Abstract
Adenovirus binds to mammalian cells via interaction of fiber with the coxsackie-adenovirus receptor (CAR). Redirecting adenoviral vectors to enter target cells via new receptors has the advantage of increasing the efficiency of gene delivery and reducing nonspecific transduction of untargeted tissues. In an attempt to reach this goal, we have produced bifunctional molecules with soluble CAR (sCAR), which is the extracellular domain of CAR fused to peptide-targeting ligands. Two peptide-targeting ligands have been evaluated: a cyclic RGD peptide (cRGD) and the receptor-binding domain of apolipoprotein E (ApoE). Human diploid fibroblasts (HDF) are poorly transduced by adenovirus due to a lack of CAR on the surface. Addition of the sCAR-cRGD or sCAR-ApoE targeting protein to adenovirus redirected binding to the appropriate receptor on HDF. However, a large excess of the monomeric protein was needed for maximal transduction, indicating a suboptimal interaction. To improve interaction of sCAR with the fiber knob, an isoleucine GCN4 trimerization domain was introduced, and trimerization was verified by cross-linking analysis. Trimerized sCAR proteins were significantly better at interacting with fiber and inhibiting binding to HeLa cells. Trimeric sCAR proteins containing cRGD and ApoE were more efficient at transducing HDF in vitro than the monomeric proteins. In addition, the trimerized sCAR protein without targeting ligands efficiently blocked liver gene transfer in normal C57BL/6 mice. However, addition of either ligand failed to retarget the liver in vivo. One explanation may be the large complex size, which serves to decrease the bioavailability of the trimeric sCAR-adenovirus complexes. In summary, we have demonstrated that trimerization of sCAR proteins can significantly improve the potency of this targeting approach in altering vector tropism in vitro and allow the efficient blocking of liver gene transfer in vivo.
Adenoviral vectors efficiently transduce a wide variety of cell types, which is one reason why they are prominent gene transfer vehicles in the field of gene therapy. However, systemic administration of vector leads to widespread distribution in tissue, which is not favorable if the desired target is a specific tissue or cell type. In addition, transduction of nontarget cells may have undesirable side effects. For example, it has been demonstrated that adenoviral vector transduction of dendritic cells in mice augments the immune response against vector, leading to more rapid elimination of transduced cells by cytotoxic T lymphocytes (20). Successful vector targeting strategies may overcome these problems by directing the entire vector dose to the appropriate site. This may improve the safety profile of the vector and permit the use of lower vector doses, which would be less toxic and potentially less immunogenic. Additionally, adenoviral vector targeting may permit transduction of cell types that are refractory to adenovirus infection. For example, carcinoma cells, which are targets for numerous gene therapy applications, are typically inefficiently transduced by adenoviral vectors, and alterations in receptor tropism have been shown to enhance gene delivery in vitro and efficacy in vivo with local delivery (5, 39). Furthermore, improved transduction efficiencies of certain tissues, such as the vascular endothelium, would expand the clinical utility of adenoviral-vector-mediated gene therapy in these settings (18, 28).
Adenovirus tropism is determined by attachment to specific cell surface molecules (31, 42). Many adenovirus serotypes, except those in subgroup B, bind to a cell surface molecule called the coxsackie-adenovirus receptor (CAR). Human CAR is a 365-amino-acid transmembrane protein which has an apparent molecular mass of 46 kDa and consists of a short leader, a 222-amino-acid extracellular domain, a membrane-spanning helical domain, and a 107-amino-acid intracellular domain (2, 3). The extracellular region contains two immunoglobulin (Ig)-related structural domains termed IgV and IgC2 (13). CAR is widely expressed in vivo, accounting for the widespread distribution of systemically administered adenoviral vector in tissue. Adenovirus serotype 3 (Ad3), which belongs to subgroup B, has been shown to bind to a different, as-yet-unidentified receptor (38). Entry of group C adenoviruses such as Ad5 into cells involves two distinct virus-cell interactions. First, high-affinity binding of Ad5 to the cell occurs via interaction of the viral fiber protein with CAR. Subsequently, the virus penton base protein binds to cellular αv integrins to mediate internalization. This step promotes virus internalization and subsequent gene transfer. More recently, a third cell surface interaction with heparan sulfate glycosaminoglycans has been demonstrated through competition analysis with group C adenoviruses Ad2 and Ad5 but did not influence entry of Ad3, a group B virus (6). As these studies demonstrate, multiple cell surface receptor binding events are needed to promote efficient viral entry, and these must be taken into consideration when strategies that modify the cell entry process are developed.
Several strategies have been used to alter the receptor tropism and binding specificity of the adenoviral particles. These include replacement of the fiber knob domain with a knob from another serotype which binds to an alternative receptor (25, 37) and insertion of peptides at the carboxyl terminus (29, 45) or in the exposed HI loop of the fiber knob (18, 24). Except for the fiber knob switch, the simple addition of peptide ligands into the fiber knob results in the expansion of the viral tropism, which may be suitable for gene therapy applications that require local delivery, such as vascular gene therapy or local delivery directly into a solid tumor mass. However, systemic delivery of adenovirus that specifically transduces a certain cell type or tissue will require a combination of elimination of the natural receptor interaction with the introduction of a novel receptor tropism by using high-affinity targeting ligands. The first step in detargeting has been accomplished through mutagenesis of specific amino acids within the fiber knob that are involved in CAR interaction. Several groups have identified critical amino acids involved in receptor interaction (19, 22, 23, 36). The potency of these mutations in vivo for successful detargeting is currently under evaluation, but recent studies suggest that fiber mutations alone will not be sufficient (26) and may need to be combined with penton base mutations that eliminate interaction with integrins, the second receptor involved in efficient adenoviral cell entry.
Recently, success in eliminating the natural receptor tropism with the introduction of a new tropism has been achieved with multicomponent systems utilizing bifunctional proteins. An advantage of this approach is that it does not require the time-consuming generation of genetically modified adenoviral vectors. These systems simultaneously block interaction with CAR and redirect receptor specificity toward specific cell types by using bifunctional antibodies (14, 40), antibody-ligand conjugates (10, 34, 44), or soluble CAR-ligand fusion proteins (7). To date, the most advanced of these multicomponent systems for in vivo targeting is the antibody-ligand conjugate comprising an antifiber Fab fragment chemically conjugated to fibroblast growth factor (FGF) (16, 34). Systemic delivery of adenovirus with this reagent has resulted in reduced toxicity and improved efficacy in vivo with mouse tumor models. However, this system may not be amenable to efficient scale-up and development, as it involves the production of three separate components, and it may benefit by conversion into a fusion protein.
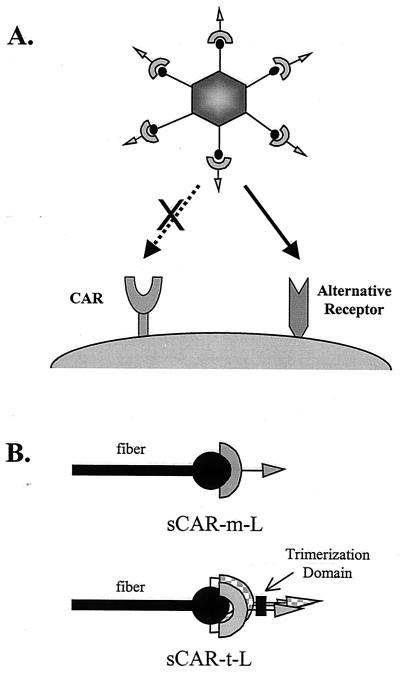
We have developed a multicomponent system for adenoviral targeting that employs the extracellular domain of CAR (soluble CAR [sCAR]). This approach involves the generation of a bifunctional protein consisting of sCAR genetically fused to targeting ligands. The sCAR fusion protein binds to the fiber knob of an adenoviral vector to block interaction with the cell surface CAR and subsequently redirects the vector to an alternative receptor (Fig. 1). This report describes the characterization of sCAR bifunctional proteins both in vitro and in vivo and demonstrates that the potency of this approach can be significantly improved by trimerization of sCAR, thereby enhancing its interaction with the fiber knob and resulting in successful liver detargeting in vivo. However, when normal mice were used, in vivo retargeting was not demonstrated with either of the two peptide ligands evaluated. Size determinations of these complexes suggest that they may be sterically hindered from entry into normal vascular beds. Further studies are in progress to evaluate the use of trimeric-sCAR-ligand fusion proteins for targeting adenovirus in vivo using mouse tumor models.
FIG. 1.
Targeting adenoviral vectors using sCAR bifunctional fusion proteins. (A) The sCAR targeting strategy consists of adding a protein molecule comprising sCAR genetically fused to a targeting ligand, represented by the arrow, to an adenoviral vector. Interaction of the bifunctional sCAR targeting molecule with the fiber knob blocks the normal tropism of the vector and will redirect the viral complex to an alternate receptor. (B) The interaction of the sCAR targeting molecule with the fiber knob can be improved via trimerization using a heterologous trimerization domain. The proposed interaction of the sCAR-m-ligand (sCAR-m-L) and sCAR-t-L targeting molecules with the fiber is shown schematically.
