Abstract
We constructed two versions of an RCASBP-based retroviral shuttle vector, RSVP (RCASBP shuttle vector plasmid), containing either the zeocin or blasticidin resistance gene. In this vector, the drug resistance gene is expressed in avian cells from the long terminal repeat (LTR) promoter, whereas in bacteria the resistance gene is expressed from a bacterial promoter. The vector contains a bacterial origin of replication (ColE1) to allow circular viral DNA to replicate as a plasmid in bacteria. The vector also contains the lac operator sequence, which binds to the lac repressor protein, providing a simple and rapid way to purify the vector DNA. The RSVP plasmid contains the following sequence starting with the 5" end: LTR, gag, pol, env, drug resistance gene, lac operator, ColE1, LTR. After this plasmid was transfected into DF-1 cells, we were able to rescue the circularized unintegrated viral DNA from RSVP simply by transforming the Hirt DNA into Escherichia coli. Furthermore, we were able to rescue the integrated provirus. DNA from infected cells was digested with an appropriate restriction enzyme (ClaI) and the vector-containing segments were enriched using lac repressor protein and then self-ligated. These enriched fractions were used to transform E. coli. The transformation was successful and we did recover integration sites, but higher-efficiency rescue was obtained with electroporation. The vector is relatively stable upon passage in avian cells. Southern blot analyses of genomic DNAs derived from successive viral passages under nonselective conditions showed that the cassette (drug resistance gene-lac operator-ColE1) insert was present in the vector up to the third viral passage for both resistance genes, which suggests that the RSVP vectors are stable for approximately three viral passages. Together, these results showed that RSVP vectors are useful tools for cloning unintegrated or integrated viral DNAs.
The retroviral life cycle depends on the conversion of the RNA genome found in virions into DNA and the subsequent integration of the DNA into the host cell genome. There are a number of different types of experiments for which the molecular cloning of either unintegrated or integrated viral DNA is a critical step. In most cases, cloning the viral DNA involves standard recombinant DNA techniques: viral DNA is inserted into a plasmid or a prokaryotic viral vector. However, there is an alternative approach. It is possible to introduce elements into retroviral vectors that allow DNA forms of the viral genome to replicate in prokaryotic hosts. These types of vectors are referred to as shuttle vectors and can simplify the recovery of viral DNA. At a minimum the shuttle vector must have a plasmid origin of replication and a gene (promoter and coding region) that can be selected in a bacteria.
Most retroviral genomes cannot accommodate the insertion of significant amounts of additional genetic information. In these cases, viral sequences must be removed to provide a place for whatever additional information is inserted. Such viruses are, by definition, replication defective. The missing viral genetic information must be supplied in trans, either by a helper cell or a helper virus. There is one exception. Avian leukosis viruses can accept approximately 2.5 kb of additional information: the naturally occurring avian leukosis virus derivative Rous sarcoma virus (RSV) contains, in addition to a full complement of viral genes, the src oncogene (21). We have prepared a family of replication-competent retroviral vectors, collectively called the RCAS vectors, that derive from the Schmidt-Ruppin strain of RSV. Basically, in the RCAS vectors, the src gene has been deleted and a unique restriction site has been left at the site of the deletion to simplify the insertion of foreign DNA. To permit the cloning and amplification of the RCAS vector DNA, the viral genome was linked to pBR322-derived plasmid sequences (10); however, the plasmid sequences lie outside the viral replicon and the viral vector brings none of the prokaryotic plasmid sequences with it when it replicates in avian cells. It is possible to insert prokaryotic plasmid sequences into the ClaI site of an RCAS vector. The following two versions have been created: p779NC327AC28F, which contains a pBR origin and an ampicillin resistance gene (unpublished observations), and a derivative, pANV-A, which contains a pBR origin, a simian virus 40 promoter, and a neomycin resistance gene linked to the Tn5 promoter (15).
The p779NC327AC28F plasmid has two disadvantages. First, there is no selection possible when the virus is propagated in avian cells. Second, the prokaryotic sequences are rapidly lost during virus propagation (unpublished observations). pANV-A can be selected in both prokaryotic and eukaryotic hosts, and the viral genome was reported to be stable through one round of viral replication (15).
