Abstract
The translation of polioviral mRNA occurs through an internal ribosomal entry site (IRES). Several RNA-binding proteins, such as polypyrimidine tract-binding protein (PTB) and poly(rC)-binding protein (PCBP), are required for the poliovirus IRES-dependent translation. Here we report that a poliovirus protein, 3Cpro (and/or 3CDpro), cleaves PTB isoforms (PTB1, PTB2, and PTB4). Three 3Cpro target sites (one major target site and two minor target sites) exist in PTBs. PTB fragments generated by poliovirus infection are redistributed to the cytoplasm from the nucleus, where most of the intact PTBs are localized. Moreover, these PTB fragments inhibit polioviral IRES-dependent translation in a cell-based assay system. We speculate that the proteolytic cleavage of PTBs may contribute to the molecular switching from translation to replication of polioviral RNA.
Picornaviruses are important pathogens of both humans (e.g., poliovirus, rhinovirus, and hepatitis A virus) and animals (e.g., foot-and-mouth disease virus [FMDV] and encephalomyocarditis virus [EMCV]) (75). Their genome consists of a positive-sense RNA that acts as a template for both translation and replication. On entering the host cell, the RNA directs the translation of a single polyprotein, which is cleaved during and after translation by virus-encoded proteases into functional proteins. In poliovirus, three virus-encoded proteases, 2Apro (specific to Tyr-Gly), 3Cpro (Gln-Gly), and 3CDpro (Gln-Gly), are involved in protein processing during virus replication (34, 50, 54, 60, 65-67, 80, 81). These proteases also play a role in host cell alteration; for example, it is well known that poliovirus 2Apro cleaves eIF4G, which results in shutting off the cap-dependent translation of host mRNAs (6, 19, 21, 51). Recent studies suggest that 2Apro and 3Cpro cleave poly(A)-binding protein (PABP), which augments the translation of the poly(A)-tailed mRNAs (42, 47). 3Cpro induces morphological changes in host cells by cleaving microtubule-associated protein (MAP-4) (41). 3Cpro inhibits the transcription of host mRNAs by proteolytically cleaving transcription factors, such as TATA-binding protein (TBP), TFIIIC, Oct-1, and CREB (15, 18, 85, 86). A recent report suggests that 3Cpro cleaves La autoantigen and that this results in the redistribution of La in the cytoplasm, which results in the enhanced translation of viral mRNAs (77).
After poliovirus RNA has been released into the cytoplasm of infected cells, translational initiation of the poliovirus RNA is initiated by the binding of ribosomes to a specialized region in the 5" nontranslated region (5"NTR), called the internal ribosomal entry site (IRES) (20, 71, 82). IRES-dependent translation of poliovirus requires IRES-specific cellular factors as well as canonical initiation factors for efficient translation (2, 10). Known cellular factors required for polioviral IRES include polypyrimidine tract-binding protein (PTB) (28, 33, 37, 38), La protein (9, 17), and poly(rC) binding protein 2 (PCBP2) (12, 25).
PTB (also known as p57 and hnRNP I) is a member of the hnRNP family and shuttles between the nucleus and the cytoplasm in a transcription-sensitive manner (58). PTB was identified originally as a protein binding to the polypyrimidine tracts (Py tracts) of adenoviral major-late and α-tropomyosin pre-mRNAs and was proposed as a splicing factor (27, 70). Binding of PTB to the Py tract near the branch point of intron was shown to modulate the alternative splicing of certain pre-mRNAs (55, 79, 83). Independently, PTB was shown to interact specifically with the IRESs of several picornaviruses (2, 10), including poliovirus (30, 33, 35), hepatitis A virus (13, 14), FMDV (56, 62), and EMCV (39, 44).
Experiments on the depletion and repletion of PTB from rabbit reticulocyte lysate (RRL) indicated that PTB is required for the efficient translation of and FMDV mRNA and a mutant EMCV mRNA (44, 62). Intriguingly, PTB is required for translation of a mutant EMCV mRNA, but not for wild-type EMCV (43). This may suggest that PTB plays a role in maintaining the proper conformation of the mutant EMCV IRES (43). Supplementation of RRL with PTB enhances the translation of polioviral mRNA (37, 38). Moreover, immunodepletion of PTB from HeLa cell lysate resulted in inhibition of polioviral IRES-dependent translation. Repletion of purified PTB to the immunodepleted lysate did not restore the polioviral IRES-dependent translation (33). The authors suggested that an unidentified translation factor(s) might be removed from the lysate in the depletion process (33). The effect of PTB on polioviral IRES-dependent translation was investigated by using artificial dicistronic mRNAs containing the PTB gene as the first cistron, the poliovirus IRES at the intercistronic region, and the chloramphenicol acetyltransferase (CAT) reporter gene as the second cistron (28). When transfected into HeLa cells, which contain a limited amount of PTB, it was found that the additional expression of PTB stimulated the activity of polioviral IRES 2.5-fold (28). These results suggest that PTB is required for, or at least enhances, the IRES activity of picornaviral IRESs.
Three isoforms of PTB have been reported (26, 27, 70). The prototype of PTB (PTB1) consists of 531 amino acids, with a molecular mass of 57 kDa. PTB2 and PTB4 have insertions of 19 and 26 amino acids (aa), respectively, after the amino acid at position 298 of PTB1 (26, 70). Four loosely conserved RNA-recognition motifs (RRMs) are distributed throughout the PTB molecule (see Fig. 3A) (46). Biochemical studies and yeast two-hybrid analysis of PTB revealed the following. First, PTB exists in oligomeric as well as monomeric forms. The central part of PTB, including RRM2, plays a key role in its oligomerization (63, 72). Second, PTB contains several RNA-binding domains, among which the C-terminal part of PTB spanning RRM3-4 exerts the strongest RNA-binding activity (16, 63, 72). Third, the N-terminal part of PTB is responsible for the enhancement of RNA-binding activity by HeLa cell cytoplasmic factor(s) (63).