MATERIALS AND METHODS
Plasmids and construction of sCAR fusion constructs.
Human cDNA encoding CAR was cloned by reverse transcription-PCR. The published CAR cDNA sequence (GenBank accession number Y07593) was used to design oligonucleotide primers for amplification. Total RNA was isolated from cultured HeLa cells with RNAzol (Tel-Test, Friendswood, Tex.), and 1 μg was reverse transcribed with a reverse transcription-PCR system (Perkin-Elmer, Norwalk, Conn.). Primers were designed to amplify the entire coding sequence of the full-length CAR cDNA starting from the start codon, ATG, and ending with the termination codon, TAG. For cloning purposes, the sense primer contained a BamHI restriction site in addition to a Kozak consensus sequence (5"-GTAGGATCCGCCACCATGGCGCTCCTGCTG-3"). The antisense primer contained an EcoRI restriction site (5"-GCGCGAATTCCTATACTATAGACCCATCCT-3"). A 1.1-kb amplified product of the expected size was obtained and cloned into pcDNA3.1 (Invitrogen, San Diego, Calif.) to generate the plasmid pcDNAhCAR. The nucleotide sequence of the extracellular domain was as expected.
Construction of recombinant plasmids encoding sCAR.
To generate a plasmid expressing human sCAR with a six-histidine (six-H) purification tag at the carboxy terminus, sequences encoding the extracellular domain of CAR were amplified by PCR from the pcDNAhCAR construct containing the entire CAR cDNA, described above. The sense primer contained an NheI site upstream of the ATG codon and had the sequence 5"-ACTAGCTAGCGCCGCCACCATGGCGCTC-3". The antisense primer contained the end of the sequence for the extracellular domain of CAR immediately followed by an XhoI site (to allow insertion of targeting ligands), a six-H tag, a TAG codon, and an XbaI restriction site and had the sequence 5"-GTGATCTAGACTAGTGATGATGGTGATGGTGCTCGAGAGCTTTATTTGAAGGAGGG-3". The PCR product was analyzed by agarose gel electrophoresis, digested with NheI and XbaI, and ligated into the corresponding sites of the expression vector pCI-neo (Promega Corporation, Madison, Wis.) to generate pCI-neo-sCARa.
To permit insertion of targeting ligands at the end of sCAR, the following modifications were made to pCI-neo-sCARa. First, the NotI site in the multiple cloning region was destroyed by digestion with NotI followed by blunting with T4 DNA polymerase and religating. Second, a pair of complementary oligonucleotides were synthesized and annealed to form a DNA duplex. The annealed oligonucleotides contained XhoI-compatible overhangs at both ends and contained a linker domain sequence, to provide better presentation of the targeting ligand to its receptor, followed by a NotI restriction site, for insertion of ligands, and a factor Xa cleavage site, to permit removal of the six-H tag. The sequences of the oligonucleotides were 5"-TCGAACCATCAGCCTCCGCATCTGCTTCCGCCCCTGGATCCGCGGCCGCCATTGAGGGCCGCC-3" and 5"-TCGAGGCGGCCCTCAATGGCGGCCGCGGATCCAGGGGCGGAAGCAGATGCGGAGGCTGATGGT-3". The annealed oligonucleotides were ligated into the XhoI site of pCI-neo-sCARa to generate pCI-neo-sCARb.
To construct expression plasmids encoding targeting ligands at the carboxy terminus of sCAR, pairs of complementary oligonucleotides were synthesized and annealed to form a DNA duplex encoding the desired targeting ligands. The DNA duplexes were designed to contain NotI-compatible overhangs on both ends so that the fragment could be inserted into the NotI site of pCI-neo-sCARb. Two different targeting ligands were fused to the carboxy terminus of sCAR. One ligand consists of the nine-amino-acid sequence CDCRGDCFC and is termed cyclic RGD (cRGD) (33). The oligonucleotides that were synthesized to generate cRGD were 5"-GGCCTGCGATTGCCGTGGTGATTGCTTTTGCGC-3" and 5"-GGCCGCGCAAAAGCAATCACCACGGCAATCGCA-3". The resulting plasmid was designated pCI-neo-sCAR-cRGD. The second targeting ligand consists of two tandem copies of amino acids 141 to 155, (LRKLRKRLLRDADDL)2, derived from apolipoprotein E (ApoE), which binds to members of the low-density lipoprotein (LDL) receptor family (11, 32). The oligonucleotides that were synthesized to generate the ApoE targeting ligand were 5"-GGCCCTGCGCAAGCTGCGTAAGCGGCTCCTCCGCGATGCCGATGACCTGGC-3" and 5"-GGCCGCCAGGTCATCGGCATCGCGGAGGAGCCGCTTACGCAGCTTGCGCAG-3". The resulting plasmid was designated pCI-neo-sCAR-ApoE.
Plasmids encoding trimerized sCAR were constructed as follows. First a pair of complementary oligonucleotides were synthesized and annealed to form a DNA duplex. The duplex contained XhoI-compatible overhangs on each end and also encoded a glycine-serine linker element followed by a NotI site and then a factor Xa cleavage site. The duplex was ligated into the XhoI site of pCI-neo-sCARa, described above, to generate pCI-neo-sCAR-Not. The oligonucleotides were designed so that the 5" XhoI site was destroyed but the 3" site remained intact. The sequences of the complementary oligonucleotides were 5"-TCGAAGGAGGAGGAGGAAGTGGAGGAGGAGGAAGTGGAGGAGGAGGAAGTGGATCCGCGGCCGCCATTGAGGGCCGCC-3" and 5"-TCGAGGCGGCCCTCAATGGCGGCCGCGGATCCACTTCCTCCTCCTCCACTTCCTCCTCCTCCACTTCCTCCTCCTCCT-3". A second pair of complementary oligonucleotides were synthesized and annealed to form a DNA duplex encoding a trimerization domain derived from the isoleucine variant of the yeast GCN4 leucine zipper molecule (17). The duplex contained NotI-compatible overhangs on both ends. The oligonucleotides were designed so that the 5" NotI site was destroyed but the 3" site remained intact. The sequences of the oligonucleotides were 5"-GGCCATGAAACAAATTGAAGACAAGATTGAAGAAATTCTATCAAAAATTTATCACATTGAAAACGAAATTGCCAGAATTAAAAAACTAATTGGCGAAGC-3" and 5"-GGCCGCTTCGCCAATTAGTTTTTTAATTCTGGCAATTTCGTTTTCAATGTGATAAATTTTTGATAGAATTTCTTCAATCTTGTCTTCAATTTGTTTCAT-3". The duplex was ligated into the NotI site of pCI-neo-sCAR-Not to generate pGS-triCARa. Next, pGS-triCARa was digested with both NotI and XhoI, and the ends were blunted with mung bean nuclease and ligated to generate pGS-triCARb, which encodes sCAR followed by a glycine-serine linker, the isoleucine variant GCN4 trimerization domain, and a six-H tag.
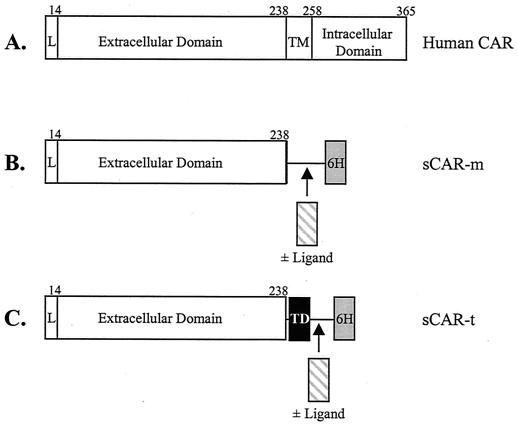
Plasmids encoding trimerized versions of sCAR containing an ApoE targeting ligand were constructed as follows. First, a pair of complementary oligonucleotides were synthesized and annealed to form a DNA duplex encoding the ApoE targeting ligand consisting of two tandem copies of amino acids 141 to 155 from ApoE. The duplex contained a NotI site near the 5" end and a XhoI site near the 3" end. The sequences of the oligonucleotides were 5"-GCGGCCGCGCTGCGCAAGCTGCGTAAGCGGCTCCTCCGCGATGCCGATGACCTGCTGCGCAAGCTGCGTAAGCGGCTCCTCCGCGATGCCGATGACCTGCTCGAGCACCAT-3" and 5"-ATGGTGCTCGAGCAGGTCATCGGCATCGCGGAGGAGCCGCTTACGCAGCTTGCGCAGCAGGTCATCGGCATCGCGGAGGAGCCGCTTACGCAGCTTGCGCAGCCGGCCGC-3". The duplex was digested with both NotI and XhoI and ligated into pGS-triCARa which had been digested with both NotI and XhoI, to generate pGS-triCARa-ApoE. Next, a linker element was inserted upstream of the targeting ligand as follows. A pair of complementary oligonucleotides were synthesized and annealed to generate a DNA duplex encoding a serine-alanine linker with NotI sites near each end. The sequences of the oligonucleotides were 5"-GGCCCCATCAGCCTCCGCATCTGCTTCCGCCCCTGGATCCGC-3" and 5"-GGCCGCGGATCCAGGGGCGGAAGCAGATGCGGAGGC TGATGGGGCCGC-3". The duplex was digested with NotI and ligated into the NotI site of pGS-triCARa-ApoE to generate pGS-triCAR-ApoE. A plasmid encoding a trimerized version of sCAR containing the cRGD targeting ligand was constructed as follows. First, a pair of complementary oligonucleotides were synthesized and annealed to form a DNA duplex encoding the cRGD ligand. The sequences of the oligonucleotides were as follows: 5"-GGCCGCGTGCATTGCCGTGGTGATTGCTTTTGCGCCC-3" and 5"-TCGAGGGCGCAAAAGCAATCACCACGGCAATCGCACGC-3". At the 5" end, the duplex contained an overhang compatible with a NotI site and the overhang on the 3" end was compatible with a XhoI site. The duplex was ligated into the plasmid pGS-triCAR-ApoE, which had been digested with both NotI and XhoI, to generate pGS-triCAR-RGD. The monomeric and trimeric sCAR proteins (sCAR-m and sCAR-t) with and without a targeting ligand are shown in Fig. 2.
