Abstract
Simian T-cell leukemia viruses (STLVs) are the simian counterparts of human T-cell leukemia viruses (HTLVs). A novel, divergent type of STLV (STLV-L) from captive baboons was reported in 1994, but its natural prevalence remained unclear. We investigated the prevalence of STLV-L in 519 blood samples from wild-living nonhuman primates in Ethiopia. Seropositive monkeys having cross-reactive antibodies against HTLV were found among 22 out of 40 hamadryas baboons, 8 of 96 anubis baboons, 24 of 50 baboons that are hybrids between hamadryas and anubis baboons, and 41 of 177 grivet monkeys, but not in 156 gelada baboons. A Western blotting assay showed that sera obtained from seropositive hamadryas and hybrid baboons exhibited STLV-L-like reactivity. A PCR assay successfully amplified STLV sequences, which were subsequently sequenced and confirmed as being closely related to STLV-L. Surprisingly, further PCR showed that nearly half of the hamadryas (20 out of 40) and hybrid (19 out of 50) baboons had STLV-L DNA sequences. In contrast, most of the seropositive anubis baboons and grivet monkeys carried typical STLV-1 but not STLV-L. These observations demonstrate that STLV-L naturally prevails among hamadryas and hybrid baboons at significantly high rates. STLV-1 and -2, the close relative of STLV-L, are believed to have jumped across simian-human barriers, which resulted in widespread infection of HTLV-1 and -2. Further studies are required to know if STLV-L is spreading into human populations.
The human T-cell leukemia virus (HTLV) is separated into two serologically and genetically distinct types (HTLV-1 and HTLV-2). Both types have a simian relative: HTLV-1 is related to simian T-cell leukemia virus type 1 (STLV-1) and HTLV-2 is related to STLV-2 (4). STLV-1 infects a wide range of wild nonhuman primates (NHPs). In fact, natural infection with STLV-1 is found among macaques, guenons, mangabeys, baboons, and apes in Asia and Africa (12, 21). In contrast, STLV-2 has been solely identified in the pygmy chimpanzee (Pan panisucus). As STLV-2 strains were isolated from captive pygmy chimpanzees, its natural occurrence remains unknown (6, 15). HTLVs and STLVs are collectively called primate T-cell leukemia virus (PTLV).
Phylogenetic characterization of PTLV strains, especially PTLV-1s, showed that they share complex evolutionary relationships that often correlate with the geographical origin of their host rather than their host's phylogeny. These complex relationships are interpreted as evidence for cross-species transmissions between different NHPs as well as between humans and NHPs (4, 5, 31, 35). For instance, a group of HTLV-1 strains found in Central Africa are more closely related to African STLV-1 than to other human strains. Furthermore, recent studies identified additional divergent HTLV-1 isolates of this type, hence indicating that zoonoses (cross-species transmission between human and animal) of PTLV-1 are rather frequent (3, 16, 23). Of particular relevance is the fact that zoonoses also play a pivotal role in the epidemic of another human retrovirus, human immunodeficiency virus (HIV). Like HTLV-1 and -2, HIV is separated into two distinct types (HIV-1 and -2). These two types of HIV are believed to have arisen in Africa by independent zoonoses; HIV-1 is likely to have originated from Central African chimpanzees and HIV-2 seems to have come from the West African sooty mangabey (8). Therefore, a better understanding of the natural prevalence of these pathogens among wild NHPs may help not only to elucidate the evolutionary origins of these human retroviruses but also to prevent a further epidemic of a new human retrovirus.
Recently, a third type of the STLV group (STLV-L) was isolated from captive hamadryas baboons (Papio hamadryas) in Belgium (7). The prototype strain of STLV-L (PH969) was serologically more closely related to PTLV-2 than to PTLV-1, but it was genetically divergent both from PTLV-1 and -2 (28). So far, STLV-L infection has not been identified among wild NHPs. We took advantage of an opportunity to conduct a serological survey for STLV among wild NHPs in Ethiopia. Ethiopia neighbors Eritrea, which is the country from which the original monkey infected with STLV-L was exported to Belgium. This country is inhabited not only by hamadryas baboons but also different monkey species. It is noteworthy that there are several natural colonies of baboons that are hybrids between hamadryas and anubis baboons, which have been established and separated for a long time from both the parental subspecies (25). We aimed to determine whether the hamadryas baboon is truly infected with STLV-L under natural condition and whether STLV-L infection is prevalent among other NHP species. We demonstrate here that hamadryas baboons are naturally infected with STLV-L at a high rate. Our results also show that STLV-L infection is confined to hamadryas baboons and hybrid baboons.
MATERIALS AND METHODS
Specimens.