We have revisited the RCAS shuttle vector problem and have prepared two new vectors that have advantages over the published vectors. First, the new vectors make use of drug resistance markers that are small (zeocin resistance and blasticidin resistance). Zeocin is a member of the bleomycin/phleomycin family of antibiotics and is a basic, water-soluble, copper-chelated glycopeptide isolated from Streptomyces verticillus (2). When zeocin enters the cell, the copper cation is reduced from Cu2+ to Cu+ and zeocin becomes activated. Zeocin will then bind DNA and cleave it, causing cell death (2). A zeocin resistance protein, the product of the Streptoalloteichus hindustanus ble gene, has been isolated from S. hindustanus, binds to zeocin, and inhibits its DNA cleavage activity (4, 6). The S. hindustanus ble gene has been used as a selectable marker for prokaryotes and eukaryotes (3-5, 14). Blasticidin S is a nucleoside antibiotic isolated from Streptomyces griseochromogenes that is a potent protein synthesis inhibitor for both prokaryotes and eukaryotes (24, 25). Blasticidin resistance is conferred by the expression of one of two blasticidin S deaminase genes, either the BSD gene of Aspergillus terreus (12) or bsr of Bacillus cereus (11). These enzymes convert blasticidin S to an inactive deaminohydroxy derivative (11). In eukaryotic cells, the RSVP vectors express the drug resistance genes via a spliced message, so that an internal promoter is not needed. In addition, we have added a completely symmetric lac operator, which makes it a simple matter to enrich for viral DNA (16, 17). This symmetric lac operator, which is an inverted repeat of a 15-bp segment from the left half of the natural operator sequence (5"-TGTGGAATTGTGAGCGCTCACAATTCCACA-3"), has been reported to bind the lac repressor 10-fold more tightly than the natural lac operator sequence (19). It is possible with this system not only to recover unintegrated circular viral DNA from infected cells but also to easily clone integration sites. The blasticidin and zeocin cassettes are approximately 1.2 kb long, so it should be possible to introduce additional information into the shuttle vectors. We have also demonstrated that the RSVP vectors are sufficiently stable that they can be passaged three times without a substantial loss of the prokaryotic plasmid sequences.
MATERIALS AND METHODS
Plasmid construction.
A cassette containing multiple cloning sites (MCS), a drug resistance gene (either zeocin or blasticidin), a lac operator sequence, and a ColE1 replication origin was introduced into the plasmid 327AC (10) as described below. The plasmid 327AC was digested with EcoRI and HindIII, and the digested DNA was ligated with two complementary 40-bp oligonucleotides that included the following cloning sites: ClaI, BglII, AvrII, NotI, XbaI, and HindIII. The resulting plasmid was called 327ACMC. The blasticidin resistance gene was PCR amplified from the pcDNA6/V5-His/lacZ plasmid (InVitrogen, Carlsbad, Calif.) by using the forward primer Bsd-Not, which anneals to the region upstream of the EM-7 promoter, and the reverse primer Bsd-Xba, which spans the termination codon for the blasticidin resistance gene. The sequence of Bsd-Not was 5"-ATCAgcggccgcATCAGCACGTGTTGAC-3". The Bsd-Not primer contains a NotI restriction site in the overhang, which is indicated by lowercase letters. The sequence of Bsd-Xba was 5"-ACtctagaTTAGCCCTCCCACACATAACC-3". The XbaI restriction site is indicated by lowercase letters, and the stop codon is underlined. The PCR product was cleaved with NotI and XbaI and inserted into the vector 327ACMC that had been digested with NotI and XbaI, which generated 327(Bsd). Next, two cDNA oligonucleotides (LacO-Xba, CTAGATGTGGAATTGTGAGCGCTCACAATTCCACAggatccTA; LacO-Hind, AGCTTAAggatccTGTGGAATTGTGAGCGCTCACAATTCCACAT), containing the lac operator (bold) and a new BamHI site (lowercase), were annealed and inserted into the XbaI/HindIII site of 327(Bsd), which generated 327(Bsd/Lac). The lac operator was sequenced to check its integrity. To create unique MCS, two cDNA oligonucleotides (MCS-Cla, CGATACTAGTCGTACGATGCATGC; MCS-Not, GGCCGCATGCATCGTACGACTAGTAT), containing SpeI, SplI, and NsiI sites, were annealed and inserted into the ClaI/NotI site of 327(Bsd/Lac), which generated 327(MCS/Bsd/Lac). The ColE1 origin in the cassette was PCR amplified from the pcDNA6/V5-His/lacZ plasmid (InVitrogen) using the forward primer Ori-BamH (TTCGGATCCATGTGAGCAAAAGGCCAGCAA) and the reverse primer Ori-Hind (GTCAAGCTTacgcgtCCCGTAGAAA AGATCAAAGGA). The reverse primer created an MluI site (lowercase letters) which was used in subsequent cloning steps. The PCR product was cleaved with BamHI and HindIII and inserted into the BamHI/HindIII site of 327(MCS/Bsd/Lac). The resulting plasmid was called 327ca(Bsd).