FIG. 2.
Full-length CAR and sCAR constructs. A schematic of the full-length human CAR protein compared with sCAR-m and sCAR-t is shown. (A) Structure of the full-length CAR protein, including the leader sequence (L), the extracellular domain, the transmembrane domain (TM), and the intracellular domain; (B) sCAR-m, comprising the extracellular domain of CAR, a serine-alanine linker, and a six-H tag with and without a targeting peptide ligand inserted between the sCAR and a six-H tag domains; (C) sCAR-t comprising the extracellular domain of CAR, a serine-alanine linker, the GCN4 heterologous trimerization domain (TD), and a six-H tag with and without a peptide targeting ligand. Amino acids are numbered above each chimeric fusion protein. The cloning and construction of the six CAR constructs used in these studies are described in Materials and Methods.
Production and purification of sCAR proteins.
To generate sCAR protein, an expression plasmid encoding the protein was introduced into COS-7 cells by electroporation. Prior to electroporation, the cells were resuspended in RPMI, 20% fetal bovine serum (FBS), and 10 mM HEPES at a final concentration of 107 cells per milliliter. A 500-μl aliquot of cells was placed in a 4-mm electroporation cuvette, and 20 μg of plasmid DNA was added to the cells. The mixture was kept on ice for 10 min. Electroporation was performed with a BTX instrument with the settings R3, 2,100 μF, and 264 V. Following electroporation, the cuvette was placed on ice for 30 min, and then the cells were placed in a 10-cm tissue culture dish with Dulbecco's modified Eagle medium plus 10% FBS. The sCAR proteins were continuously secreted into the medium over the course of at least 1 week. However, sCAR-t fused to targeting ligands was not secreted into the medium; consequently, sCAR-t was purified from cell lysates.
To isolate sCAR-m with or without a targeting ligand and sCAR-t proteins without a targeting ligand, the medium was removed from the cells at various times after electroporation and replaced with fresh medium. Typically, medium was collected every other day for a week. The medium was dialyzed against phosphate-buffered saline (PBS) to remove glutamine and any other components, which may interfere with six-H tag binding to the nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, Inc., Valencia, Calif.). Ni-NTA resin was added to the medium and mixed for 2 h at 4°C. The resin with bound protein was pelleted by centrifugation and washed several times with 50 mM NaH2PO4-0.5 M NaCl-60 mM imidazole, pH 8.0. Fusion proteins were eluted from the resin with 50 mM NaH2PO4-0.5 M NaCl-400 mM imidazole, pH 8.0. Removal of imidazole, buffer exchange, and concentration of purified proteins were performed with Centricon Plus-20 concentrators (Millipore Corporation, Bedford, Mass.). The purified proteins were stored at −70°C in PBS plus 10% glycerol.
To generate sCAR-t fused to a targeting ligand, an expression plasmid encoding the fusion protein was introduced into COS-7 cells by electroporation. The day after electroporation, the medium was removed from the cells and replaced with fresh medium. The next day, the cells were washed with PBS and then lysed by adding 2 ml of radioimmunoprecipitation buffer (5 mM Tris [pH 8.0], 0.15 M NaCl, 1% deoxycholate, 1% NP-40) plus 1 mM phenylmethylsulfonyl fluoride. The cells were sheared by passage through a 21-gauge needle. The cell lysate was incubated on ice for 30 min and then centrifuged at 8,160 × g for 20 min to remove cell debris. The supernatant was transferred to a clean tube, and Ni-NTA resin was added (150 μl per 10-cm plate). The tube was rocked at 4°C for 1 h. The resin with bound sCAR fusion protein was collected by centrifugation at 3,500 rpm for 5 min. The supernatant was removed, and the resin was transferred to a clean tube and washed twice with 800 μl of wash buffer (50 mM NaPO4, 500 mM NaCl, 40 mM imidazole, 10% glycerol), a third time with wash buffer containing 60 mM imidazole, and a fourth time with wash buffer containing 80 mM imidazole. The sCAR fusion protein was eluted from the resin by using 100 to 400 μl of elution buffer (50 mM NaPO4, 500 mM NaCl, 400 mM imidazole, 10% glycerol).
Production of anti-CAR antiserum.
The extracellular domain of CAR was also expressed in bacteria. Recombinant sCAR-m was purified from bacterial occlusion bodies by metal ion affinity chromatography on Ni-NTA-Sepharose (Qiagen) following standard protocols. Purified sCAR-m was used for immunization of New Zealand White rabbits according to standard protocols (Loftstrand Labs, Ltd., Gaithersburg, Md.). The purified IgG fraction was isolated with the ImmunoPure IgG protein A purification kit (Pierce Inc., Rockford, Ill.) as recommended by the manufacturer. Immunoreactivity was confirmed by Western immunoblot analysis against the purified sCAR-m protein.
Western immunoblot analysis.
For Western immunoblot analysis, a 300-ng aliquot of each sCAR protein was subjected to sodium dodecyl sulfate (SDS)-4 to 12% polyacrylamide gel electrophoresis (PAGE). Following electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane. The membrane was blocked for 1 h at room temperature in TBS (10 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA) containing 5% dried milk and then washed three times, 5 min each, at room temperature with wash buffer (TBS plus 0.1% Tween). Next, the membrane was incubated overnight at 4°C with either a 1:1,000 dilution of a purified IgG fraction from rabbit antiserum raised against human CAR or a 1:1,000 dilution of rabbit polyclonal antibody recognizing the six-H tag (Santa Cruz Biotechnology, Santa Cruz, Calif.). The primary antibody was diluted with TBS plus 5% dried milk and 0.1% Tween. The membranes were washed three times, 10 min each, at room temperature with wash buffer. Next, the membranes were incubated with a 1:4,000 dilution of an anti-rabbit horseradish peroxidase-conjugated antibody (Amersham, Arlington Heights, Ill.) for 1 h at room temperature. The secondary antibody was diluted with TBS plus 5% dried milk and 0.1% Tween. After the washing, the membrane was immersed in Super Signal (Pierce) for 5 min at room temperature and then exposed to film.
Cross-linking of sCAR proteins.
Preparations of sCAR-m and sCAR-t were incubated with bis(sulfosuccinimidyl)suberate (BS3) (Pierce) for cross-linking analysis. BS3 was dissolved in 5 mM sodium citrate (pH 5.0) at 20 mM. A 3-μg aliquot of either sCAR-m or sCAR-t was incubated for 30 min at room temperature in 5 mM BS3. Next, 1 μl of 1 M Tris (pH 7.5) was added to the reaction mixture to yield a final concentration of 50 mM Tris, and the mixture was incubated another 15 min at room temperature. An aliquot of the mixture containing 2.1 μg of sCAR was analyzed by SDS-4 to 12% PAGE under denaturing conditions.
Fiber receptor binding assay.
As described previously, the interaction of Ad5 fiber protein (5F) with CAR was evaluated with a fiber receptor-binding assay (38). Briefly, 5F was radioactively labeled in vitro by using the T7 coupled transcription-translation reticulocyte lysate system (Promega). A 1-μg aliquot of plasmid DNA encoding fiber was incubated with the system components, including 40 μCi of l-[35S]methionine in a total volume of 50 μl for 20 h at 30°C. A 1-μl aliquot of the labeled protein mixture was analyzed by nondenaturing SDS-4 to 15% PAGE and fluorography. The 35S-labeled fiber protein was applied to HeLa cell monolayers in a six-well dish at a density of 106 cells per well for 1 h at room temperature. In some cases, either sCAR-m or sCAR-t was added to the cells concomitantly with the labeled fiber protein to assess the ability of sCAR to inhibit fiber binding to HeLa cells. 35S-labeled cell-bound proteins were analyzed by SDS-4 to15% PAGE and fluorography under nondenaturing conditions.