FIG. 3.
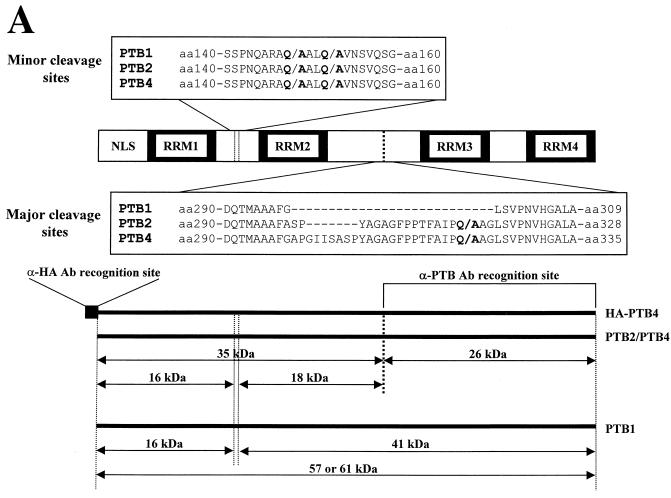
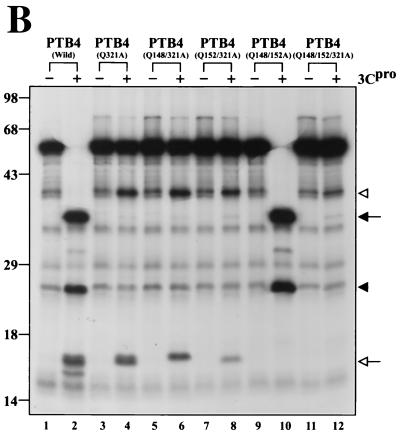
Identification of 3Cpro cleavage sites in PTBs. (A) Schematic diagram of 3Cpro cleavage sites in PTBs. The amino acid sequences around the major and minor cleavage sites in PTBs are shown in the single-letter motif. The NLS and RRMs in PTB are indicated by open and solid boxes, respectively, and the sizes of the cleaved products are shown in kilodaltons. Ab, antibody. (B) [35S]methionine-labeled PTB4 and its mutants were incubated with (+) or without (−) 3Cpro at 37°C for 3 h. Protease reactions were stopped by the addition of 2× Laemmli sample buffer, and the samples were analyzed by SDS-PAGE followed by autoradiography.
After the synthesis of viral proteins, the input virion RNA should serve as a template for the synthesis of negative-strand RNA (5, 73). The switch from translation to replication should occur properly, since the continuous translation of polioviral mRNA blocks the replication of viral RNA (1, 8, 24). Recent studies suggest that the 5"-terminal cloverleaf (CL) RNA structure in poliovirus is required for both positive- and negative-strand RNA synthesis (3, 7, 73). CL RNA forms a ternary ribonucleoprotein complex with poly(rC) binding protein (PCBP) and the uncleaved precursor of viral protease-polymerase 3CD (12, 22, 23, 25, 69, 78, 84). Moreover, the interactions of PCBP2 with the poliovirus 5"NTR are modulated by the viral protein 3CDpro (22). It has also been suggested that the change of PCBP2 binding sites from domain IV to domain I of the polioviral 5"NTR results in a molecular switching from translation to the replication of viral RNA (22, 24, 25).
In order to further understand the mechanism of the molecular switching, we analyzed the effect of modifications upon cellular proteins known to enhance polioviral IRES-dependent translation during poliovirus infection. Isoforms of PTB were cleaved as poliovirus infection proceeds. In vitro and in vivo analyses indicate that 3Cpro is responsible for the cleavage of PTB isoforms, which occurs at multiple cleavage sites (with amino acid residues Ala-X-X-Gln/Ala) of PTB. The C-terminal parts, as well as possibly the N-terminal parts, of the cleaved products were redistributed to the cytoplasm from the nucleus, where most of the intact PTBs are localized. Expression of the PTB fragments inhibited the translation of polioviral RNAs in a cell-based assay system. We speculated that the cleavage of PTB by polioviral protease 3Cpro and/or 3CDpro contributes, at least in part, to the molecular switching from translation to replication.
MATERIALS AND METHODS
Cells and cell cultures.
HeLa, H1-HeLa, 293T (H1-HeLa and 293T cells were gifts from E. Wimmer), and Chang liver cells were cultured in Dulbecco's modified Eagle medium (DMEM; GIBCO BRL) supplemented with 10% fetal bovine serum. A HeLa-derived cell line, designated HeLa Tet-Off, which expresses the chimeric tet-responsive transcriptional activator (tTA), was purchased from Clontech. The cell line was cultured in DMEM supplemented with 10% fetal bovine serum, 2 μg of tetracycline (Sigma) per ml, and 100 μg of G418 (Calbiochem) per ml. To generate stably transformed HeLa cells expressing hemagglutinin (HA)-tagged PTB4, HeLa Tet-Off cells were cotransfected with pTRE/HA-PTB4 containing the HA-PTB4 gene under the control of tTA-responsive promoter and pPUR (Clontech), which contains the puromycin resistance gene. Transformants were selected with puromycin, and puromycin-resistant stable cell clones were screened for the expression of HA-PTB after withdrawal of tetracycline from the medium. The expression of HA-PTB4 was monitored by Western blotting with mouse anti-HA monoclonal antibody (Santa Cruz Biotechnology, Inc.). Among several positive clones identified, we selected those that showed highly regulated expression of HA-PTB4. Thus, in the subsequent experiments, we used a clone named HeLa12/HA-PTB4 that expressed HA-PTB4.