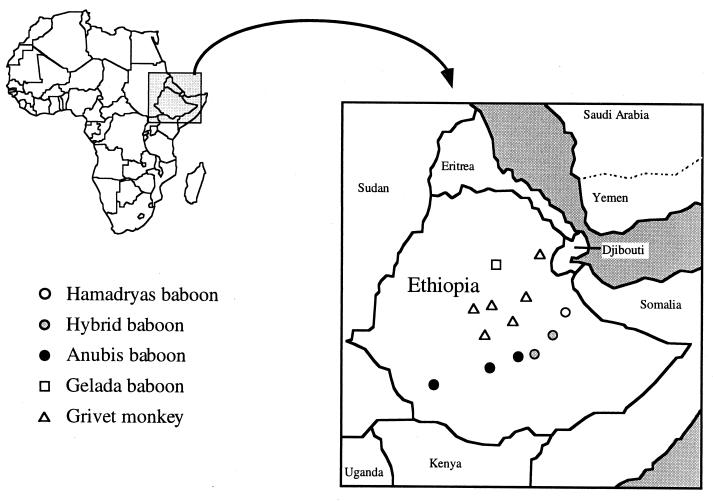
The blood samples used in this study were collected from 519 wild NHPs in Ethiopia for genetic studies between 1975 and 1985 by the Primates Research Institute of Kyoto University. When the blood samples were taken, they were divided into blood cells and plasma and stored at −80°C. The 519 animals consisted of 40 hamadryas baboons (Papio hamadryas hamadryas), 96 anubis baboons (Papio hamadryas anubis), 50 baboons that were hybrids between hamadryas baboons and anubis baboons (25), 156 gelada baboons (Theropithecus gelada) (26), and 177 grivet monkeys (Cercopithecus aethiops) (24). The locations at which the monkeys were sampled are shown in Fig. 1.
FIG. 1.
Sources and locations of the blood samples collected in Ethiopia between 1975 and 1985.
Serological assays.
Plasma samples were first screened for HTLV antibodies with a particle agglutination (PA) kit (Serodia-HTLV-1; Fujirebio, Tokyo, Japan). Weakly reactive plasma samples were further tested with a different PA kit which is somewhat sensitive for both types of HTLV-1 and -2 (Serodia-HTLV-1 and -2; Fujirebio). Several positive samples were further tested with two Western blotting (WB) kits (HTLV blot 2.4 [Genelabs Diagnostics, Singapore] and PROBLOT HTLV [Fujirebio]) according to the manufacturers' instructions. The former kit was capable of distinguishing between HTLV-1 and -2 antibodies. All the serological assays were carried out according to the manufacturer's instructions. Four PA-positive samples of the hamadryas baboons and two PA-positive samples from hybrid baboons were tested by indirect immunofluorescence assay (IFA) using MT1 cells, an HTLV-1-infected T-cell line, as antigen (9).
PCR.
We carried out nested PCR to amplify two proviral regions (pX and LTR) on chromosomal DNA extracted from the blood cells. DNA was extracted either with Easy-DNA Kit (Invitrogen, Carlsbad, Calif.) or QIAamp Blood DNA Mini Kit (QIAGEN, Hilden, Germany) according to the instruction manuals. We carried out the nested PCR with special care to avoid cross-contamination. Throughout this study, all negative controls gave negative signals. The nucleotide sequences of the oligonucleotide primers, the positions in the prototype strains and the expected fragment sizes are shown in Table 1 (34). Oligonucleotide primers for amplification of type L strains were designed based on the reported STLV-L strain (PH969) (30). The nested-PCR conditions are described elsewhere (18, 33).
TABLE 1.