Adenoviral vectors.
All adenoviral vectors used in these studies were based on Ad5 and were third-generation vectors (Av3) in which the E1, E2a, and E3 regions were deleted (15). The production and characterization of Av3nBg, which encodes a nucleus-localizing β-galactosidase, were described previously (15). Av3Luc contains the firefly luciferase gene, which was derived from pGEMluc (Promega). Av3GFP contains the green fluorescent protein (GFP) gene, which was derived from pEGFP-N2 (Clontech, Palo Alto, Calif.). All vectors utilized the Rous sarcoma virus promoter. Vectors were propagated on AE1-2a cells (15) and purified as described by Jakubczak et al. (19). Viral particle titers (particles per milliliter) were determined as described by Mittereder et al. (30).
Adenovirus transduction.
Typically, various amounts of sCAR protein were incubated with 2 × 109 particles of Av3GFP for 30 min at room temperature in a total volume of 50 μl in PBS. Following incubation, the complex was diluted to 1 ml with the appropriate tissue culture medium plus 2% FBS and then added to six-well plates containing 106 human diploid fibroblasts (HDFs). Cells were washed with PBS prior to the addition of sCAR adenoviral vector complexes. Cells were incubated with the complexes for 2 h at 37°C, and then the medium was changed. After 24 to 48 h, the cells were observed for green fluorescence with a Nikon fluorescence microscope. In some cases, to demonstrate specificity of binding, the cells were preblocked with either 10 μg of competing 5F or an appropriate competitor per ml. The competitors used to assess the interaction of the ApoE peptide with its receptor were a specific rabbit antibody against human LDL receptor-related protein (LRP) and a nonspecific rabbit IgG1 molecule (Research Diagnostics, Inc., Flanders, N.J.). The competitors for the cRGD constructs included a specific cRGD or nonspecific RGE peptides (Gibco Life Technologies, Inc., Gaithersburg, Md.). Competitors were incubated with the HDF monolayers for 30 min at 37 or 4°C before addition of the sCAR-adenoviral-vector complexes. As described previously (38), a baculovirus expression system (Invitrogen, Carlsbad, Calif.) was used to generate 5F.
In vivo analysis of sCAR targeting molecules.
C57BL/6 male mice were purchased from Harlan Sprague Dawley (Indianapolis, Ind.). When the mice were 5 weeks of age they received Av3nBg or Av3Luc via tail vein injection at a dose of 2.5 × 1012 particles per kg of body weight, which is approximately 5 × 1010 particles per mouse. Cohorts of five mice received either unmodified adenoviral vector or vector complexed with either sCAR-m or sCAR-t targeting molecules. In cases where sCAR was added to the vector prior to injection, 3 μg of each sCAR protein was mixed with the vector for 30 min at room temperature. The vector was diluted to 2.5 × 1011 particles per ml with Hanks balanced salt solution immediately prior to injection. Three days after vector delivery, the animals were sacrificed, and tissues, including liver, lung, heart, kidney, and spleen, were collected. The tissue samples were used to analyze β-galactosidase or luciferase expression with the Galacto-Light Plus chemiluminescent assay (Tropix, Inc., Foster City, Calif.) or a luciferase assay (Promega). Tissue samples were collected in lysis matrix tubes containing two ceramic spheres (Bio101, Carlsbad, Calif.) and frozen on dry ice. The tissues were thawed, and 500 μl of lysis buffer from the Galacto-Light Plus kit was added to each tube. The tissue was homogenized for 30 s with a FastPrep system (Bio101). Liver samples were homogenized for an additional 30 s.
In addition, slices of liver approximately 2 to 3 mm thick were placed in 10% neutral buffered formalin. After fixation, these samples were embedded in paraffin, sectioned, and analyzed by immunohistochemistry for β-galactosidase expression. A 1:1,200 dilution of a rabbit anti-β-galactosidase antibody (ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) was used in conjunction with a Vectastain ABC kit from Vector Laboratories, Inc. (Burlingame, Calif.) to visualize positive cells.
RESULTS
Design and production of the sCAR bifunctional targeting molecules.
Given the success of the various multicomponent systems previously used for targeting adenoviral vectors both in vitro and in vivo, we have developed a multicomponent system for adenoviral targeting that employs the extracellular domain of CAR. This approach involves the generation of a bifunctional protein consisting of sCAR genetically fused to targeting ligands. As diagrammed in Fig. 1A, this system is designed to eliminate the native receptor interaction while redirecting binding to an alternative receptor on a desired target cell or tissue and is similar to that described by Dmitriev et al. (7). We hypothesized that trimerization of sCAR should improve its interaction with the fiber knob as shown in Fig. 1B. To test this hypothesis, a series of sCAR targeting molecules was generated (Fig. 2). First, the extracellular domain of the full-length CAR was cloned and combined with a short serine alanine linker and a six-H tag to generate sCAR-m. Two peptide targeting ligands were incorporated prior to the six-H tag and included a 9-amino-acid cRGD peptide and a 30-amino-acid peptide corresponding to a dimeric repeat of the receptor binding domain of ApoE. The cRGD peptide interacts with cell surface integrins and has been shown to target low-molecular-weight compounds to tumors in vivo (1) and to broaden the tropism of adenoviral vectors (8, 18). The ApoE peptide was selected for specific liver targeting, as the receptors for this ligand are located primarily on hepatocytes. Both peptides were incorporated into sCAR-m, and these constructs are referred to as sCAR-m-RGD and sCAR-m-ApoE. Trimerization of sCAR-m was achieved by incorporation of the isoleucine variant of the GCN4 leucine zipper mutant. This peptide has been characterized and shown to effectively trimerize (17). The sequence of this 31-amino-acid trimerization peptide was incorporated between the sCAR and peptide ligand domains with serine-alanine linkers separating each region to generate sCAR-t. Both targeting peptides were also incorporated into the trimeric construct, and the resulting are designated sCAR-t-RGD and sCAR-t-ApoE.
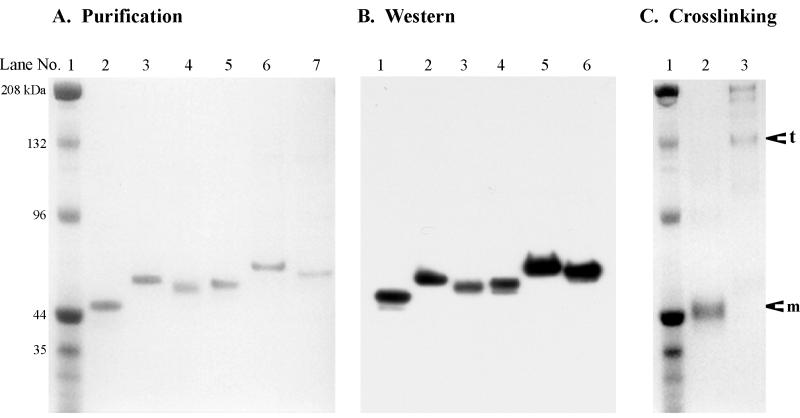
To produce the sCAR proteins, expression plasmids encoding each cDNA were electroporated into COS cells. Electroporation resulted in the production of high levels of each sCAR protein. All of the sCAR proteins included a six-H purification tag at the carboxy terminus to permit protein purification by metal affinity resin, as described in Materials and Methods. After purification, each protein was analyzed by SDS-PAGE under denaturing conditions (Fig. 3A). The results showed relatively pure proteins that migrated slightly higher than expected, possibly because of glycosylation at the two potential N-linked glycosylation sites identified within the extracellular domain (2). To confirm the identity of the proteins, Western immunoblot analysis was performed with a rabbit polyclonal antibody specific for CAR (Fig. 3B). As expected, all of the sCAR proteins were detected in this analysis. The presence of the six-H tag on each protein was also confirmed by Western immunoblot analysis with a rabbit polyclonal antibody specific for the six-H tag (data not shown).
FIG. 3.