Viruses and virus infection.
Mahoney type 1 poliovirus stocks were prepared and infections were initiated as previously described (31). To prepare human rhinovirus type 14 (HRV-14), plasmid pHRV-14 (provided by E. Wimmer) was digested with XmaI to produce the viral RNAs with T7 RNA polymerase. These RNA transcripts were used to transfect H1-HeLa monolayers (31), and HRV-14 was grown and purified as previously described (57). All virus infections were performed at a multiplicity of infection of 100. Cells were harvested at the indicated times and pelleted. For immunoblotting, the cells were resuspended in phosphate-buffered saline (PBS) and mixed with 2× Laemmli sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel analysis. The mouse anti-PTB monoclonal antibody (DH-17) was used as previously described (49). The mouse anti-influenza HA monoclonal antibody was used at a dilution of 1:500, as described by the manufacturer (Santa Cruz Biotechnology, Inc.). Horseradish peroxidase-linked anti-mouse (1:5,000 dilution) immunoglobulin G (IgG) (Amersham) was used as the secondary antibody.
Construction of plasmids.
To obtain the specific cDNA sequence of PTB4, a cDNA library was constructed with the mRNAs from the Chang liver cells with a HybriZAP-2.1 XR library construction kit (Stratagene). cDNA corresponding to the region from aa 295 to 355 of PTB4 was isolated from the Chang liver cDNA library by PCR with four primers: 5" AACGTCAAGTACAACAATGAC 3" and 5" GCGCTGCACGTCACCGTAGAC 3" for the first round of PCR and 5" GACTACACACGCCCAGACCTG 3" and 5" TGTGACTCTCTCTGGGTTGAG 3" for the second round of PCR. The amplified cDNA fragment and plasmid pTM1H/PTB1 (63) were digested with SacII and SmaI and then ligated to produce construct pTM1H/PTB4. The clone was confirmed by sequencing. To construct pTM1H/PTB2, PCR was used to amplify a cDNA fragment corresponding to the region from aa 295 to 550 of pTM1H/PTB4. The two primers used were primer 6 (5" CCCAATTCCGCGGCCTTCGCCTCTCCGTATGCAGGAGCTGGTTTCCCTCCC 3") and primer 7 (5" TGCAGGTCGACCTAGATGGTGGACTTGGAGAAGG 3"). The PCR fragment and pTM1H/PTB4 were digested with SacII and SalI and then ligated to produce pTM1H/PTB2. The clones were confirmed by sequencing. pT7-7/PTB1 has been described by Oh et al. (63). pT7-7/PTB4 was constructed by inserting DNA fragments of pTM1H/PTB4 treated with EcoRI and SalI into pT7-7 vector treated with EcoRI and SalI.
Site-directed mutagenesis was performed by PCR. All of the inserted PCR fragments were completely sequenced. To construct pSK/PTB4(1-325) containing the mutation Gln321→Ala, PCR was performed to amplify a cDNA fragment corresponding to the region from aa 1 to 325 of pTM1H/PTB4. The two primers used were primer A (5" AACAGAATTCCCCGATCCATCG 3") and primer B (5" AACGTCAGGCCTGCGGCGGCAGGAATGGCAAAGGTGGG 3"). The PCR fragment containing mutation (Q321A) was treated with polynucleotide kinase and T4 polymerase. The PCR product was inserted into pBluescript SK vector (Stratagene) cleaved with SmaI. To construct pSK/PTB4(1-325)(Q148/321A), DNA fragments were amplified from pTM1H/PTB4. The four primers used were primer A and primer 2 (5" AGGAACCCGGGCCTGGTTGGGAGAGCTGTC 3") for cDNA fragment 1, corresponding to the region from aa 1 to 146, and primer 1 (5" AGGAACGCCCGGGCCGCGGCGGCCCTGCAGGCGGTG 3") and primer B for cDNA fragment 2, corresponding to the region from aa 145 to 325. cDNA fragment 1 treated with XmaI and EcoRI and cDNA fragment 2 treated with XmaI and PflMI were inserted into pSK/PTB4(1-325)(Q321A) treated with EcoRI and PflMI. To construct pSK/PTB4(1-325)(Q152/321A), the DNA fragment was amplified from pTM1H/PTB4. The four primers used were primer A and primer 4 (5" AGGTTAGTTAACCGCCGCCAGGGCCGCCTG 3") for cDNA fragment 3, corresponding to the region from aa 1 to 155, and primer 3 (5" AGGTTAGTTAACTCGGTCCAGTCGGGG 3") and primer B for cDNA fragment 4, corresponding to the region from aa 154 to 325. cDNA fragment 3 treated with HindII and EcoRI and cDNA fragment 4 treated with HindII and PflMI were inserted into pSK/PTB4(1-325)(Q321A) treated with EcoRI and PflMI. To construct pSK/PTB4(1-325)(Q148/152/321A), the DNA fragment was amplified from pSK/PTB4(1-325)(Q152/321A). The two primers used were primer 5 (5" AGGAACGCCCGGGCCGCGGCGGCCCTGGCGGCGGTTAAC 3") and primer B for cDNA fragment 5, corresponding to the region from aa 145 to 325. cDNA fragment 1 treated with XmaI and EcoRI and cDNA fragment 5 treated with XmaI and PflMI were inserted into pSK/PTB4(1-325)(Q321A) treated with EcoRI and PflMI.