The zeocin resistance gene with the associated EM-7 promoter was PCR amplified from the pZeoSV2(+) plasmid (InVitrogen) using two primers, Zeo-Not (ATTgcggccgcTGTTGACAATTAATCATCGGC) and Zeo-Xba (GCCtctagaTCAGTCCTGCTCCTCGGCCAC). A NotI site (lowercase) in the Zeo-Not primer was introduced upstream of the EM promoter and an XbaI site (lowercase) in the Zeo-Xba primer was introduced immediately downstream of the stop codon (underlined). The resulting PCR fragment was digested with NotI and XbaI and used to replace the NotI/XbaI fragment containing the blasticidin resistance gene of 327ca(Bsd), resulting in the plasmid 327ca(Zeo).
The retroviral backbone was constructed in the plasmid 779/2795 (9). To insert the pol region from the Bryan strain of RSV, a 4.7-kb SacI-to-KpnI fragment containing the gag-pol region of 779/2795 was replaced with a SacI-to-KpnI fragment of RCASBP(A) (18), which generated the plasmid 779BP. The cassette described above was purified as a ClaI-to-MluI fragment from 327ca(Bsd) and 327ca(Zeo) and was inserted into the ClaI-to-MluI site of 779BP to generate 779BP-ca(Bsd) and 779BP-ca(Zeo), respectively. These vectors, however, contained an ampicillin resistance gene and a second replication origin derived from pBR322 that was present in the original 779/2795 plasmid. The SalI fragment containing both the ampicillin gene and the second origin was removed from the 779BP-ca(Bsd) and 779BP-ca(Zeo) plasmids to generate 779(ΔSal)BP-ca(Bsd) and 779(ΔSal)BP-ca(Zeo) plasmids. Finally, to introduce a 3" splice acceptor site upstream from the drug-resistant genes in these vectors, the splice acceptor-containing segment was taken from RCASBP(A) as a SalI-ClaI fragment. This fragment was used to replace the corresponding SalI-ClaI segment in 779(ΔSal)BP-ca(Bsd) and 779(ΔSal)BP-ca(Zeo), which generated RSVP(A)B and RSVP(A)Z, respectively.
Cells, transfection, and infection.
DF-1, a continuous line of chicken fibroblasts, was derived from EV-O embryos (7, 20). The cells were maintained in Dulbecco's modified Eagle medium (GIBCO BRL, Gaithersburg, Md.) supplemented with 5% fetal bovine serum, 5% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin (Quality Biological, Inc., Gaithersburg, Md.) per ml and incubated at 39°C with 5% CO2. Cells were passaged 1:3 at confluence with trypsin DeLarco (pH 6.8). Plasmid DNA was introduced into DF-1 cells by calcium phosphate precipitation (13). Precipitates containing 10 μg of DNA per 100-mm-diameter plate were incubated with subconfluent DF-1 cells for 4 h at 39°C and then with medium containing 15% glycerol for 5 min at 39°C. Cells were washed twice with phosphate-buffered saline and incubated in growth medium for 48 h. The transfected cells were passaged two to three times to let the viruses spread throughout the culture. Culture medium containing the virus was harvested and subjected to low-speed centrifugation to remove cellular debris. A portion of the infectious virions was used to infect fresh DF-1 cells. Selection for drug resistance was initiated at 48 h postinfection at 10 μg/ml for blasticidin (InVitrogen) and 400 μg/ml for zeocin (InVitrogen).
Cloning unintegrated viral DNA into Escherichia coli.
The circularized unintegrated viral DNA was recovered from infected cells by the method of Hirt (8). Three hundred nanograms of the recovered DNA was used to transform chemically competent E. coli DH5α (Life Technologies, Gaithersburg, Md.) or ElectroMax DH10B (Life Technologies) by electroporation. Electroporation was performed with the BTX Electro Cell Manipulator 600 (Biotechnologies and Experimental Research, Inc., San Diego, Calif.). Recipient cells were subjected to a single 5-ms pulse at a field strength of 5.5 kV/cm using a 2-mm gap Gene Pulser cuvette electrode (Bio-Rad, Hercules, Calif.) at room temperature. After a 1-h recovery period in NZY broth at 37°C, the transformed bacteria were plated onto low-salt Luria-Bertani plates containing either 100 μg of blasticidin per ml or 25 μg of zeocin per ml.
lac repressor-mediated recovery of integrated retroviral DNA.