Biochemical characterization of sCAR proteins. sCAR proteins were generated and purified as described in Materials and Methods. (A) SDS-PAGE analysis of sCAR proteins. One microgram of each purified protein was subjected to electrophoresis on an SDS-4 to 12% polyacrylamide gel under denaturing conditions. The gel was treated with Gel Code Blue (Pierce) to visualize protein bands. Lane 1, prestained molecular weight standards (Bio-Rad Laboratories, Hercules, Calif.); lane 2, sCAR-m; lane 3, sCAR-m-ApoE; lane 4, sCAR-m-cRGD; lane 5, sCAR-t; lane 6, sCAR-t-ApoE; lane 7, sCAR-t-cRGD. (B) Western immunoblot analysis of sCAR proteins. A 300-ng aliquot of each sCAR protein was subjected to SDS-PAGE and Western immunoblot analysis under denaturing conditions. The membrane was developed with a rabbit anti-human CAR polyclonal antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody by chemiluminescence. Lane 1, sCAR-m; lane 2, sCAR-m-ApoE; lane 3, sCAR-m-cRGD; lane 4, sCAR-t; lane 5, sCAR-t-ApoE; lane 6, sCAR-t-cRGD. (C) Oligomerization of the sCAR proteins was assessed by cross-linking analysis. Both sCAR-m and sCAR-t proteins were treated with the BS3 cross-linking reagent as described in Materials and Methods. Subsequently, 4 μg of each protein was subjected to SDS-4 to 12% PAGE. The gel was treated with Gel Code Blue to visualize protein bands. Lane 1, prestained molecular weight standards; lane 2, sCAR-m treated with BS3; lane 3, sCAR-t treated with BS3. The arrows indicate the positions of sCAR-t and sCAR-m proteins.
To demonstrate that versions of sCAR containing the GCN4 trimerization domain formed trimers, cross-linking analysis was carried out (Fig. 3C). Preparations of sCAR-m and sCAR-t were incubated with BS3 and evaluated by SDS-PAGE under denaturing conditions. sCAR-m migrated identically whether treated with BS3 or not (compare Fig. 3A, lane 2, with Fig. 3C, lane 2). However, sCAR-t (containing the GCN4 trimerization peptide) migrated at the position expected for trimers following treatment with the BS3 cross-linking reagent (Fig. 3C, lane 3). Small amounts of monomers, dimers, and other multimers greater than trimers were also observed, although the trimer was the predominant species found. These results indicate that insertion of the GCN4 trimerization peptide successfully trimerized the sCAR molecule.
sCAR inhibits fiber binding to HeLa cells.
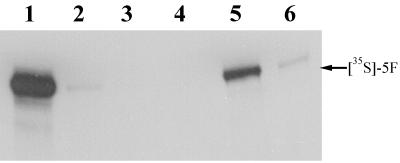
The ability of sCAR to bind the adenovirus fiber protein was assessed by examining its ability to block binding of radioactively labeled fiber to HeLa cell monolayers. 5F was produced in vitro and labeled with [35S]methionine with a coupled transcription-translation reticulocyte lysate system. Fiber protein produced in this manner forms the native trimeric structure (38). Binding of [35S]5F trimers to HeLa cells was assessed by analyzing cell lysates by SDS-PAGE and fluorography. Fiber binding was blocked by addition of purified 5F (Fig. 4, lane 2) as well as by either sCAR-m or sCAR-t (Fig. 4, lanes 3 to 6). Importantly, sCAR-t inhibited fiber binding to cells more effectively than sCAR-m, as it required less protein to completely block fiber interaction. These data indicate that the sCAR proteins produced in COS-7 cells were biologically active and were capable of binding to the adenovirus fiber knob and also show that sCAR-t has an enhanced ability to interact with fiber. Adenoviral transduction can also be blocked by using sCAR-m and sCAR-t, with sCAR-t again being more potent and requiring lower protein concentrations (data not shown).
FIG. 4.
Fiber-cell binding assay. 5F was radioactively labeled in vitro as described in Materials and Methods. [35S]5F was applied to HeLa cell monolayers in either the presence or absence of sCAR-m or sCAR-t, and then the labeled cell-bound proteins were analyzed by nondenaturing SDS-4 to 12% PAGE and fluorography. [35S]5F was added to HeLa cells in the presence of no competitor (lane 1), 15 μg of unlabeled 5F competitor (lane 2), 3 μg of sCAR-t (lane 3), 15 μg of sCAR-t (lane 4), 3 μg of sCAR-m (lane 5), or 15 μg of sCAR-m (lane 6).
Improved potency of sCAR targeting molecules via trimerization.
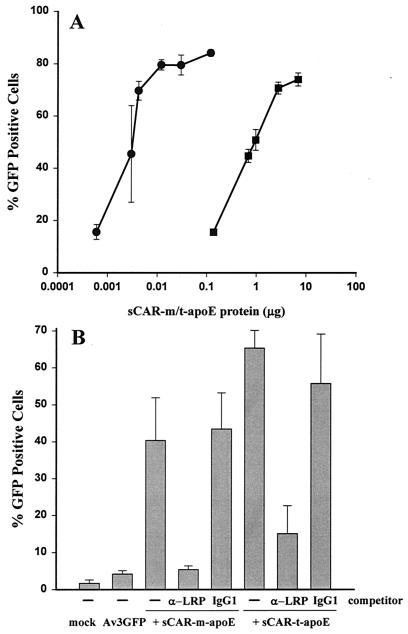
Both sCAR-m and sCAR-t constructs displaying the ApoE or cRGD targeting peptides were analyzed for their ability to restore transduction to HDF cells, which are negative for cell surface expression of CAR (38). Av3GFP was mixed with various amounts of either sCAR-t or sCAR-m containing the ApoE ligand and added to cells. The day after transduction, the cells were harvested and analyzed by flow cytometry for GFP expression. Since HDF cells express little or no CAR on their surfaces, they are poorly transduced by adenoviral vectors (38). Transduction with Av3GFP alone yielded approximately 8.3% GFP-positive cells. However, vector complexed with either sCAR-m-ApoE or sCAR-t-ApoE efficiently transduced HDF cells (Fig. 5A). The percentage of transduction was dependent upon the amount of sCAR protein added to the vector and ranged from 20 to 80% positive cells. Notably, sCAR-t was approximately 100-fold more potent than sCAR-m.
FIG. 5.
LRP-mediated adenoviral transduction of HDF cells. (A) Av3GFP was mixed with increasing amounts, from 0.6 ng to 7 μg, of either sCAR-m (squares) or sCAR-t (circles) targeting molecules fused with the ApoE peptide targeting ligand. The vector-sCAR protein mixture was allowed to associate for 30 min at 37°C before addition of the complex to HDF monolayers at a particle/cell ratio of 5,000. After 24 h, the adenovirus-mediated expression of GFP was analyzed by flow cytometry. Each point is the average of triplicate determinations ± the standard deviation. (B) The specificity of receptor interaction was demonstrated through competition analysis with an antibody against LRP as a direct competitor. Av3GFP was mixed with the amount of sCAR fusion protein that resulted in approximately 50% GFP-positive cells and was then added to HDF monolayers in the presence or absence of the indicated competitor. Approximately 900 and 3.5 ng of the sCAR-m-ApoE and sCAR-t-ApoE, respectively, were complexed with 5 × 108 particles of Av3GFP prior to addition to HDF monolayers at 5,000 particles per cell in the presence or absence of an anti-LRP or nonspecific IgG1 antibody competitor. Adenovirus-mediated GFP expression was determined by flow cytometry 24 h after transduction. The data are the averages of triplicate determinations ± standard deviations.
To confirm that transduction of HDF cells occurred via binding of the ApoE peptide to its receptor, LRP, the presence of LRP on the cell surface was verified by immunofluorescence (data not shown), and then competition analysis was carried out (Fig. 5B). Transduction of HDFs by Av3GFP complexed with sCAR-ApoE, either monomeric or trimeric, was significantly reduced by an anti-LRP antibody. A considerably higher concentration of competitor was required to inhibit transduction via sCAR-t-ApoE (20 versus 50 μg). This may be due to the higher affinity of the trimerized ApoE peptide for its receptor. As a control, it was shown that the same concentration of purified mouse IgG1 had no effect on transduction.
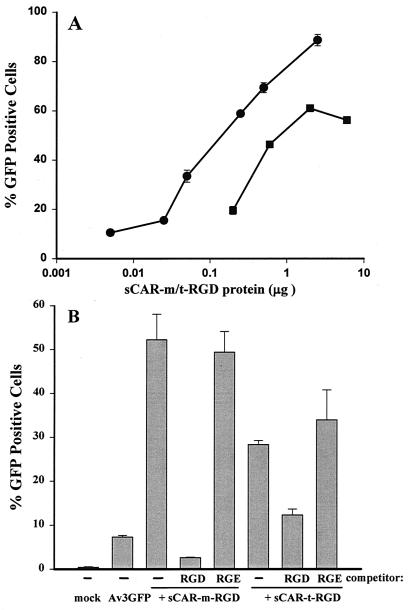
Next, the cRGD targeting ligand was analyzed in a similar fashion for its ability to mediate adenoviral transduction of HDFs via cell surface integrins. Adenovirus complexed with either sCAR-m-cRGD or sCAR-t-cRGD efficiently transduced HDF cells (Fig. 6A). The percentage of transduction was dependent upon the amount of sCAR protein added and ranged from 20 to 80% positive cells; in this case, sCAR-t was approximately 10-fold more potent than sCAR-m.