pTM1H/PTB4(Q321A) was constructed by inserting a DNA fragment from pSK/PTB4(1-325)(Q321A) treated with StuI and EcoRI into the vector pTM1H/PTB2 treated with StuI and EcoRI. pTM1H/PTB4(Q148/321A) was constructed by inserting a DNA fragment from pSK/PTB4(1-325)(Q148/321A) treated with EcoRI and StuI into the vector pTM1H/PTB2 treated with EcoRI and StuI. pTM1H/PTB4(Q152/321A) was constructed by inserting a DNA fragment from pSK/PTB4(1-325)(Q152/321A) treated with EcoRI and SacII into the vector pTM1H/PTB4(Q321A) treated with EcoRI and SacII. pTM1H/PTB4(Q148/152A) was constructed by inserting a DNA fragment from pSK/PTB4(1-325)(Q148/152/321A) treated with EcoRI and SacII into the vector pTM1H/PTB4 treated with EcoRI and SacII. pTM1H/PTB4(Q148/152/321A) was constructed by inserting a DNA fragment from pSK/PTB4(1-325)(Q148/152/321A) treated with EcoRI and StuI into the vector pTM1H/PTB2 treated with EcoRI and StuI.
pTRE/HA-PTB4 was constructed by inserting DNA fragments from pCMV/HA-PTB4 treated with SalI and Klenow fragment into the vector pTRE (Clontech) treated with BamHI-EcoRI-Klenow fragment. pCMV-HA was made by inserting the HA sequence into the multicloning site of pΔEGFP-N1 (49a). To construct pCMV/HA-PTB4, pCMV/HA was digested with XbaI and then treated with Klenow fragment. Plasmid pTM1H/PTB4 was digested with EcoRI-SalI-Klenow fragment. PTB4 insert was ligated into the vector to generate pCMV/HA-PTB4.
To construct pEGFP/PTB4 (RRM1-2-3-4; aa 1 to 557), pEGFP/PTB4-RRM1-2 (aa 1 to 324), and pEGFP/PTB4-RRM3-4 (aa 325 to 557), pTM1H/PTB4 was treated with EcoRI-SalI-Klenow for PTB4(1-557), with EcoRI-StuI-Klenow for PTB4-RRM1-2 (aa 1 to 324), and with StuI for PTB4-RRM3-4 (aa 325 to 557). Each fragment was ligated into pEGFP-C1 (Clontech) treated with SalI and Klenow fragment. To construct pEGFP/PTB4-RRM1-2ΔN (aa 58 to 324), pEGFP/PTB4-RRM1-2 (aa 1 to 324) was treated with XbaI-EcoRI-Klenow and self-ligated to remove aa 1 to 57. To construct pEGFP/PTB1-RRM2-3-4 (aa 154 to 530), a DNA fragment was amplified from pTM1H/PTB1(1-530). The two primers used were primers 3 and 7. The cDNA fragment treated with HindII was inserted into pEGFP-C1 treated with HindIII and Klenow fragment. The clone was confirmed by sequencing.
To construct pEGFP/PTB4/RFP, pEGFP/PTB4 was treated with BsaI-Klenow fragment-NdeI for GFP/PTB4(1-552). The GFP/PTB4(1-552) fragment was inserted into pDsRed1-N1 (Clontech) treated with XhoI-Klenow fragment-NdeI.
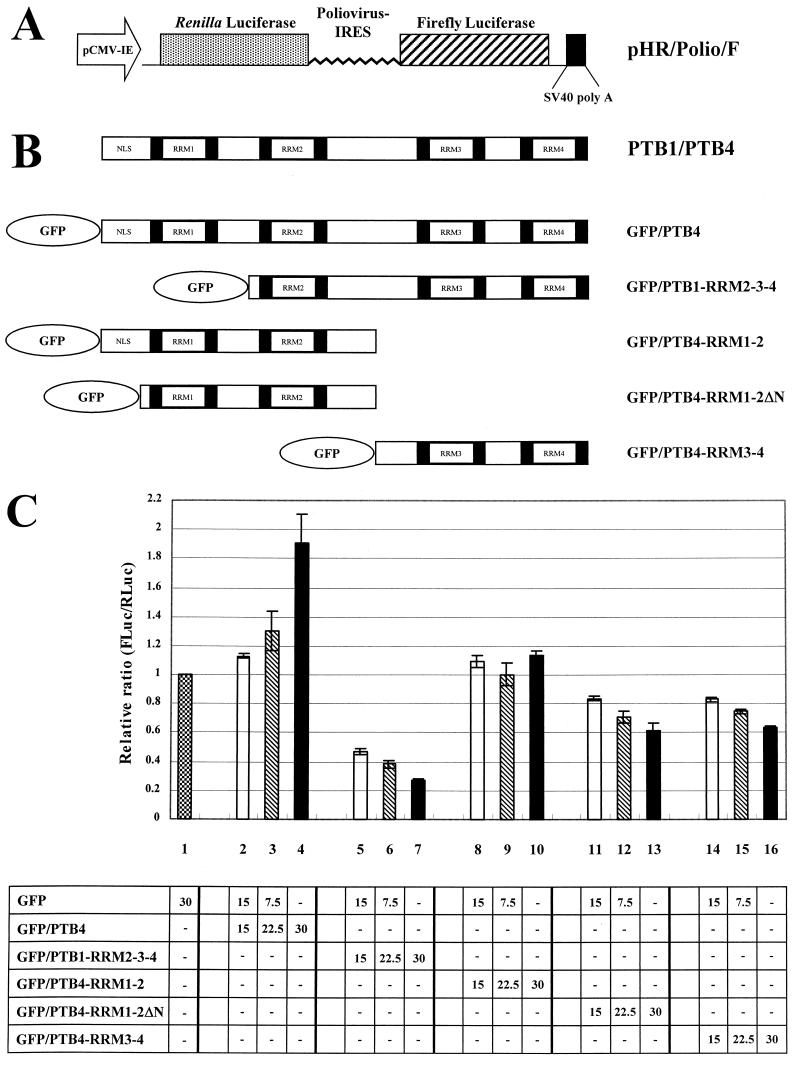
To construct dicistronic reporter plasmid (pHR/Polio/F), we used pHRF for the vector and pECPL for the insert. pHR/Polio/F contains a hairpin structure followed by a Renilla luciferase, poliovirus IRES, and firefly luciferase. The stem-loop structure contains a stable double-stranded RNA region (−91.6 kcal/mol). To construct pHRF, the duplex inserts were made by annealing heat-denatured two single-stranded oligonucleotides (5" CCCCTAGCTAGCCTCGAGCGGATCCGTGGATCCACGGATCCGCCATGGCATG 3" and 5" CATGCCATGGCGGATCCGTGGATCCACGGATCCGCTCGAGGCTAGCTAGGGG 3"). The duplex insert digested with NheI and NcoI was inserted into pRF (49a). Plasmid pECPL contains the 5"NTR of poliovirus between the CAT and firefly luciferase reporter genes. It was constructed by replacing the BiP IRES sequences of pECBL (49) with the poliovirus IRES sequence of pMPS1-ECAT (39). pHR/Polio/F was constructed by inserting the DNA fragment from pECPL treated with NotI and SalI into the vector pHRF treated with NotI and SalI.
