HIS3 ORF [13]) in BY4733 cells containing pF1ΔP but not in cells containing frameshifted pF1fsΔP (data not shown). To determine the ability of piH2F to initiate replication in different environments, piH2F was transformed into cells containing pF1ΔP alone, pF1ΔP and the F2U RNA replicon, or pF1ΔP and a plasmid expressing the URA3 gene directly. These three piH2F-containing cell lines were applied to individual galactose-containing inducing plates appropriately selective for plasmids and the F2U replicon and allowed to grow for 4 days. To identify cells expressing F2H via RNA replication, these plates were replica plated onto selective dextrose-containing noninducing plates that either lacked both histidine and uracil, lacked only histidine, or lacked neither (Fig. 4A).
FIG. 4.
Simultaneous expression of two replicons. Induction was assayed by replica plating cells from galactose synthetic medium onto three different glucose synthetic media, as indicated above the photos. Plates are numbered at the lower right. In panel A, the URA3 replicon was induced in cells containing pF1ΔP and piU2F, which lacked the HIS3 gene or contained it on transfected replicon H2F or a plasmid (pRS313 [29]), as indicated at the left. In panel B, the HIS3 replicon was induced in cells containing pF1ΔP and piH2F, which lacked the URA3 gene or contained it on transfected replicon U2F or a plasmid (YEp24 [6]), as indicated at the left. Plates were incubated for 4 days at 30°C and then photographed. Plates showing only the residual cells transferred during replica plating are indicated as background (Bkg).
Overall, replica plating resulted in faithful transfer of cells from individual colonies on the master plate to a series of selective plates. Where most cells were able to grow, they reproduced the colony on the master plate, as shown in the control plates in Fig. 4 (plates A3, A6, A9, B3, B6, and B9). Where only a few cells were able to grow, they gave rise to small colonies in the printed cells of the master plate colony, giving the transferred print a “punctate” appearance. This was very similar to the situation observed in row 7 of Fig. 1B, where only a few of a large number of cells spotted onto a plate formed colonies. Where no cells were able to form colonies, only the printed cells of the master plate colony were observed (Fig. 4, plates A1, B1, and B4). When the F2H replicon was induced and selected from piH2F in cells containing pF1ΔP and a plasmid expressing URA3, the colonies on the master plate were reproduced on plates lacking both uracil and histidine and on plates lacking histidine (Fig. 4A, plates A7 and A8 versus plate A9). Similarly, the master plate colonies were reproduced on plates lacking histidine when F2H was induced and selected in cells containing only pF1ΔP (Fig. 4, plate A2 versus plate A3). However, these colonies failed to grow and left only printed cells on plates lacking uracil and histidine (plate A1) because they lacked the URA3 gene. Growth was also obtained under both selective conditions when F2H was induced and selected in cells replicating F2U; however, the transferred colonies had a smaller, more punctate appearance, indicating that the induction frequency was somewhat reduced (Fig. 4, plates B4 and B5 versus plate B6). In selected instances, Northern blot analysis confirmed the presence of F2H (data not shown). Thus, piH2F was able to launch F2H replication, even in cells that contained another replicon, F2U.
The converse experiments revealed that piU2F was less efficient than piH2F at inducing colony formation. Here, piU2F was transfected into cells containing either pF1ΔP alone, pF1ΔP and the F2H RNA replicon, or pF1ΔP and a plasmid expressing the HIS3 gene directly. These three cell lines were applied to appropriately selective individual galactose-containing plates and replica plated onto selective dextrose-containing plates lacking both histidine and uracil, lacking only uracil, or lacking neither (Fig. 4B). In these experiments, all selective plates displaying growth also displayed a punctate appearance (Fig. 4, plates B2 versus B3, B5 versus B6, and B7 and B8 versus B9), indicating less efficient induction and selection of the F2U replicon compared to the F2H replicon on the equivalent plates in panel A. Indeed, the induction efficiency was so low that only the printed cells of the master plate colony were observed when F2U was induced in cells containing F2H (Fig. 4, plate B4). The most punctate plate was B5, where F2U was perhaps able to initiate replication only in cells that lacked F2H (estimated as about 40%; unpublished results). Indeed, quantitative analysis of induction and selection of colonies from piU2F and piH2F in the absence of a competing replicon showed that piH2F supported colony formation at a frequency about 25 times higher than that of piU2F.
DISCUSSION
The ability to replicate the positive-sense RNA animal virus FHV in the model eukaryote S. cerevisiae is unique. Development of this system for the genetic and biochemical analysis of the virus and its relationship with the host offers useful opportunities. This system coupled reporter expression to viral replication of FHV RNA2 via an inducible polII transcript. These results were consistent with the pathway for URA3 expression from piU2F depicted in Fig. 2 and inconsistent with the conversion of RNA to DNA by an endogenous reverse transcriptase activity that has been documented to occur in yeast at a frequency of 10−9 (12). Background colony formation resulting from the translation of unreplicated RNA or from DNA recombination was undetected, making this an excellent scheme for the tightly regulated expression of toxic gene products, for example. The induction frequency was insufficient for certain negative selection schemes but more than sufficient for screens involving replica plating (Fig. 4). Introduction of RNA replicons was much easier by this method than by transfection of spheroplasts with in vitro transcripts (26). As intended, this system will facilitate the identification of inhibitors of RNA replication, be they proteins, RNAs, drugs, or mutations in viral or host genes. The effect of such inhibitors can be easily quantitated as reduced or inhibited colony formation.