lac repressor-mediated recovery was carried out essentially as described previously (16, 17). Either 200 or 100 μg of genomic DNA from the RSVP(A)B-infected DF-1 cells or RSVP(A)Z-infected DF-1 cells, respectively, was digested with ClaI. The reaction mixture was adjusted to 150 mM NaCl, 10 mM EDTA, 50 μg of bovine serum albumin per ml, and 10% (vol/vol) glycerol in a final volume of 600 μl. The digested DNA was incubated with 6 μg of lac repressor protein (kindly provided by P. Lu) for 30 min at room temperature. The DNA-lac repressor protein mixture was then filtered through nitrocellulose which had been pretreated with 0.4 M potassium hydroxide and washed twice with water. The nitrocellulose filter was washed three times with 1.5 ml of wash buffer (150 mM NaCl, 10 mM EDTA) and eluted twice with 1 ml of elution buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA, 10 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) for 30 min at 37°C. The enriched DNA was extracted with phenol-chloroform and precipitated with ethanol. The precipitated DNA was ligated with the Rapid DNA ligation kit (30 U/200 μl; Roche, Indianapolis, Ind.) for 18 h at 16°C. The ligated DNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 8 μl of water. Two microliters of DNA was used for transformation as described above.
Sequencing of the integrated viral DNA.
DNA sequences were determined by cycle sequencing with a primer specific for the U5 region of RCASBP using a PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.). Sequencing reactions were analyzed with an automated 373A DNA sequencer (Applied Biosystems). The sequence of the primer was 5"-ACCACATTGGTGTGCACCTGG-3".
Southern blot.
Genomic DNA was isolated from the infected cells using the cell culture DNA Midi kit (Qiagen, Valencia, Calif.) following the manufacturer's recommendations. For Southern blot analyses, 15 μg of genomic DNAs was digested with an appropriate restriction enzyme (EcoRI) and separated by electrophoresis on 1% agarose-Tris-acetate-EDTA buffer gels. DNA samples were denatured with alkali, neutralized, and transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) by capillary blotting with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were baked at 80°C under vacuum for 2 h. The hybridization probe (encompassing env to the long terminal repeat [LTR] region) was a 1.2-kb EcoRI fragment purified from RCASBP(A) and labeled with [α-32P]dCTP using Prime-It II (Stratagene, La Jolla, Calif.). Prehybridizations and hybridizations were carried out overnight at 42°C. Hybridization solution contained 50% formamide, 1× Denhardt solution, 0.5% sodium dodecyl sulfate, 7.5% dextran sulfate, and 5× SSC. After stringent washes, membranes were autoradiographed for 1 day.
RESULTS
Construction of replication-competent RSVP.
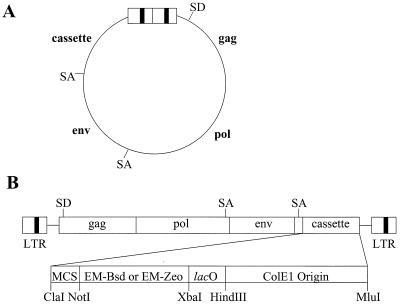
We constructed RSVP by inserting a cassette (MCS-drug resistance gene-lac operator-ColE1) into the replication-competent avian retroviral vector 779BP. 779BP is a derivative of 779/2795 that was constructed by replacing the pol region of 779/2795 with the corresponding pol region of the Bryan high-titer strain of RSV (18, 23). In terms of the retroviral vector portion carried by the plasmid, 779/2795 is the same as RCAN(A) and 779BP is the same as RCANBP(A). The details of this construction are given in Materials and Methods. The final form of the plasmids is shown in Fig. 1. The cassette contains one of two drug resistance genes, either blasticidin or zeocin resistance. The RSVP vectors contain a splice acceptor (SA) sequence immediately upstream of the cassette. The drug resistance gene is expressed in avian cells as a subgenomic mRNA from the viral promoter within the LTR, whereas in bacteria the same gene is expressed from the EM-7 bacterial promoter. The EM-7 promoter is synthetic and was isolated from the plasmids carrying the blasticidin and zeocin resistance genes (see Materials and Methods). To provide a simple and rapid purification of the vector DNA, the perfectly symmetric lac operator sequence was inserted in the cassette. Finally, the cassette contains a bacterial origin of replication (ColE1) to allow closed circular forms of viral DNA to replicate as plasmids in bacteria.
FIG. 1.
Schematic drawing showing the structure of the RSVP vector. (A) RSVP. The viral genes gag, pol, and env are shown (not to scale). The positions of the splice donor (SD) and SA are also shown. (B) Schematic representation of the cassette. EM-Bsd, EM-7 promoter-blasticidin resistance gene; EM-Zeo, EM-7 promoter-zeocin resistance gene; lacO, lac operator.
Recovery of unintegrated viral DNA in E. coli.