Purification of PTB1, PTB4, and polioviral 3Cpro.
The purification steps used to purify the recombinant PTB1 and PTB4 are described elsewhere (63). Polioviral 3Cpro was expressed from plasmid pET3Che (kindly provided by E. Wimmer) as a C-terminal histidine-tagged protein in BL21(DE3) cells and purified to homogeneity on Ni-nitrilotriacetic acid (NTA) resin (Qiagen) under nondenaturing conditions (42). All purified proteins were dialyzed against HT buffer (16.2 mM HEPES-KOH [pH 7.5], 36 mM KCl, 160 mM KOAc, 1.24 mM MgOAc, 1.6 mM dithiothreitol, 2.8 mM β-mercaptoethanol).
In vitro transcription and translation.
Plasmid DNAs were purified by the polyethylene glycol precipitation method (76) and linearized with the appropriate restriction enzymes. The linearized DNAs were then treated with phenol-chloroform and ethanol precipitated. Transcription reactions were performed with T7 RNA polymerase (Boehringer Mannheim) at 37°C for 90 min, according to the manufacturer's instructions. The concentration of the RNA transcripts was determined by using a UV spectrophotometer. Plasmids pTM1H/PTB1, pTM1H/PTB2, pTM1H/PTB4, pTM1H/PTB4(Q321A), pTM1H/PTB4(Q148/321A), pTM1H/PTB4(Q152/321A), pTM1H/PTB4(Q148/152A), and pTM1H/PTB4(Q148/152/321A) were digested with SalI and used to generate the PTB1/2/4 and PTB4 mutant mRNAs, respectively. In vitro translations in micrococcal nuclease-treated RRL (Promega) were performed in 20-μl volumes of reaction mixtures containing 30 nM mRNA. Translation reactions were carried out at 30°C for 1 h in the presence of [35S]methionine (NEN) and were stopped by adding 0.6 mg of cycloheximide per ml and 1 mM cold methionine.
PTB cleavage assays and peptide sequencing of the cleaved products.
In vitro cleavage assays of PTBs were performed under essentially the same conditions regardless of the substrate source. We used HeLa cells as a source of three isoforms of PTB. The cell lysates were prepared by sonication and used as substrates in cleavage reactions with protease in vitro. Reaction mixtures containing 20 μg of cell lysate, various amounts of protease, and HT buffer were incubated at 37°C for 3 h. Purified PTB1 or PTB4 proteins (1.0 μg) were incubated with the indicated quantities of protease for 4 h at 37°C. [35S]methionine-labeled PTBs and mutants were incubated with 1.0 μg of 3Cpro for 1 h. All reactions were stopped by addition of 2× Laemmli sample buffer. Samples were analyzed on SDS-polyacrylamide gels by immunoblotting or autoradiography. To determine the N-terminal sequences of the PTB1 or PTB4 cleavage products, the cleavage assays were allowed to proceed overnight. Proteins were electrophoresed on SDS-12% polyacrylamide gel and then transferred to polyvinylidene difluoride membranes. The Coomassie blue-stained protein bands were microsequenced with an automatic protein sequencer (Applied Biosystems) at the Korea Basic Science Institute (KBSI).
Fluorescence microscopy analysis.