Oligonucleotide primers for DNA amplification
| Region | Specificity | Primer | Sequence (5" → 3") | Orientation | Positions | Location | Size (bp)a | [Mg2+] (mM)b |
|---|---|---|---|---|---|---|---|---|
| pX | Types 1 and 2 | ATLpX1 | CCCACTTCCCAGGGTTTGGACAGAGTCTT | Sense | 7432-7461c | |||
| ATLpX5 | GGAGGGGAGTCGAGGGATAAGG | Antisense | 7637-7658c | Outer | 226 | 6 | ||
| SK43 | CGGATACCCAGTCTACGTGT | Sense | 7466-7485c | |||||
| SK44 | GAGCCGATAACGCGTCCATCG | Antisense | 7604-7624c | Inner | 158 | 6 | ||
| Type L | AV45Ld | GGACGCGTTGTCAGCTC | Sense | 7432-7448e | ||||
| AV46Ld | GGGGGAGAGCTGGTAGAGGTA | Antisense | 7714-7734e | Outer | 302 | 4 | ||
| AV42Ld | CTCCCCTCCTTCCCCAC | Sense | 7474-7490e | |||||
| AV43Ld | CCACATGGTGTATACGTTTTGG | Antisense | 7671-7692e | Inner | 218 | 4 | ||
| AV51Ld | ACAATTGCCTCGAGCTCACCC | Sense | 7601-7621e | |||||
| AV82Ld | GAGGCACACGACGGA | Antisense | 7696-7713e | Inner | 112 | 6 | ||
| LTR | Type 1 | ATLTR1 | TGACACTGACCATGAGCCCCAAAT | Sense | 130-153c | |||
| ATLTR2 | TCGTATCCCGGACGAGCCCCCAA | Antisense | 886-908c | Outer | 778 | 6 | ||
| ATLTR11 | ACTAAGGCTCTGACGTCTCCCCC | Sense | 228-250c | |||||
| ATLTR12 | CGGTACTTGGCCGTGGGCCAAGCCG | Antisense | 790-814c | Inner | 586 | 6 | ||
| Type L | LTR-1L | CAACAACCACCAACTAGGGG | Sense | 8260-8279e | ||||
| LTR-2L | AATGTTACAGGCGCTGG | Antisense | 8841-8857e | Outer | 597 | 3 | ||
| LTR-3L | CAAATAGCTGAATCATCCGTCTG | Sense | 8281-8303e | |||||
| LTR-4L | GCAACAGAAGTGCTACTTTCG | Antisense | 8797-8817e | Inner | 536 | 3 |
Size indicates the expected fragment size.
[Mg2+], MgCl2 concentrations for PCR.
Nucleotide positions correspond to the full genome sequence of ATK, the prototype strain of HTLV-1 (J02029).
These primers, originally developed as a generic PCR system for PTLV, were modified to specifically amplify STLV-L.
Nucleotide positions corresponds to the full genome sequence of PH969, the prototype strain of PTLV-L (Z29673).
Cloning and DNA sequencing.
The amplified fragments were blunt ended by T4 DNA polymerase and subcloned into the HincII site of pUC119. The nucleotide sequences were determined in both directions by using an automated DNA sequencer (373A auto sequencer; Applied Biosystems, Foster City, Calif.) and a commercial kit (Taq DyeDeoxy Terminator Cycle Sequencing Kit, Applied Biosystems). We usually sequenced two clones for each sample.
Phylogenetic analysis.
For construction of phylogenetic trees, both the new and previously reported nucleotide sequences were aligned by using the computer software CLUSTAL W (27) and minor modifications. Pairwise genetic distances were estimated for each resampling by Kimura's two-parameter method (13). All phylogenetic trees in the present study were constructed by the neighbor-joining (NJ) method (20), which is considered to be the most reasonable algorithm in various phylogenetic inference methods. In order to ascertain the robustness of the constructed NJ trees, bootstrapping was done to generate 1,000 resamplings of the original sequence alignments. The trees were visualized with the computer program TREEVIEW (19).
Nucleotide sequence accession numbers.
The new nucleotide sequences in the present study have been deposited in GenBank under accession no. AF378160-2 (pX region) and AY33490-2 (LTR).
RESULTS
In an attempt to understand the evolutionary origins of STLV, we carried out serological and molecular analyses on five different monkey groups from Ethiopia. A total of 519 plasma samples were screened using the PA assay. Cross-reactive antibodies against HTLV were observed in 95 (18.3%) of the samples. These 95 seropositive monkeys included 8 out of 96 (8.3%) anubis baboons, 22 out of 40 (55.0%) hamadryas baboons, 24 out of 50 (48.0%) hybrid baboons, and 41 out of 177 (23.2%) grivet monkeys. None of the 156 gelada baboons was seropositive for STLV (Table 2). This observation was surprising. First, our previous study did not indicate any positivity among the same hamadryas baboons. Second, the number of seropositive hybrid baboons were much higher in the present study than in the previous one. Since the previous study employed IFA for the serological screening assay (11) with an HTLV-1-infected cell line as the antigen, we considered this finding to be a result of the broad specificity of the PA assay used in the present study. Indeed, we conducted IFA on four hamadryas and two hybrid baboons, but none of these samples were seropositive (data not shown). Thus, we speculate that there is a divergent PTLV-related retrovirus (such as STLV-2 or STLV-L) that is PA positive but IFA negative.
TABLE 2.
Prevalence of STLV-1 and -L among seropositive Ethiopian monkeys
| Common name | Total no. of samples | No. of positivesa | PCR resultsb
|
|
|---|---|---|---|---|
| STLV-1 | STLV-L | |||
| Hamadryas baboon | 40 | 22 | 0 (0.0%) | 20 (50.0%) |
| Hybrid baboon | 50 | 24 | 4c (8.0%) | 19c (38.0%) |
| Anubis baboon | 96 | 8 | 8 (8.3%) | 0 (0.0%) |
| Gelada baboon | 156 | 0 | NDd | ND |
| Grivet monkey | 177 | 41 | 35 (20.0%) | 0 (0.0%) |
Data are the numbers of seropositive samples judged by PA.