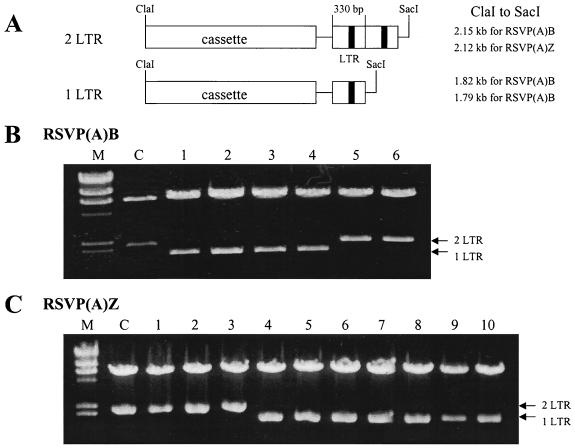
To rescue unintegrated retroviral DNA, the RSVP vectors were transfected into DF-1 cells and the transfected cells were passaged two times to let the viruses spread throughout the culture. RSVP(A)Z virus confers resistance to zeocin and the RSVP(A)B virus confers resistance to blasticidin. Because the viruses are replication competent, it was not possible to determine the titer in DF-1 cells. However, the RSVP vectors appear to replicate as efficiently after transfection (or infection) as the corresponding RCASBP(A) vectors. The viral supernatant was harvested and used to infect fresh DF-1 cells. To show that unintegrated viral DNAs present in these infected DF-1 cells were due to the successful viral infection and not because of carryover of RSVP plasmids from the transfected DF-1 cells, the circularized unintegrated retroviral DNAs were obtained from infected cells by the method of Hirt. The DNA was used to transform E. coli, selecting either blasticidin- or zeocin-resistant colonies. To demonstrate that the viral DNAs rescued from infected cells contained both of the circular forms of viral DNA (one-LTR and two-LTR circles), restriction digests were carried out. The starting plasmid, used to initiate infection, was a two-LTR circle. The only SacI site in RSVP is located between the LTR and the gag gene, and the ClaI site is between env and the LTR. Thus, digestion with ClaI and SacI should yield two fragments, a 6.8-kb vector DNA fragment and either a 2.1-kb fragment if the proviral DNA contains two LTRs or a 1.8-kb fragment if it contains one LTR (Fig. 2A). For RSVP(A)B, of the 13 recovered plasmids, 4 contained only one LTR, 2 contained two LTRs (Fig. 2B), and 7 were the products of autointegration events (data not shown) (15, 22). Similarly, for RSVP(A)Z, of the 20 recovered plasmids, 10 contained one LTR, 3 contained two LTRs (Fig. 2C), and 7 were autointegration products (data not shown). This result suggests that proviral DNAs observed in infected cells were due to successful replication of RSVP RNA and transfer of viral particles and not due to carryover of RSVP from the initial transfection.
FIG. 2.
Rescue of unintegrated retroviral DNA. Low-molecular-weight DNA was isolated by HIRT extraction and the extracted DNA was used to transform E. coli DH5α. The selection was for either blasticidin- or zeocin-resistant colonies. (A) Diagram showing the difference between two-LTR and one-LTR DNA segments. (B and C) Plasmids were digested with ClaI and SacI. The positions of DNAs containing two LTRs and one LTR are indicated. C, parental RSVP vector; M, size marker (λ DNA digested with HindIII).
Although we were able to get appropriate clones, the recovery was relatively low with chemically competent cells. We tried using electroporation to increase the transformation efficiency (see Materials and Methods for details). When the same amount of DNA sample was used to transform E. coli by electroporation, we obtained 238 and 802 transformants from RSVP(A)B and RSVP(A)Z, respectively. These results suggest that the efficiency of unintegrated plasmid recovery was 15 to 40 times higher with electroporation.
Rescue of integrated viral DNA.