After infection or transfection, the culture medium was removed, and the cells washed three times with PBS. The cells on the coverslip were fixed with 3.5% (wt/vol) paraformaldehyde (Sigma) at room temperature for 12 min. After being washed three times with PBS, the cells on the coverslip were permeabilized in 0.1% Triton X-100 at room temperature for 2 min and washed three times with PBS. For PTB immunostaining, the samples were blocked in solution (PBS containing 1% bovine serum albumin [BSA]) for 30 min at room temperature and then incubated with anti-PTB antibody for 1 h at room temperature. After being washed with PBS, the samples were treated with Hoechst 33258 for 2 min at room temperature and washed again with PBS three times, before being reacted with rhodamine (tetramethyl rhodamine isothiocyanate [TRITC])-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. To observe the fluorescence of enhanced green fluorescence protein (EGFP) and red fluorescence protein (RFP), the samples were also treated with 0.1 μg of Hoechst 33258 per ml for 2 min at room temperature. Finally, the coverslips were washed three times with PBS, and the coverslips with adhering cells were placed on a glass slide and sealed with transparent nail polish. The images were captured with a cooled charge-coupled device camera and a Zeiss (Jena, Germany) Axioplan microscope. Data were processed with Adobe photoshop software (Mountain View, Calif.).
Measurement of polioviral IRES activities in vivo.
293T cells were cultured and transfected by the electroporation procedure. Cells were cotransfected with 0.75 μg of reporter plasmid and the indicated amount of effector plasmids. The total amount of effector plasmids was maintained by adding control plasmid pEGFP-C1 if necessary. Approximately 48 h after transfection, the cells were harvested and lysed by freeze-thawing. Cell extracts were analyzed for Renilla luciferase and firefly luciferase activities by using the dual-luciferase reporter assay system (Promega).
RESULTS
Cleavage of PTB in poliovirus-infected HeLa and HeLa12/HA-PTB4 cell lines.
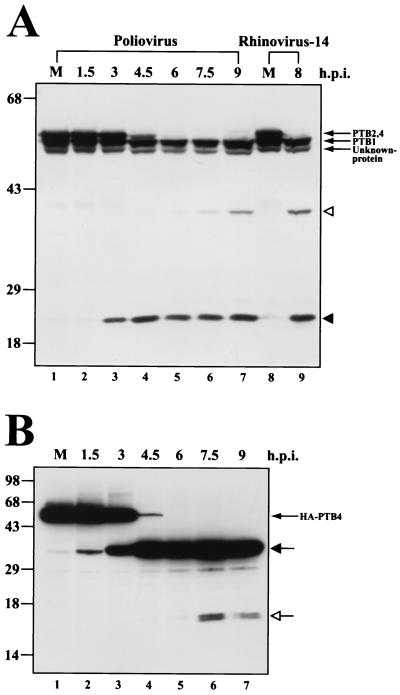
The cleavages of PTB proteins were observed in HeLa and HeLa12/HA-PTB4 cell lines infected with poliovirus or HRV-14. Figure 1A shows an immunoblot result with total cell lysates from poliovirus- or HRV-14-infected HeLa cells harvested at various times postinfection with a monoclonal antibody (DH-17; kindly provided by E. Wimmer) that recognizes the C-terminal part of PTB. Monoclonal antibody DH-17 revealed two upper bands (∼61 and ∼60 kDa), which are not resolved in Fig. 1A, and one lower band (∼57 kDa), as well as a band of unidentified protein, the smallest band, in mock-infected HeLa cells. A PTB cleavage product of about 26 kDa was detected 3 h after infection (Fig. 1A, lane 3), and a weak cleavage product of about 41 kDa was slightly observed 4.5 h after infection (Fig. 1A, lane 4). The amount of the cleavage products increased gradually as the virus infection proceeded. Two slower-migrating bands indicated as PTB2 and -4 were completely cleaved 6 h after infection (Fig. 1A, lane 5). A similar result was observed with the rhinovirus infection (Fig. 1A, lane 9). To reduce the complexity of the PTB cleavage patterns due to isoforms of PTB, we examined the cleavage pattern of PTB by using a cell line expressing N-terminal HA-tagged PTB4. HA-PTB4 was induced by removal of tetracycline from the medium, and the poliovirus was then inoculated. Cells were harvested, and HA-PTB4 protein was monitored by Western blotting with a monoclonal antibody against HA. One major band, indicated as HA-PTB4, was detected from mock-infected HeLa12/HA-PTB4 cells (Fig. 1B, lane 1). Upon poliovirus infection, the N-terminal cleavage product (∼35 kDa) of HA-PTB4 protein was detected 1.5 h after infection (Fig. 1B, lane 2), and HA-PTB4 was completely cleaved 6 h after infection (Fig. 1B, lane 5). At this time, ∼16-kDa double bands began to appear. This result indicates that PTB4 had been initially cleaved centrally, as indicated by the appearance of an ∼35-kDa polypeptide 1.5 h postinfection, and then the N-terminal part was further processed to smaller polypeptides, as indicated by the appearance of an ∼16-kDa polypeptide 6 h postinfection.
FIG. 1.