Type-specific PCR was done to differentiate between STLV-1 and -L. The numbers in parentheses indicate the percentage of positivity.
One sample was dually positive in STLV-1- and STLV-L-specific PCR.
ND, not done because no seropositive samples were found.
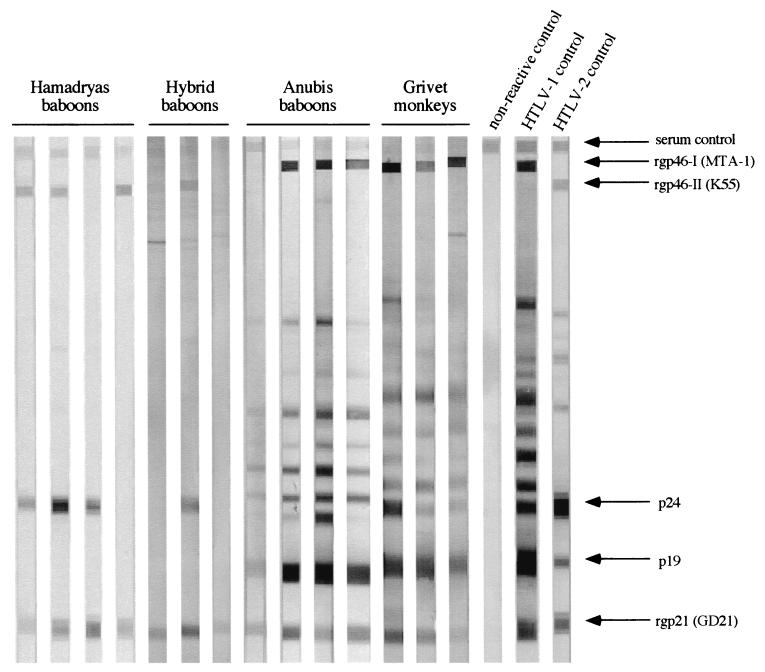
To test this possibility, we carried out further serological assays with two different WB kits. One of them, which is specific for PTLV-1, did not show any clear positive reactivity in two plasma samples of the PA-positive hamadryas baboons (data not shown). The other WB kit that can differentiate PTLV-1 and -2 indicated strong reactivity against the K55, p24, and GD21 proteins in two out of the four PA-positive hamadryas and one of the three tested hybrid baboons (Fig. 2). The WB pattern of these baboons is very similar to the WB patterns of captive baboons infected with STLV-L (29). In contrast, the tested seropositive anubis baboons and grivet monkeys exhibited a typical WB profile of STLV-1. Thus, these results suggested that STLV-L-like viruses are present in the wild hamadryas and hybrid baboons.
FIG. 2.
WB profiles of antibodies against HTLV antigens from 14 Ethiopian monkeys. rgp46-I (MTA-1) and rgp46-II (K55) are envelope recombinant proteins unique to HTLV-1 and -2, respectively, while rgp21 (GD21) is a common yet specific HTLV-1 and -2 epitope recombinant envelope GD21 protein. p24 and p19 are HTLV Gag proteins.
To confirm the presence of STLV, we carried out nested PCR to detect the proviral DNA. At first, we used primer sets that are able to detect PTLV-1 and HTLV-2. We were able to amplify STLV sequences in the samples with an STLV-1-like WB profile from anubis baboons and grivet monkeys, but not in the samples with an STLV-L-like WB profile from hamadryas and hybrid baboons. This result led us to assume that these PCR-negative baboons are infected with STLV-L. Subsequently, we attempted a nested PCR with primers that were specific for STLV-L. To obtain these primers, we started with primers for PTLV-1, -2, and -L (30) and then modified a few nucleotides to make them specific for STLV-L (28). Nested PCR for STLV-L yielded positive signals on the seropositive samples of the hamadryas baboons. In addition, the hybrid baboons with an STLV-L-like WB pattern were also found to be positive with this STLV-L-specific PCR system. We also confirmed that the primer sets for STLV-L were not able to amplify the STLV-1 and -2 proviral DNA, suggesting that the amplified proviruses were STLV-L.