FIG. 6.
Integrin-mediated adenoviral transduction of HDFs. (A) Av3GFP was mixed with increasing amounts, from 5 ng to 6 μg, of either sCAR-m (squares) or sCAR-t (circles) targeting molecules fused to the cRGD peptide targeting ligand. The vector-sCAR protein mixture was allowed to associate for 30 min at 37°C before addition of the complex to HDF monolayers at a particle/cell ratio of 5,000. After 24 h, the adenovirus-mediated expression of GFP was analyzed by flow cytometry. Each point is the average of triplicate determinations ± the standard deviation. (B) The specificity of receptor interaction was demonstrated through competition analysis using an RGD peptide as a competitor. Av3GFP was mixed with the amount of sCAR fusion protein that resulted in approximately 30 to 50% GFP-positive cells and was then added to HDF monolayers in the presence or absence of the indicated competitor. Approximately 1 and 0.1 μg of sCAR-m-cRGD and sCAR-t-cRGD, respectively, were complexed with 5 × 108 particles of Av3GFP prior to addition to HDF monolayers at 5,000 particles per cell in the presence or absence of an RGD or nonspecific RGE peptide competitor. Adenovirus-mediated GFP expression was determined by flow cytometry 24 h after transduction. The data are the averages of triplicate determinations ± standard deviations.
The specificity of the RGD peptide targeting ligand interaction with cell surface integrins was verified through competition analysis with RGD peptides (Fig. 6B). Transduction of HDFs by Av3GFP complexed with sCAR-RGD, either monomeric or trimeric, was significantly reduced by the addition of a cRGD peptide but not by the control RGE peptide (Fig. 6B), indicating that transduction was indeed mediated by cell surface integrins. Thus, both the ApoE and RGD targeting peptides fused to sCAR proteins specifically redirected adenoviral vectors to alternative receptors in vitro. Additionally, trimerization of these targeting molecules significantly increased the potency of these bifunctional proteins.
sCAR-t efficiently blocks liver transduction in vivo.
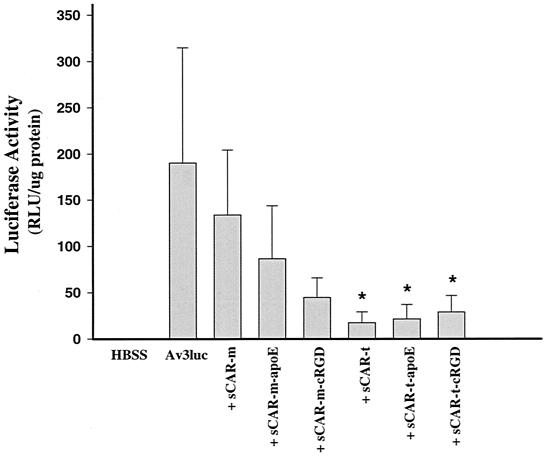
To evaluate the sCAR multicomponent system for systemic delivery in vivo, normal mice were treated with adenoviral vectors complexed with the various sCAR targeting molecules. The following experiment was designed to determine whether sCAR-m or sCAR-t without a targeting ligand could block adenovirus-mediated transduction of liver in mice and to evaluate the function of the ApoE and RGD peptide targeting ligands to restore liver gene transfer. Av3nBg and Av3Luc were administered to C57BL/6 mice via tail vein injection at a dose of 2.5 × 1012 particles per kg. Cohorts of five mice received either unmodified adenoviral vector or vector complexed with either sCAR-m or sCAR-t targeting molecules. In cases where sCAR was added to the vector prior to injection, 3 μg of sCAR-m or sCAR-t was mixed with the adenoviral vector for 30 min at room temperature, corresponding to fiber monomer-to-sCAR molar ratios of 1:60 and 1:20, respectively. Three days after vector delivery, the animals were necropsied and livers were collected. Adenovirus-mediated gene expression in the liver was analyzed by measurement of luciferase activity (Fig. 7). Systemic delivery of Av3Luc resulted in efficient liver transduction, with high levels of luciferase gene expression found in the liver. When it was complexed with sCAR-m without a targeting ligand, there was not a significant reduction in liver gene expression, suggesting that sCAR-m was not effective at blocking liver uptake. However, trimerization of sCAR enabled the in vivo application of this technology for blocking liver uptake and adenovirus-mediated transduction. Both targeting peptides were evaluated for their ability to restore liver gene transfer. Both the ApoE and cRGD peptides were expected to bind to hepatocytes in vivo, as both the LRP and integrins should be present on mouse hepatocytes. Interestingly, sCAR-t with either ligand did not restore liver gene transfer as expected and resulted in reduction in luciferase gene expression. The gene expression results were confirmed directly by analyzing adenoviral copy number in the liver by using real-time PCR (data not shown).
FIG. 7.
Inhibition of liver transduction by the sCAR-t targeting molecules. Av3Luc was administered to C57BL/6 mice via tail vein injection at a dose of 2.5 × 1012 particles per kg. Cohorts of five mice received either unmodified vector or vector complexed with the sCAR-m or sCAR-t targeting molecules. A 3-μg portion of each sCAR fusion protein was mixed with vector for 30 min at room temperature prior to administration. Three days after vector delivery, the mice were sacrificed and livers were analyzed for luciferase gene expression. The average luciferase activity level plus the standard deviation is shown for each treatment group. RLU, relative light units. ∗, significantly different from virus-only control according to an unpaired, two-tailed t test (P < 0.01).
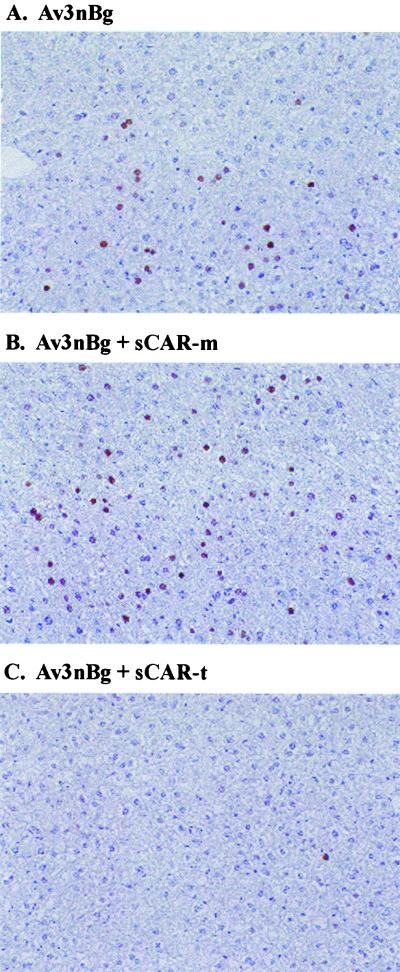
To visualize what cell types in the liver were transduced, immunohistochemistry for β-galactosidase expression was carried out in mice treated with Av3nBg (Fig. 8). In mice that received unmodified Av3nBg, approximately 10 to 15% of hepatocytes expressed β-galactosidase, as indicated by the brown nuclear staining (Fig. 8A). sCAR-m had no effect on the percentage of hepatocytes expressing the β-galactosidase transgene (Fig. 8B). In contrast, sCAR-t effectively blocked liver transduction so that less than 1% of hepatocytes were positive for β-galactosidase expression (Fig. 8C). Similar results were obtained with sCAR-t-ApoE and sCAR-t-RGD (data not shown).
FIG. 8.
Immunohistochemical localization of β-galactosidase in mouse livers. Av3nBg was administered to C57BL/6 mice via tail vein injection at a dose of 2.5 × 1012 particles per kg. Cohorts of five mice received either unmodified vector (A) or vector complexed with sCAR-m (B) or sCAR-t (C) targeting molecules. A 3-μg aliquot of each sCAR targeting molecule was mixed with vector for 30 min at room temperature prior to administration. Three days after vector delivery, the mice were sacrificed and livers were analyzed for β-galactosidase expression by immunohistochemistry. Representative photomicrographs are shown.
One hypothesis to explain these in vivo results is that the effective size of the adenovirus-sCAR-t complexes influences the bioavailability and thus reduces access to normal vascular beds, such as in the liver. To explore this proposed mechanism, the morphology of the sCAR complexes was examined by electron microscopy; this confirmed that adenovirus complexed with sCAR-t protein had approximately the same diameter, 80 to 120 nm, and that complexing did not result in aggregation of the adenoviral particles (data not shown).