Cleavage of PTBs in poliovirus-infected cells. (A) HeLa cells (lanes 1 to 7) and H1-HeLa cells (lanes 8 and 9) were infected with poliovirus (lanes 2 to 7) or HRV-14 (lane 9). Samples were then harvested at the indicated times, Aliquots were subjected to SDS-PAGE and immunoblotted with monoclonal antibody against PTB. The numbers on top of panel A indicate the number of hours postinfection (h.p.i). The PTB fragments generated by poliovirus infection are indicated by solid and open arrowheads. Only the C-terminal parts of PTBs are revealed by Western blot analysis. (B) HA-tagged PTB4 was expressed in the stable HeLa cell line HeLa12/HA-PTB4 by withdrawal of tetracycline from culture medium. Forty-eight hours after induction, the cells were infected with poliovirus and analyzed by immunoblotting with anti-HA monoclonal antibody. The positions of the major and the minor cleavage products are indicated by solid and open arrows, respectively (Fig. 3A). Only the N-terminal parts of HA-PTB4 are revealed by Western blot analysis.
Poliovirus 3Cpro cleaves PTB isoforms.
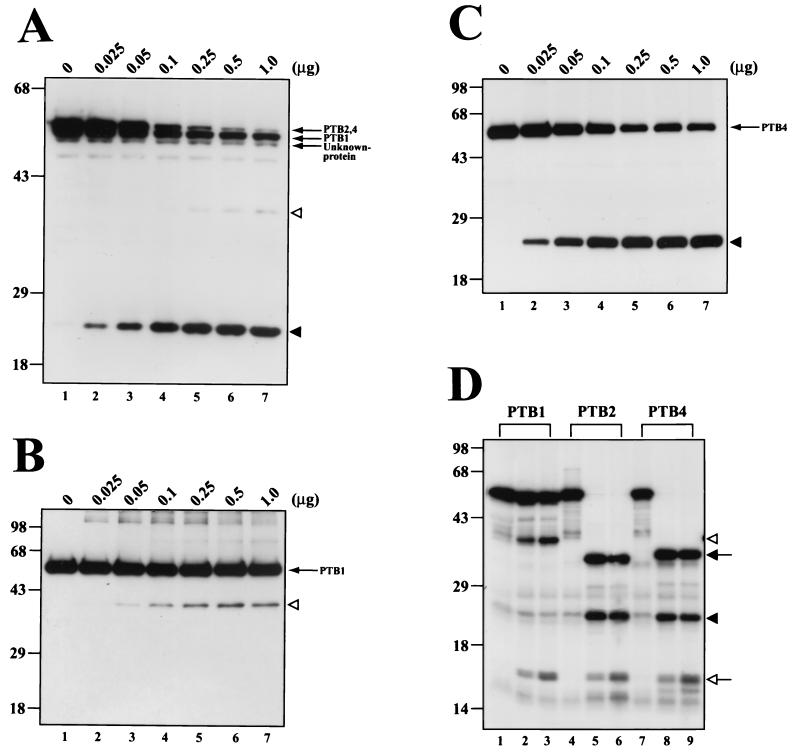
Poliovirus encodes two proteases, 2Apro and 3Cpro, that proteolytically process viral polyprotein (34, 50, 54, 60, 65-67, 80, 81) and modulate cellular functions (15, 18, 41, 42, 47, 51, 77, 85, 86). We tested whether one of these proteases is responsible for the cleavage of PTB. Polioviral polyproteins spanning P1-P2, containing 2Apro, and P1-P2-P3, containing both 2Apro and 3Cpro, were translated in HeLa extract, and the integrity of PTB was monitored by Western blot analysis. The PTB cleavage product was detected when P1-P2-P3 was produced, but not when P1-P2 was produced (data not shown). This result indicates that 3Cpro and/or 3CDpro is responsible for the cleavage of PTB. Cleavage of PTB isoforms by 3Cpro was investigated by incubating purified 3Cpro with HeLa cell extracts. After incubation of HeLa cell extract for 3 h with different amounts of 3Cpro, the PTB patterns were monitored by Western blot analysis with monoclonal antibody against PTB. As shown in Fig. 2A, isoforms of PTB were cleaved by the addition of 3Cpro in a dose-dependent manner. The cleavage pattern of PTB isoforms by 3Cpro was very similar to the cleavage pattern of PTBs induced by poliovirus infection (compare lanes 1 to 7 in Fig. 1A with lanes 1 to 7 in Fig. 2A).
FIG. 2.
In vitro cleavage of PTB proteins by recombinant 3Cpro. (A) Different amounts of purified recombinant 3Cpro were incubated with HeLa cell extracts (20 μg) at 37°C for 3 h. (B and C) Purified recombinant PTB1 or PTB4 (1.0 μg) was incubated with the indicated quantities of protease at 37°C for 4 h. Reactions were stopped by the addition of 2× Laemmli sample buffer and were analyzed by SDS-PAGE followed by immunoblotting with anti-PTB monoclonal antibody (A, B, and C). (D) Five microliters of [35S]methionine-labeled PTB isoforms was incubated with 1.0 μg of 3Cpro for 1 h (lanes 2, 5, and 8) or for 3 h (lanes 3, 6, and 9). Negative control samples incubated for 3 h without addition of 3Cpro are shown in lanes 1, 4, and 7. Samples were resolved by SDS-PAGE, and the protein bands were detected by autoradiography. The positions of major cleavage products of PTB are indicated by a solid arrow and solid arrowhead, and the minor cleavage products are indicated by an open arrow and open arrowhead.