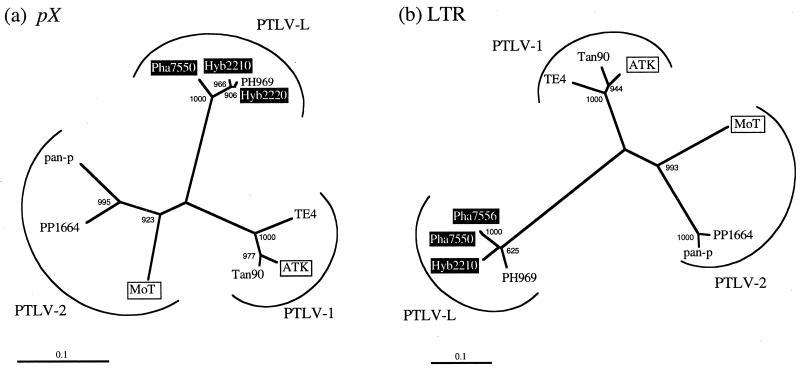
We next determined the nucleotide sequences of pX and LTR. Two amplified fragments were sequenced from each baboon group (hamadryas and hybrid baboons). A BLAST search indicated that the new DNA sequences showed the highest homology to that of PH969, the prototype strain of STLV-L (2, 28). We first compared the pX region of PTLV. Sequence similarities of the new four sequences to PH969 were 94.4 to 99.4%, while those to HTLV-1 and HTLV-2 were 70.6 to 72.2% and 73.9 to 74.4%, respectively (Fig. 3). A phylogenetic tree based on the pX region indeed revealed that the hamadryas and hybrid STLV made a monophyletic cluster together with the prototypic STLV-L strain PH969 (Fig. 4a). Next, we examined the LTR sequences for confirmation of the findings on the pX region (Fig. 4b). The homologies between the new STLV-L and PH969, HTLV-1, and HTLV-2 were 91.1, 49.9, and 53.5%, respectively. A phylogenetic analysis based on LTR also showed that these new strains belong to STLV-L with statistical support. These findings indicate that wild hamadryas baboons are infected with STLV-L.
FIG. 3.
Alignment of partial pX sequences of a new STLV strain from a hamadryas baboon (Pha7550) with prototype strains of STLV-L (PH969), HTLV-1 (ATK), and HTLV-2 (MoT). Dots indicate the same nucleotide as in Pha7550. The DNA sequences except for Pha7550 were obtained from the DNA data bank. The accession numbers of the DNA sequences are J02029 (ATK), M10060 (MoT), and Z29673 (PH969). Nucleotide positions correspond to the full genome sequence of PH969.
FIG. 4.
Phylogenetic trees of PTLV based on part of the pX gene (nucleotide positions 7491 to 7670 in PH969) (a) and part of the LTR region (8304 to 8796 in PH969) (b). The new isolates (white letters in black boxes) were compared with prototype strains representing three types of PTLV. Pha7550 and Pha7556 were derived from hamadryas while Hyb2210 and Hyb2220 were from hybrid baboons. ATK and MoT are human strains while the others are simian strains. The trees were constructed by the NJ method. The scales at the bottom of the trees indicate the number of nucleotide substitutions per site. The lengths of the branches are proportional to the genetic distance estimated by Kimura's two-parameter method. The numbers at the nodes are bootstrap values. The other DNA sequences used for construction of this tree were obtained from the DNA data bank. The accession numbers of the DNA sequences are J02029 (ATK), M10060 (MoT), Z29673 (PH969), Z32851 (PP1664), S74220 (pan-p), Z46900 (TE4), and AF074966 (Tan90).
To examine the prevalence of STLV-L in detail, we carried out type-specific nested PCR for all the positive samples in the PA assay. We found proviral STLV-L in 20 of the 22 seropositive hamadryas baboons. We also detected proviral DNA by type-L-specific PCR in 19 of the 24 hybrid baboons. In contrast, no positive signals were obtained for STLV-L in seropositive anubis baboons or seropositive grivet monkeys. The seropositivity of both the anubis baboons and grivet monkeys was attributed solely to STLV-1 infection as shown by type-1-specific nested PCR (Table 2). We also found proviral STLV by type-1-specific nested PCR in four seropositive hybrid baboons. One of them was also positive in the type-L-specific PCR, suggesting a dual infection with STLV-1 and -L. These results indicate that STLV-L infections are confined to baboons with a blood relation to hamadryas baboons and that their prevalence is extremely high.
DISCUSSION
Simian retroviruses pose a serious threat to public health. Two human retroviruses (HIV and HTLV, the causative agents of AIDS and malignant leukemia, respectively) are believed to have come from NHPs (4, 5, 8, 31). Therefore, a thorough understanding of natural prevalence of these pathogens among wild NHPs may help us not only to elucidate the evolutionary origins of these human retroviruses but also to prevent an epidemic of a new human retrovirus. The present study showed that wild hamadryas baboons in Ethiopia have cross-reactive antibodies against HTLV with a WB profile that is very similar to that of STLV-L-infected captive baboons originally reported and they harbor viral DNAs that are closely related to the prototypic strain of STLV-L (PH969). Although we did not attempt virus isolation from our specimens, the existence of STLV-L was shown to be an infectious by Goubau et al. (7). Thus, our study confirms that STLV-L prevails among hamadryas baboons and extends that the virus also prevails among wild baboons.