This system also provides a powerful way to select for improved FHV RNA2 replication. For example, we have identified mutants of FHV RNA1 that can replicate themselves but support only very low levels of FHV RNA2 replication (L. Eckerle and L. A. Ball, submitted for publication). By plating billions of cells containing a selectable RNA2 replicon together with the mutant FHV RNA1, we can select for cells that can form colonies and rapidly identify mutations that restore RNA2 replication. Similarly, although some combinations of RNA1 and RNA2 from different nodaviruses produce infectious reassortants, this is not true for all combinations (15). This system can be used to identify those combinations that support limited cross-replication (K. L. Johnson, B. D. Price, and L. A. Ball, unpublished data), and we can select for improvements in replication and potentially identify virus-specific replication signals. Finally, this system may provide a way to identify putative cis elements that recruit an RNA to the replication complex. In conclusion, selectable reporters combined with the introduction of RNA templates by induction of DNA-dependent transcription greatly increases the utility of this unique virus/host system and will lead to an enhanced understanding of the mechanism of FHV RNA replication.
Acknowledgments
We thank Cindy Luongo, Sean Whelan, and the members of the Ball lab for valuable comments on the manuscript and the UAB Comprehensive Cancer Center DNA sequencing facility for determination of DNA sequences.
This work was supported by NIH grants R37 GM35072 to P.A. and R01 AI18270 to L.A.B. and by Center for AIDS Research Developmental Award AI27767 to B.D.P. and K. L. Johnson.
REFERENCES
- 1.Albariño, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. 2001. Characterization of novel RNA species generated during flock house virus RNA replication. J. Virology 289:269-282. [DOI] [PubMed] [Google Scholar]
- 2.Ball, L. A. 1992. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 66:2335-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. USA 91:12443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A., J. M. Amann, and B. K. Garrett. 1992. Replication of Nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 66:2326-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, L. A., and K. L. Johnson. 1999. Reverse genetics of nodaviruses. Adv. Virus Res. 53:229-244. [DOI] [PubMed] [Google Scholar]
- 6.Botstein, D., S. C. Falco, S. E. Stewart, M. Brennan, S. Scherer, D. T. Stinchcomb, K. Struhl, and R. W. Davis. 1979. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 8:17-24. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Bratty, J., G. Ferbeyre, C. Molinaro, and R. Cedergren. 1996. Stimulation of mitotic recombination upon transcription from the yeast GAL1 promoter but not from other RNA polymerase I, II and III promoters. Curr. Genet. 30:381-388. [DOI] [PubMed] [Google Scholar]
- 9.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasmahapatra, B., R. Dasgupta, K. Saunders, B. Selling, T. Gallagher, and P. Kaesberg. 1986. Infectious RNA derived by transcription from cloned cDNA copies of the genomic RNA of an insect virus. Proc. Natl. Acad. Sci. USA 83:63-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dearing, S. C., P. D. Scotti, P. J. Wigley, and S. D. Dhana. 1980. A small RNA virus isolate from the grass grub Costelytra zealandica (Coleoptera: Scarabaeidae). N. Z. J. Zool. 7:267-269. [Google Scholar]
- 12.Derr, L. K., J. N. Strathern, and D. J. Garfinkel. 1991. RNA-mediated recombination in S. cerevisiae. Cell 67:355-364. [DOI] [PubMed] [Google Scholar]
- 13.Fasullo, M. T., and R. W. Davis. 1988. Direction of chromosome rearrangements in Saccharomyces cerevisiae by use of his3 recombinational substrates. Mol. Cell. Biol. 8:4370-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friesen, P. D., P. Scotti, J. Longworth, and R. R. Rueckert. 1980. Black beetle virus: propagation in Drosophila line 1 cells and an infection-resistant subline carrying endogenous black beetle virus-related particles. J. Virol. 35:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher, T. M. 1987. Synthesis and assembly of nodaviruses. Ph.D. thesis. University of Wisconsin, Madison.
- 16.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 71:7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, K. N., K. L. Johnson, R. Dasgupta, T. Gratsch, and L. A. Ball. 2001. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 82:1855-1866. [DOI] [PubMed] [Google Scholar]
- 19.Kaesberg, P., R. Dasgupta, J. Y. Sgro, J. P. Wery, B. H. Selling, M. V. Hosur, and J. E. Johnson. 1990. Structural homology among four nodaviruses as deduced by sequencing and X-ray crystallography. J. Mol. Biol. 214:423-435. [DOI] [PubMed] [Google Scholar]
- 20.Kroner, P., and P. Ahlquist. 1992. RNA-based viruses, p. 23-34. In S. J. Gurr, M. J. McPherson, and D. J. Bowles. (ed.), Molecular plant pathology, vol. 1. IRL, Oxford, United Kingdom. [Google Scholar]
- 21.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 22.Lundblad, V. 1994. Saccharomyces cerevisiae, p. 13.0.3-13.0.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York. N.Y. [Google Scholar]
- 23.Newman, T. C., M. Ohme-Takagi, C. B. Taylor, and P. J. Green. 1993. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5:701-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima, Y. 1982. Regulatory circuits for gene expression: the metabolism of galactose and phosphate, p. 159-180. In J. Strathern, E. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- 25.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, M., and F. Winston. 1984. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol. Gen. Genet. 193:557-560. [DOI] [PubMed] [Google Scholar]
- 28.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA 87:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spingola, M., L. Grate, D. Haussler, and M. Ares, Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimatsu, T., and F. Nagawa. 1989. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244:1346-1348. [DOI] [PubMed] [Google Scholar]
- 32.Zhong, W., R. Dasgupta, and R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]