DISCUSSION
In this article, we describe the development of a multicomponent system to target adenoviral vectors. The bifunctional protein used consists of the extracellular domain of CAR fused to peptide targeting ligands. The results showed successful alteration of the tropism of adenoviral vectors in tissue culture by using these sCAR fusion proteins. While the studies described in this article were in progress, a similar targeting strategy using the extracellular domain of CAR was described by Dmitriev et al. (7). Cell binding and transduction experiments in vitro with sCAR-m fused to epidermal growth factor provide evidence for the alteration of the receptor tropism of an Ad5-based luciferase vector on various human cancer cell lines. In the present study, we significantly improved the potency of the sCAR targeting molecule via trimerization and demonstrated that sCAR-t targeting molecules were more efficient than the corresponding monomeric protein. In addition, the results obtained in vivo extend these studies by demonstrating that the trimeric construct was extremely effective at blocking liver gene transfer.
The trimeric targeting approach described here is facilitated by the multivalent interaction of the sCAR interaction with fiber knob and that of the peptide ligand with its receptor. The presence of the heterologous trimerization domain brings three sCAR monomers in closer proximity with the knob, which allows efficient interaction with each fiber monomer (Fig. 1B). This concept is further supported by the recent kinetic analysis of fiber knob interacting with CAR D1, which proposed a multivalent interaction for CAR D1 with the trimeric knob (27). The sCAR-m targeting molecules both in vitro and in vivo were not as efficient at affecting adenoviral transduction, presumably because of inefficient interaction with fiber knob and/or the presence of noncomplexed fiber. Dmitriev et al. (7) separated unbound sCAR-m from adenovirus and found reduced gene transfer efficiency in vitro after purification, supporting the concept that sCAR-m does not bind tightly. A multivalent interaction would improve the affinity of sCAR interaction with the fiber knob and shift the equilibrium towards binding. Kinetic on-off rates of sCAR-m may be sufficiently high to cause dissociation, particularly in vivo.
The development of multicomponent systems for adenoviral targeting has several advantages. First, the use of these systems circumvents the need to generate modified adenoviral vectors for each new target. In contrast, targeting strategies based on insertion of targeting ligands into a capsid protein, such as fiber, penton, or hexon, require the production of new adenoviral vectors for each ligand to be tested. Second, the use of bifunctional proteins for in vivo targeting simultaneously eliminates the normal tropism of the vector and redirects it to an alternative receptor. A longer-term approach is to incorporate both detargeting and retargeting modifications genetically into the adenoviral genome. Genetic detargeting strategies can effect the normal tropism and sometimes effect viral viability (26). Several groups have identified fiber knob mutations that inhibit CAR interaction (19, 23, 26, 36). Efficient growth of adenoviral vectors containing fiber mutation requires further manipulation of either the packaging cells or the virus that will allow production of vector. This issue has been addressed by the production of pseudoreceptor cell lines that provide an alternative cell binding pathway, which allows the efficient growth of detargeted adenoviral vectors (9, 12). The sCAR targeting strategy does not require manipulation of the virus or the packaging cells, so vectors can be grown to normal titers on standard packaging cells.
Additionally, the sCAR targeting strategy may be compatible with the use of relatively large targeting ligands. Empirical evidence indicates that only short targeting peptides can be inserted into the fiber knob without disrupting the structure and viability of the virus. For example, there may be an upper limit of 45 to 85 amino acids that can be successfully inserted into the HI loop of fiber knob, and this limit may be ligand dependent. Similarly, only relatively short peptides, such as an RGD-containing peptide or a short stretch of lysine residues, have been successfully added to the carboxy terminus of fiber (41, 43). To date, only moderate-length peptides have been tested as fusion proteins with sCAR. Dmitriev et al. (7) utilized a 53-amino-acid EGF peptide successfully. In our study, both a small (9-amino-acid) RGD peptide and a 30-amino-acid peptide derived from ApoE were effectively used to alter the receptor tropism of adenoviral vectors in vitro. However, there are several concerns associated with the use of small peptide ligands for systemic delivery of adenoviral vectors. These include the relatively low affinity and specificity of the peptide interaction compared with those of larger proteins, such as FGF or single-chain antibodies. It may be feasible to fuse larger proteins, such as a single-chain antibody (scFv), to sCAR-t. Typically, an scFv consists of 125 to 150 amino acids and has a relatively high affinity and specificity for its target. Several recently published studies demonstrate the feasibility of targeting other viruses, such as Moloney murine leukemia virus, by using an scFv (21). sCAR-t also provides a platform and a convenient method of rapidly screening a variety of ligands for subsequent analysis in adenoviral targeting.
Two targeting ligands were evaluated in this study and were chosen because of their ability to bind to important targets. First, the ApoE ligand interacts with members of the LDL receptor family, which are found on hepatocytes, and it was thought that this ligand would be useful in liver-directed gene therapy applications. Two tandem copies of the receptor-binding region of ApoE were genetically fused to sCAR-m and sCAR-t. Efficient targeting of LRP-positive cells was demonstrated in vitro with the ApoE peptide ligand; however, this peptide did not restore adenoviral liver gene transfer in vivo. When complexed with lipoproteins in vivo, the ApoE peptide was capable of mediating their clearance in vivo primarily to the liver and spleen (32) but did not allow adenoviral uptake into these tissues. Second, the cRGD peptide binds to cell surface integrins, which are found on activated tissues and tumor endothelium. Efficient targeting of integrin-positive cells was demonstrated in vitro, but similar to the ApoE peptide, cRGD did not restore liver gene transfer in vivo. The cRGD peptide, when incorporated into the fiber HI loop, was found to alter the biodistribution of a systemically delivered adenoviral vector with enhanced transgene expression found in the liver, kidney, spleen, and lung (35). We expected to find a similar biodistribution when the adenovirus was delivered systemically complexed with the sCAR-t-cRGD protein; however, this was not the case.
Trimerization of sCAR improved the potency of targeting in vitro and in blocking liver transduction in vivo. However, the mechanism by which sCAR-t inhibited liver gene transfer is unclear and may also be related to the reason why the targeting peptides did not restore liver transduction. Several hypotheses may be proposed to explain these results. First, the binding of sCAR-t to adenoviral capsids may alter and/or prevent the virion's normal hepatic uptake. The in vitro results presented here demonstrate efficient cellular entry and gene expression with either the ApoE or cRGD sCAR targeting molecule, suggesting that hepatic uptake via the LDL receptor or integrins should be unaffected. Second, the inability to retarget in vivo may be related to the stability of the sCAR-adenovirus complexes. However, it is unlikely that the instability of these complexes accounts for these observations, for several reasons. First, sCAR-t without a targeting ligand successfully detargeted the vector from the liver, suggesting that sCAR remained bound to the virus following tail vein administration to mice. Additionally, the stability of the sCAR-adenovirus complexes was evaluated by the ability to withstand centrifugation banding in cesium chloride followed by freezing, thawing, and subsequent incubation in mouse serum for 60 min without a loss in activity compared to freshly formed complex (data not shown). Third, an alternative hypothesis to explain this phenomenon may be that the sCAR-t-adenovirus complexes are effectively too large to pass through the small hepatic fenestrations found in mouse livers. The sCAR-t specifically binds to the fiber knob and may provide an apparent shield on the particle that increases the effective diameter or agglutinates the adenovirus particles. To rule out agglutination, the morphology of the sCAR-t-adenovirus complexes was analyzed by electron microscopy, which showed that they are individual particles comparable to adenovirus alone. It appears likely then that binding of sCAR-t to adenoviral particles may increase the effective diameter and prevent passage through the hepatic fenestrations. This size hypothesis has been proposed previously to explain, in part, the decrease found in adenoviral liver gene transfer by using the Fab-FGF reagent, which results in complex diameters of approximately 300 nm (34). Additionally, Nguyen et al. have proposed that the larger diameter of lentiviral vectors, approximately 120 nm, explains the inability of these vectors to transduce mouse hepatocytes in vivo (T. H. Nguyen, J. Oberholzer, and D. Trono, abstract from the Fourth Annual Meeting of the American Society of Gene Therapy, Mol. Ther. 3:S321, 2001). Although we have demonstrated that sCAR-t without a targeting ligand can efficiently block liver transduction in vivo, an important next step will be to demonstrate in vivo targeting. Further studies will evaluate the sCAR targeting strategy in mouse tumor models, where it is proposed that these tumors will have abnormal, “leaky” vasculatures with discontinuous endothelium and larger pore sizes (4). These anatomical features may allow access of the adenovirus-sCAR complexes.
Acknowledgments
We thank Christine Mech for completing the immunohistochemical analysis of β-galactosidase, Russette Lyons and Donna Goldsteen for assistance with the in vivo studies, and Scott Pattison for critical review of the manuscript.