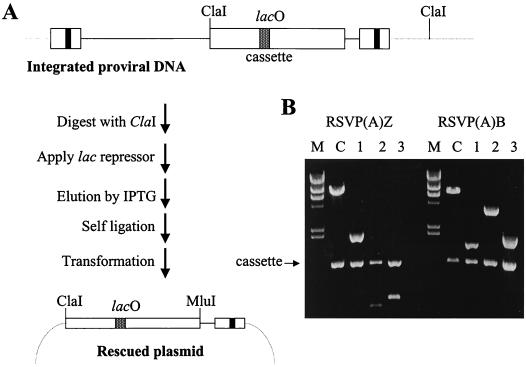
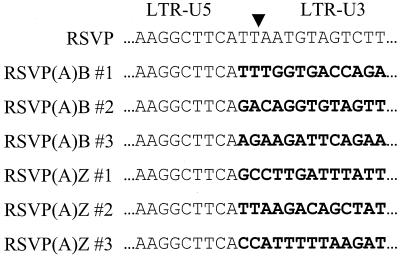
To rescue integrated viral DNA, genomic DNA was isolated from infected cells and digested with ClaI. Since one ClaI site comes from the RSVP vector, the second ClaI site must come from the adjacent cellular DNA (Fig. 3A). The ClaI-digested DNAs were enriched for viral sequences by binding to the lac repressor protein (see Materials and Methods) and then self-ligated. These enriched fractions were used to transform E. coli. Restriction cleavage analyses with ClaI and MluI showed that 3 of the 16 RSVP(A)B transformants and 3 of the 7 RSVP(A)Z transformants contained cellular DNA (Fig. 3B). For these plasmids, double digestion with ClaI and MluI generated a band representing the cassette insert. In addition, the digestion yielded another band of variable size due to the variable position of the nearest ClaI cleavage site in the cellular DNA. To demonstrate whether these inserts were derived from the cellular DNA, rescued plasmids were directly sequenced with a primer derived from the U5 sequence of the viral vector. As expected, all plasmids showed the integration-specific sequence feature that a CA dinucleotide pair was present at the site of joining to host DNA (Fig. 4). RSVP(A)B clones 2 and 3 and RSVP(A)Z clones 1 and 3 appear to contain novel inserts derived from the cellular DNA. However, sequencing results revealed that RSVP(A)B clone 1 and RSVP(A)Z clone 2 contained inserts identical to part of the RSVP vector sequence, indicative of autointegration (22).
FIG. 3.
Rescue of integrated retroviral DNA. (A) Schematic diagram of lac repressor-mediated recovery (see Materials and Methods for details). (B) Plasmids were digested with ClaI and MluI. The position of the cassette insert is indicated. C, parental RSVP vector; M, size marker (λ DNA digested with HindIII).
FIG. 4.
Sequences of integrated retroviral DNA. The inverted triangle indicates the boundary between the two LTRs. Sequences flanking the LTR are shown in bold.
As described above, we tried electroporation to obtain additional integration sites. When the same amount of enriched DNA sample was used to transform E. coli by electroporation, we were able to get 1,760 and 886 transformants from RSVP(A)B and RSVP(A)Z, respectively; thus, the efficiency of enriched plasmid recovery was more than 100 times higher with electroporation.
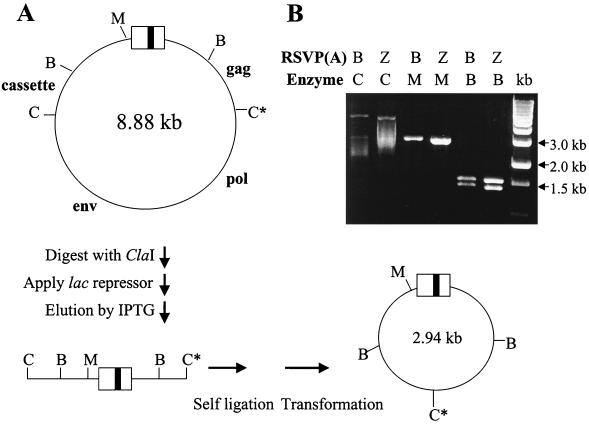
When we recovered integrated viral DNA, in addition to the plasmids containing integration sites we also obtained plasmids of approximately 3 kb in more than half of the clones. Further analyses of these plasmids revealed that they were by-products derived from the circularized unintegrated viral DNAs. There is a second ClaI site within the gag gene (Fig. 5A). In plasmids grown in E. coli, this site is subject to dam methylation. However, there is no dam methylation in avian cells. When genomic DNA isolated from the infected DF-1 cells was digested with ClaI, both ClaI sites in the circular unintegrated viral DNAs were cleaved and the cassette (lacO)-containing DNA segment was enriched by binding to the lac repressor protein. These 3-kb plasmids were generated by self-ligation and were recovered by transformation into E. coli. After transformation and growth in E. coli, the ClaI site is again subject to dam methylation and thus digestion with ClaI did not cut the 3-kb mini plasmids. Digestion with MluI cut the mini plasmids once and digestion with BamHI generated two fragments, of 1.54 and 1.39 kb, as expected (Fig. 5B). The recovery of the 3-kb mini plasmid provides additional evidence that the unintegrated viral DNAs were generated by successful viral infection rather than carryover of initial RSVP plasmids from the transfected DF-1 cells.
FIG. 5.
Major by-product obtained in the recovery of integrated viral DNA. (A) Schematic drawing of the generation of the plasmid. M, MluI; B, BamHI; C, ClaI; C*, ClaI site within gag gene subjected to dam methylation in E. coli but not in avian cells. (B) Restriction enzyme analyses of the plasmid. Digestion with ClaI did not cut the plasmid due to dam methylation. Digestion with MluI cut the plasmid once and digestion with BamHI generated two fragments, of 1.54 and 1.39 kb, as expected. kb, DNA ladder in kilobases.