Ethiopia is also inhabited by anubis baboons, another subspecies of baboons living in southwest Ethiopia, in addition to hamadryas baboons, which are found mostly in northeast Ethiopia. Hybridization between these subspecies frequently occurs in boundary areas in which these two baboons are cohabiting (25). In addition to hamadryas baboons, we tested the prevalence of STLV-L among anubis baboons and hybrid baboons. We also tested gelada, which are known to be close to baboons (genus Papio). We observed that STLV-L is present in hamadryas and hybrid baboons, but not in anubis baboons and gelada. Seropositive anubis baboons were exclusively infected with STLV-1, and no seropositive samples were obtained from gelada. It is important to note that the hamadryas baboons we tested are a pure population, as determined by genetic studies (25). Thus, these results suggest that STLV-L in hybrid baboons originated from hamadryas baboons.
In addition to baboons, African green monkeys (AGMs) are known to naturally harbor STLV-1 (12, 22). And it was reported that cross-species transmission has likely occurred between baboons and AGMs (14, 17). Thus, we determined whether grivet monkeys, a subspecies of AGMs, are the original natural reservoir of STLV-L from which hamadryas baboons could have acquired STLV-L. Similar analyses were done, but we could not find any evidence for STLV-L infection in grivet monkeys and observed that the seropositivity of the monkeys was completely due to STLV-1 infection. Thus, it is unlikely that hamadryas STLV-L originated from AGMs or that cross-species transmission occurred between hamadryas and AGMs. Thus, STLV-L infections seem to be confined to hamadryas and hamadryas-like (hybrid) baboons. And, how hamadryas baboons acquired STLV-L remains unknown.
The present study revealed that the prevalence of STLV-L was remarkably high in wild baboons. This implies that there has been and will continue to be frequent accidental physical contact between humans and feral baboons infected with STLV-L, which could result in STLV-L spreading into the human population. This seems feasible in view of the fact that STLV-L grows in human cord blood cells in vitro (7). Human and monkey contact is not negligible; for instance, the expansion of human habitats often causes more frequent encounters with baboons and other NHPs. Some other human retroviruses, which cause fatal diseases such as HIV, are indeed believed to come from NHPs (8, 10). Moreover, recent reports have shown that not a single episode but multiple episodes of cross-species transmission of STLV-1 have occurred in Africa, implying that retroviral zoonoses occur frequently (3, 16, 23). In this regard, it is interesting that Virelenik et al. indicated that some Ethiopian individuals (0.43% of the population) exhibit HTLV-2 positive reactivity (32). This result was based on a WB assay that was similar to the one used in the present study. The occurrence of HTLV-2 in Ethiopia may be unrelated to the occurrence of STLV-L in the baboons. However, given the high prevalence of STLV-L in Ethiopia, the possibility that HTLV-2-like seroreactivity of the Ethiopians was due to infection with STLV-L-like retrovirus cannot be ruled out.
Baboons have been considered potential donors for organ transplantation (xenotransplantation), although this proposal has been strongly criticized because of the potential of transmitting new infectious disease to humans (1). In fact, the U.S. Food and Drug Administration recommends further research before proceeding with NHP xenotransplantation. In this regard, the present findings indicate that STLV-L should be considered another pathogen of baboons that could jump to humans and stress the need for further investigation of captive baboons.
ADDENDUM IN PROOF
After submission of the manuscript, the presence of STLV-L was also identified in Cameroonian monkeys by two independent research groups (S. Van Dooren, M. Salemi, X. Pourrut, M. Peeters, E. Delaporte, M. Van Ranst, and A.M. Vandamme, J. Virol. 75:11939-11941, 2001, and L. Meertens, R. Mahieux, P. Maucl re, J. Lewis, and A. Gessain, J. Virol. 76:259-268, 2002).