REFERENCES
- 1.Arap, W., R. Pasqualini, and E. Ruoslahti. 1998. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 279:377-380. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., A. Krithivas, L. Celi, G. Droguett, M. S. Horwitz, T. Wickham, R. L. Crowell, and R. W. Finberg. 1998. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J. Virol. 72:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. S., E. di Tomaso, D. M. McDonald, R. Jones, R. K. Jain, and L. L. Munn. 2000. Mosaic blood vessels in tumors: frequency of cancer cells in contact with flowing blood. Proc. Natl. Acad. Sci. USA 97:14608-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cripe, T. P., E. J. Dunphy, A. D. Holub, A. Saini, N. H. Vasi, Y. Y. Mahller, M. H. Collins, J. D. Snyder, V. Krasnykh, D. T. Curiel, T. J. Wickham, J. DeGregori, J. M. Bergelson, and M. A. Currier. 2001. Fiber knob modifications overcome low, heterogeneous expression of the coxsackievirus-adenovirus receptor that limits adenovirus gene transfer and oncolysis for human rhabdomyosarcoma cells. Cancer Res. 61:2953-2960. [PubMed] [Google Scholar]
- 6.Dechecchi, M. C., A. Tamanini, A. Bonizzato, and G. Cabrini. 2000. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology 268:382-390. [DOI] [PubMed] [Google Scholar]
- 7.Dmitriev, I., E. Kashentseva, B. E. Rogers, V. Krasnykh, and D. T. Curiel. 2000. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J. Virol. 74:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, J. T., C. R. Miller, M. Kim, I. Dmitriev, G. Mikheeva, V. Krasnykh, and D. T. Curiel. 1999. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat. Biotechnol. 17:470-475. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, J. T., B. E. Rogers, M. E. Rosenfeld, S. I. Michael, M. Feng, and D. T. Curiel. 1996. Targeted gene delivery by tropism-modified adenoviral vectors. Nat. Biotechnol. 14:1574-1578. [DOI] [PubMed] [Google Scholar]
- 11.Dyer, C. A., D. P. Cistola, G. C. Parry, and L. K. Curtiss. 1995. Structural features of synthetic peptides of apolipoprotein E that bind the LDL receptor. J. Lipid Res. 36:80-88. [PubMed] [Google Scholar]
- 12.Einfeld, D. A., D. E. Brough, P. W. Roelvink, I. Kovesdi, and T. J. Wickham. 1999. Construction of a pseudoreceptor that mediates transduction by adenoviruses expressing a ligand in fiber or penton base. J. Virol. 73:9130-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freimuth, P., K. Springer, C. Berard, J. Hainfeld, M. Bewley, and J. Flanagan. 1999. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 73:1392-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman, C. K., B. E. Rogers, J. T. Douglas, B. A. Sosnowski, W. Ying, G. P. Siegal, A. Baird, J. A. Campain, and D. T. Curiel. 1997. Targeted gene delivery to Kaposi's sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 57:1447-1451. [PubMed] [Google Scholar]
- 15.Gorziglia, M. I., C. Lapcevich, S. Roy, Q. Kang, M. Kadan, V. Wu, P. Pechan, and M. Kaleko. 1999. Generation of an adenovirus vector lacking E1, e2a, E3, and all of E4 except open reading frame 3. J. Virol. 73:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, D. L., A. M. Gonzalez, M. A. Printz, J. Doukas, W. Ying, M. D'Andrea, D. K. Hoganson, D. T. Curiel, J. T. Douglas, B. A. Sosnowski, A. Baird, S. L. Aukerman, and G. F. Pierce. 1999. Fibroblast growth factor 2 retargeted adenovirus has redirected cellular tropism: evidence for reduced toxicity and enhanced antitumor activity in mice. Cancer Res. 59:2608-2614. [PubMed] [Google Scholar]
- 17.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 18.Hay, C. M., H. DeLeon, J. D. Jafari, J. L. Jakubczak, C. A. Mech, P. L. Hallenbeck, S. K. Powell, G. Liau, and S. C. Stevenson. 2001. Enhanced gene transfer to rabbit jugular veins by an adenovirus containing a cyclic RGD motif in the HI loop of the fiber knob. J. Vasc. Res. 38:315-323. [DOI] [PubMed] [Google Scholar]
- 19.Jakubczak, J. L., M. L. Rollence, D. A. Stewart, J. D. Jafari, D. J. Von Seggern, G. R. Nemerow, S. C. Stevenson, and P. L. Hallenbeck. 2001. Adenovirus type 5 viral particles pseudotyped with mutagenized fiber proteins show diminished infectivity of coxsackie B-adenovirus receptor-bearing cells. J. Virol. 75:2972-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jooss, K., Y. Yang, K. J. Fisher, and J. M. Wilson. 1998. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J. Virol. 72:4212-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khare, P. D., L. Shao-Xi, M. Kuroki, Y. Hirose, F. Arakawa, K. Nakamura, and Y. Tomita. 2001. Specifically targeted killing of carcinoembryonic antigen (CEA)-expressing cells by a retroviral vector displaying single-chain variable fragmented antibody to CEA and carrying the gene for inducible nitric oxide synthase. Cancer Res. 61:370-375. [PubMed] [Google Scholar]
- 22.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 1999. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J. Virol. 73:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 74:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 72:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasnykh, V. N., J. T. Douglas, and V. W. van Beusechem. 2000. Genetic targeting of adenoviral vectors. Mol. Ther. 1:391-405. [DOI] [PubMed] [Google Scholar]
- 26.Leissner, P., V. Legrand, Y. Schlesinger, D. A. Hadji, M. van Raaij, S. Cusack, A. Pavirani, and M. Mehtali. 2001. Influence of adenoviral fiber mutations on viral encapsidation, infectivity and in vivo tropism. Gene Ther. 8:49-57. [DOI] [PubMed] [Google Scholar]
- 27.Lortat-Jacob, H., E. Chouin, S. Cusack, and M. J. van Raaij. 2001. Kinetic analysis of adenovirus fiber binding to its receptor reveals an avidity mechanism for trimeric receptor-ligand interactions. J. Biol. Chem. 276:9009-9015. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, G. A., G. Zhu, Y. Li, I. Kovesdi, T. J. Wickham, and V. P. Sukhatme. 1999. Efficient adenoviral gene transfer to kidney cortical vasculature utilizing a fiber modified vector. J. Gene Med. 1:103-110. [DOI] [PubMed] [Google Scholar]
- 29.Michael, S. I., J. S. Hong, D. T. Curiel, and J. A. Engler. 1995. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 2:660-668. [PubMed] [Google Scholar]
- 30.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemerow, G. R. 2000. Cell receptors involved in adenovirus entry. Virology 274:1-4. [DOI] [PubMed] [Google Scholar]
- 32.Nikoulin, I. R., and L. K. Curtiss. 1998. An apolipoprotein E synthetic peptide targets to lipoproteins in plasma and mediates both cellular lipoprotein interactions in vitro and acute clearance of cholesterol-rich lipoproteins in vivo. J. Clin. Investig. 101:223-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasqualini, R., E. Koivunen, and E. Ruoslahti. 1997. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 15:542-546. [DOI] [PubMed] [Google Scholar]
- 34.Printz, M. A., A. M. Gonzalez, M. Cunningham, D. L. Gu, M. Ong, G. F. Pierce, and S. L. Aukerman. 2000. Fibroblast growth factor 2-retargeted adenoviral vectors exhibit a modified biolocalization pattern and display reduced toxicity relative to native adenoviral vectors. Hum. Gene Ther. 11:191-204. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds, P., I. Dmitriev, and D. Curiel. 1999. Insertion of an RGD motif into the HI loop of adenovirus fiber protein alters the distribution of transgene expression of the systemically administered vector. Gene Ther. 6:1336-1339. [DOI] [PubMed] [Google Scholar]
- 36.Santis, G., V. Legrand, S. S. Hong, E. Davison, I. Kirby, J. L. Imler, R. W. Finberg, J. M. Bergelson, M. Mehtali, and P. Boulanger. 1999. Molecular determinants of adenovirus serotype 5 fibre binding to its cellular receptor CAR. J. Gen. Virol. 80:1519-1527. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson, S. C., M. Rollence, J. Marshall-Neff, and A. McClelland. 1997. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J. Virol. 71:4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, S. C., M. Rollence, B. White, L. Weaver, and A. McClelland. 1995. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J. Virol. 69:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, K., J. Fueyo, V. Krasnykh, P. N. Reynolds, D. T. Curiel, and R. Alemany. 2001. A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin. Cancer Res. 7:120-126. [PubMed] [Google Scholar]
- 40.Watkins, S. J., V. V. Mesyanzhinov, L. P. Kurochkina, and R. E. Hawkins. 1997. The "adenobody' approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 4:1004-1012. [DOI] [PubMed] [Google Scholar]
- 41.Wickham, T. J., D. Haskard, D. Segal, and I. Kovesdi. 1997. Targeting endothelium for gene therapy via receptors up-regulated during angiogenesis and inflammation. Cancer Immunol. Immunother. 45:149-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 43.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 44.Wickham, T. J., D. M. Segal, P. W. Roelvink, M. E. Carrion, A. Lizonova, G. M. Lee, and I. Kovesdi. 1996. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J. Virol. 70:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickham, T. J., E. Tzeng, L. L. Shears, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]