Stability of vectors.
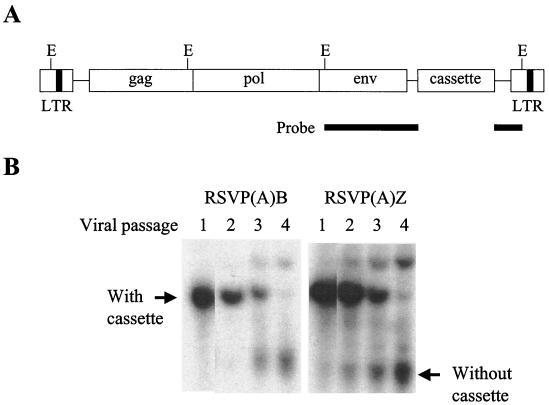
To determine that the cassette insert is reasonably stable upon viral passage in avian cells, genomic DNAs derived from virus that had been passaged under nonselective conditions were digested with the restriction enzyme EcoRI and analyzed by Southern blot. To provide equivalent hybridization to EcoRI fragments that contained the cassette and the related fragments from viral genomes that had lost the cassette, the probe was prepared from RCASBP(A) as a 1.2-kb EcoRI gel-purified DNA fragment (Fig. 6A). As can be seen in Fig. 6B, the cassette insert was present in the vector up to three viral passage for both drugs, but there was significant loss in the fourth viral passage, suggesting that RSVP vectors are stable for approximately three viral passages. The efficiency of transformation does affect the apparent stability of the insert; a relatively efficient transformation gives a larger burst of virus and fewer rounds of replication are required to get the virus stock. With this caveat, the stability of the two drug cassettes are similar to an “average” insert in RCASBP(A).
FIG. 6.
Southern transfer analyses of the stability of the RSVP vector. (A) The probe was prepared from RCASBP(A) as a 1.2-kb EcoRI fragment to provide equal opportunity to hybridize to EcoRI fragments that contained (or had lost) the cassette. EcoRI recognition sites are indicated (E) (not to scale). (B) Detection of the cassette insert in the genomic DNA derived from cells infected with the RSVP(A)B and RSVP(A)Z vectors. Genomic DNA was digested with EcoRI, resolved in an agarose gel, transferred onto a nitrocellulose membrane, and hybridized with 32P-labeled DNA prepared from the 1.2-kb EcoRI fragment of RCASBP(A). The larger band represents a 2.4-kb EcoRI fragment containing the insert, whereas the smaller band represent a 1.2-kb EcoRI fragment lacking the insert.
DISCUSSION
We have developed two related replication-competent retroviral shuttle vectors (RSVPs). The sequences that permit the viral DNA to replicate as a plasmid and to be selected in E. coli (either blasticidin resistance or zeocin resistance) are small (approximately 1.2 kb), which should make it possible to insert additional sequence in these vectors. The drug resistance genes were inserted in a fashion that permits expression of the selectable markers (via a spliced message) in avian cells; as expected, infection with the vectors confers resistance. The inserts are relatively stable during viral passage; no appreciable loss of the inserts was seen until the fourth viral passage.
One of the problems with recovering shuttle vector DNA after infection is the large amount of contaminating cellular DNA. Unintegrated viral DNA is, in general, easier to recover and clone than integrated DNA, presumably because it is possible to use physical fractionation to enrich for the closed circular forms of unintegrated viral DNA.
We particularly wanted to develop a vector system that makes it simple to recover both unintegrated and integrated DNA. To simplify enrichment of viral DNA, we included the lacO sequence in the inserted segment. DNAs containing the lacO sequence can be rapidly enriched by binding to lac repressor protein (16, 17). The DNA-protein complex is captured on a nitrocellulose filter, and then the DNA is selectively eluted by the addition of IPTG. This simple protocol allowed us to directly clone integration sites. In the initial experiments we used chemically competent cells and were able to get valid clones. However, the cloning of unintegrated viral DNA was more-than-100-fold more efficient when the DNA was introduced into E. coli by electroporation.
In addition to recovering integration sites, we want to use these vectors to aid in the recovery of viral vectors that have been adapted by passage. In previous experiments we have found that, in some cases, the initial version of a vector replicated poorly. In some cases, it is possible, with a replication-competent virus, to adapt the vector by passage (1). Variant viruses that grow better have a selective advantage. The adapted virus is obtained simply by repeated passage of the viral stock either in cultured cells or in infected embryos. However, to be used as a vector, the adapted virus must be molecularly cloned and characterized. A shuttle vector should be quite useful in adaptation protocols, provided that the insert is stable enough to be retained during the adaptation process. The zeocin and blasticidin inserts are stable for three viral passages in the RSVP vectors, even in the absence of drug selection, and should be useful for recovering adapted viruses.