Acknowledgments
This study was partly supported by a Grant-in-Aid for Cancer Research, Overseas Scientific Research and Scientific Research from the Japanese Ministry of Education, Science and Culture (1978, 1983, and 1993) and by a cooperative research grant and the Overseas Special Research Program of the Primate Research Institute, Kyoto University (1978 and 1983). M.Y. and M.K.S. were supported as research fellows by the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Allan, J. S. 1996. Xenotransplantation at a crossroads: prevention versus progress. Nat. Med. 2:18-21. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J., L. Zekeng, M. Yamashita, J. Takehisa, T. Miura, E. Ido, I. Mboudjeka, J. Tsague, M. Hayami, and L. Kaptue. 1995. HTLV type I isolated from a Pygmy in Cameroon is related to but distinct from the known central African type. AIDS Res. Hum. Retrovir. 11:1529-1531. [DOI] [PubMed] [Google Scholar]
- 4.Franchini, G., and M. J. Reitz. 1994. Phylogenesis and genetic complexity of the nonhuman primate Retroviridae. AIDS Res. Hum. Retrovir. 9:1047-1060. [DOI] [PubMed] [Google Scholar]
- 5.Gessain, A., and G. de The. 1996. Geographic and molecular epidemiology of primate T lymphotropic retroviruses: HTLV-I, HTLV-II, STLV-I, STLV-PP and PTLV-L. Adv. Virus Res. 47:377-426. [DOI] [PubMed] [Google Scholar]
- 6.Giri, A., P. Markham, L. Digilio, G. Hurteau, R. C. Gallo, and G. Franchini. 1994. Isolation of a novel simian T-cell lymphotropic virus from Pan paniscus that is distantly related to the human T-cell leukemia/lymphotropic virus types I and II. J. Virol. 68:8392-8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goubau, P., M. Van Brussel., A. M. Vandamme, H. F. Liu, and J. Desmyter. 1994. A primate T-lymphotropic virus, PTLV-L, different from human T- lymphotropic viruses types I and II, in a wild-caught baboon (Papio hamadryas). Proc. Natl. Acad. Sci. USA 91:2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 9.Hayami, M., A. Komuro, K. Nozawa, T. Shotake, K. Ishikawa, T. Yamamoto, T. Ishida, S. Honjo, and Y. Hinuma. 1984. Prevalence of antibodies to adult T-cell leukemia virus-associated antigens (ATLA) in Japanese monkeys and other nonhuman primates. Int. J. Cancer 33:179-183. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, V. M., G. Dapolito, R. Goeken, and B. J. Campbell. 1995. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr. Opin. Genet. Dev. 5:798-806. [DOI] [PubMed] [Google Scholar]
- 11.Ishida, T., K. Yamamoto, T. Shotake, K. Nozawa, M. Hayami, and Y. Hinuma. 1986. A field study of infection with human T-cell leukemia virus among African primates. Microbiol. Immunol. 30:315-321. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa, K., M. Fukasawa, H. Tsujimoto, J. Else, M. Isahakia, N. Ubhi, T. Ishida, O. Takenaka, Y. Kawamoto, T. Shotake, H. Ohsawa, B. Ivanoff, R. Cooper, E. Frost, F. Grant, Y. Spriatna, Y. Sutarman, K. Abe, K. Yamamoto, and M. Hayami. 1987. Serological survey and virus isolation of simian T-cell leukemia/T-lymphotropic virus type I (STLV-I) in non-human primates in their native countries. Int. J. Cancer 40:233-239. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Koralnik, I., E. Boeri, W. Saxinger, A. Monico, J. Fullen, A. Gessain, H. Guo, P. Markham, V. Kalyanarayan, V. Hirsh, J. Allan, K. Murthy, P. Alford, J. Slattery, S. O'Brien, and G. Franchini. 1994. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J. Virol. 68:2693-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, H., A. Vandamme, M. Brussel, J. Desmyter, and P. Goubau. 1994. New retroviruses in human and simian T-lymphotropic viruses. Lancet 344:265-266. [DOI] [PubMed] [Google Scholar]
- 16.Mahieux, R., F. Ibrahim, M. Mauclere, V. Herve, P. Michel, F. Tekaia, C. Chappey, B. Garin, E. van Der Ryst, B. Guillemain, E. Ledru, E. Delaporte, G. de The, and A. Gessain. 1997. Molecular epidemiology of 58 new African human T-cell leukemia virus type 1 (HTLV-1) strains: identification of a new and distinct HTLV-1 molecular subtype in Central Africa and in Pygmies. J. Virol. 71:1317-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahieux, R., S. J. Pecon, G. M. Chen, and A. Gessain. 1998. Evolutionary inferences of novel simian T lymphotropic virus type 1 from wild-caught chacma (Papio ursinus) and olive baboons (Papio anubis). Virology 251:71-84. [DOI] [PubMed] [Google Scholar]
- 18.Miura, T., J. Sakuragi, M. Kawamura, M. Fukasawa, E. Moriyama, T. Gojobori, K. Ishikawa, J. Mingle, V. Netty, H. Akari, M. Enami, H. Tsujimoto, and M. Hayami. 1990. Establishment of a phylogenetic survey system for AIDS-related lentivirus and demonstration of a new HIV-2 subgroup. AIDS 4:1257-1261. [DOI] [PubMed] [Google Scholar]
- 19.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-4358. [DOI] [PubMed] [Google Scholar]
- 20.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 21.Saksena, N., V. Herve, J. Durand, B. Leguenno, O. Diop, J. Digoutte, C. Mathiot, M. Muller, J. Love, S. Dube, M. Sherman, P. Benz, S. Erensoy, A. Galat-Luong, G. Galat, B. Paul, D. Dube, F. Barre-Sinoussi, and B. Poiesz. 1994. Seroepidemiologic, molecular, and phylogenetic analyses of simian T-cell leukemia viruses (STLV-I) from various naturally infected monkey species from Central and Western Africa. Virology 198:297-310. [DOI] [PubMed] [Google Scholar]
- 22.Saksena, N., V. Herve, M. Sherman, J. Durand, C. Mathiot, M. Muller, J. Love, D. Dube, and B. Poiesz. 1993. Sequence and phylogenetic analyses of a new STLV-I from a naturally infected tantalus monkey from Central Africa. Virology 192:312-320. [DOI] [PubMed] [Google Scholar]
- 23.Salemi, M., D. S. Van, E. Audenaert, E. Delaporte, P. Goubau, J. Desmyter, and A. M. Vandamme. 1998. Two new human T-lymphotropic virus type I phylogenetic subtypes in seroindeterminates, a Mbuti pygmy and a Gabonese, have closest relatives among African STLV-I strains. Virology 246:277-287. [DOI] [PubMed] [Google Scholar]
- 24.Shimada, M. K., and T. Shotake. 1997. Genetic variation of blood proteins within and between local populations of grivet monkey (Cercopithecus aethiops aethiops) in central Ethiopia. Primates 38:399-414. [Google Scholar]
- 25.Shotake, T. 1981. Population genetical study of natural hybridization between Papio anubis and P. hamadryas. Primates 22:285-308. [Google Scholar]
- 26.Shotake, T., and K. Nozawa. 1984. Blood protein variations in baboons. II. Genetic variability within and among herds of gelada baboons in the central Ethiopian plateau. J. Hum. Evol. 13:265-274. [Google Scholar]
- 27.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Brussel, M., P. Goubau, R. Rousseau, J. Desmyter, and A. Vandamme. 1997. Complete nucleotide sequence of the new simian T-lymphotropic virus, STLV-PH969 from a Hamadryas baboon, and unusual features of its long terminal repeat. J. Virol. 71:5464-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Brussel, M., M. Salemi, H. Liu, P. Goubau, J. Desmyter, and A.-M. Vandamme. 1999. The discovery of two new divergent STLVs has implications for the evolution and epidemiology of HTLVs. Rev. Med. Virol. 9:155-170. [DOI] [PubMed] [Google Scholar]
- 30.Vandamme, A., K. Laethem, H. Liu, M. van Brussel, E. Delaporte, C. de Castro Costa, C. Fleischer, G. Taylor, U. Bertazzoni, J. Desmyter, and P. Goubau. 1997. Use of a generic polymerase chain reaction assay detecting human T-lymphotropic virus (HTLV) types I, II and divergent simian strains in the evaluation of individuals with indeterminate HTLV serology. J. Med. Virol. 52:1-7. [PubMed] [Google Scholar]
- 31.Vandamme, A. M., M. Salemi, and J. Desmyter. 1998. The simian origins of the pathogenic human T-cell lymphotropic virus type I. Trends Microbiol. 6:477-483. [DOI] [PubMed] [Google Scholar]
- 32.Vrielink, H., Y. Sisay, H. W. Reesink, M. Woerdeman, C. Winkel, S. J. de Leeuw, P. N. Lelie, and, C. L. van der Poel. 1995. Evaluation of a combined lysate/recombinant antigen anti-HTLV-I/II ELISA in high and low endemic areas of HTLV-I/II infection. Transfus. Med. 5:135-137. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita, M., A. Achiron, T. Miura, J. Takehisa, E. Ido, T. Igarashi, K. Ibuki, M. Osame, S. Sonoda, E. Melamed, M. Hayami, and B. Shohat. 1995. HTLV-I from Iranian Mashhadi Jews in Israel is phylogenetically related to that of Japan, India, and South America rather than to that of Africa and Melanesia. Virus Genes 10:85-90. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita, M., R. Veronesi, M. Menna-Barreto, W. Harrington, Jr., C. Sampio, C. Brites, R. Badaro, A. Andrade-Filho, S. Ohkura, T. Igarashi, J. Takehisa, T. Miura, D. Chamone, O. Bianchini, C. Jardim, S. Sonoda, and M. Hayami. 1999. Molecular epidemiology of HTLV-I in Brazil: the predominant HTLV-Is in South America differ from HTLV-Is of Japan and Africa as well as those of Japanese immigrants and their relatives in Brazil. Virology 261:59-69. [PubMed] [Google Scholar]
- 35.Yanagihara, R. 1994. Geographic-specific genotypes or topotypes of human T-cell lymphotropic virus type I as markers for early and recent migrations of human populations. Adv. Virus Res. 43:147-186. [DOI] [PubMed] [Google Scholar]