The plasmid cassettes are small and are flanked by ClaI sites. It should be possible to introduce these prokaryotic cassettes into other viral vectors. In cases in which the vector will not generate an appropriately spliced message for the selectable markers, the cassette could be linked to an internal promoter or an internal ribosome entry site.
Acknowledgments
We are most grateful to Ponzy Lu, who generously provided the purified Lac repressor used in the experiments described here. We are also grateful to the NCI-Frederick DNA sequencing facility for help in determining sequences and to Hilda Marusiodis for help in preparing the manuscript.
This research was supported by the National Cancer Institute and the National Institute for General Medical Sciences.
REFERENCES
- 1.Barsov, E. V., W. S. Payne, and S. H. Hughes. 2001. Adaptation of chimeric retroviruses in vitro and in vivo: isolation of avian retroviral vectors with extended host range. J. Virol. 75:4973-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berdy, J. 1980. Handbook of antibiotic compounds, vol. 4, p. 459-491. CRC Press, Boca Raton, Fla.
- 3.Calmels, T., M. Parriche, H. Burand, and G. Tiraby. 1991. High efficiency transformation of Tolypocladium geodes conidiospores to phleomycin resistance. Curr. Genet. 20:309-314. [DOI] [PubMed] [Google Scholar]
- 4.Drocourt, D., T. Calmels, J.-P. Reynes, M. Baron, and G. Tiraby. 1990. Cassettes of the Streptoalloteichus hidustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 18:4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatignol, A., M. Baron, and G. Tiraby. 1987. Pleomycin resistance encoded by the ble gene from transposon Tn5 as a dominant selectable marker in Saccharomyces cerevisiae. Mol. Gen. Genet. 207:342-348. [DOI] [PubMed] [Google Scholar]
- 6.Gatignol, A., H. Durand, and G. Tiraby. 1988. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett. 230:171-175. [DOI] [PubMed] [Google Scholar]
- 7.Himly, M., D. N. Foster, I. Botoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leucosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 8.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 9.Hughes, S. H., and E. Kosik. 1984. Mutagenesis of the region between env and src of the SR-A strain of the Rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology 136:89-99. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izumi, M., H. Miyazawa, T. Kamakura, I. Yamaguchi, T. Endo, and F. Hanaoka. 1991. Blasticidin S-resistance gene (bsr): a novel selectable marker for mammalian cells. Exp. Cell Res. 197:229-233. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, M., A. Takatsuki, and I. Yamaguchi. 1994. Blasticidin S deaminase gene from Aspergillus terrus (BSD): a new drug resistance gene for transfection of mammalian cells. Biochim. Biophys. Acta 1219:653-659. [DOI] [PubMed] [Google Scholar]
- 13.Kingston, R. E., C. A. Chen, and H. Okayama. 1989. Introduction of DNA into mammalian cells. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 14.Mulsant, P., G. Tiraby, J. Kallerhoff, and J. Perret. 1988. Pleomycin resistance as a dominant selectable marker in CHO cells. Somat. Cell Mol. Genet. 14:243-252. [DOI] [PubMed] [Google Scholar]
- 15.Olsen, J. C., C. Bova-Hill, D. P. Grandgenett, T. P. Quinn, J. P. Manfredi, and R. Swanstrom. 1990. Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants. J. Virol. 64:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA 87:6019-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. USA 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petropoulos, C. J., and S. H. Hughes. 1991. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J. Virol. 65:3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadler, J. R., H. Sasmor, and J. J. Betz. 1983. A perfectly symmetric lac operator binds to the lac repressor very tightly. Proc. Natl. Acad. Sci. USA 80:6785-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O derived cell line DF-1 supports the efficient replication of avian leucosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 22.Shoemaker, C., J. Hoffman, S. P. Goff, and D. Baltimore. 1981. Intramolecular integration within Moloney murine leukemia virus DNA. J. Virol. 40:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudol, M., T. L. Lerner, and H. Hanafusa. 1986. Polymerase-defective mutant of the Bryan high-titer strain of Rous sarcoma virus. Nucleic Acids Res. 14:2391-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi, S., K. Hirayama, K. Ueda, H. Sakai, and H. Yonehara. 1958. Blasticidin S, a new antibiotic. J. Antibiot. 11a:1-5. [PubMed] [Google Scholar]
- 25.Yamaguchi, H., C. Yamamoto, and N. Tanaka. 1965. Inhibition of protein synthesis by blasticidin S. I. Studies with cell-free system from bacterial and mammalian cells. J. Biochem. 57:667-677. [PubMed] [Google Scholar]