Cleavage of PTB isoforms by 3Cpro was further confirmed with purified 3Cpro and PTB1 and -4 (see Fig. 4B and C). When purified PTB1 was incubated with purified 3Cpro, PTB1 was partially cleaved (Fig. 2B). The size of the cleavage product was about 41 kDa, as indicated by the open arrowhead in Fig. 2B, which suggests that 3Cpro cleaves PTB1 near the N-terminal end of the protein (note that the monoclonal antibody DH-17 recognizes the C-terminal part of PTB). On the other hand, PTB4 produced a smaller cleavage product of about 26 kDa, as indicated by the solid arrowhead in Fig. 2C, which indicates that the major 3Cpro target site in PTB4 is closer to the C-terminal end of the protein. Moreover, 3Cpro cleaved PTB4 better than PTB1 (compare the bands indicated by the solid arrowhead in Fig. 2C with those indicated by the open arrowhead in Fig. 2B). The same cleavage patterns of PTB isoforms by 3Cpro are also detected from PTBs in HeLa cell extracts (Fig. 2A).
FIG. 4.
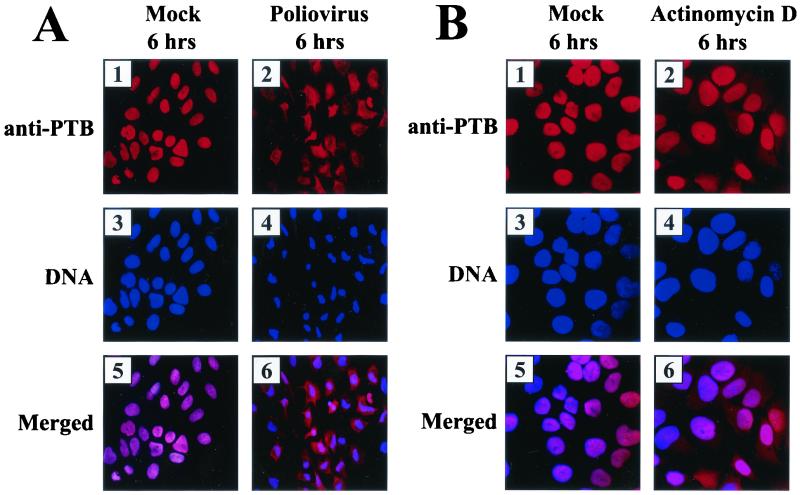
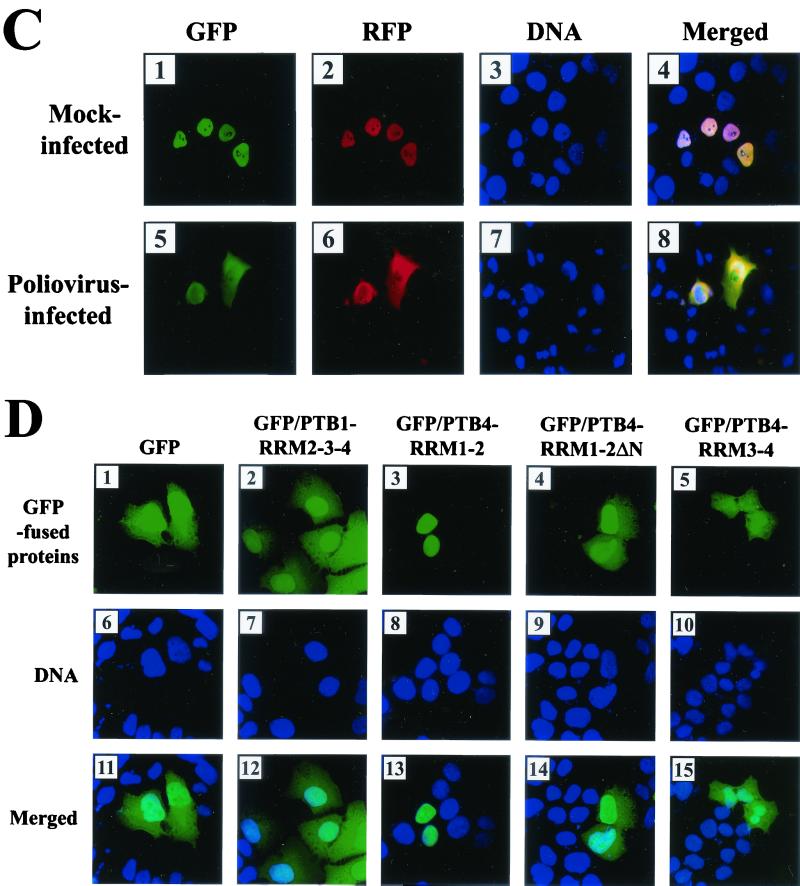
Subcellular localization of PTB and its derivatives. (A) Effect of poliovirus infection on PTB localization. HeLa cells were mock infected or infected with poliovirus for 6 h and then stained with antibody directed against PTB. The top panels show cells examined with a TRITC filter, the middle panels show Hoechst 33258 staining of the same field with a 4",6"-diamidino-2-phenylindole (DAPI) filter, and the bottom panels show the merged image of the TRITC and Hoechst images. (B) Effect of transcription blockage by actinomycin D on PTB localization. Uninfected HeLa cells were incubated without or with actinomycin D (5 μg/ml) for 6 h and then fixed and stained with anti-PTB monoclonal antibody. (C) HeLa cells were transfected with plasmids expressing GFP/PTB4/RFP and then mock infected or infected with poliovirus for 6 h from 40 h posttransfection. GFP fluorescence and RFP fluorescence were visualized with a fluorescein isothiocyanate (FITC) filter and a TRITC filter, respectively. DNA was examined with a DAPI filter after Hoechst 33258 staining. Panels 4 and 8 are merged images of FITC, TRITC, and Hoechst images. (D) Subcellular localization of PTB fragments. HeLa cells were transiently transfected with plasmids expressing GFP (panels 1, 6, and 11), GFP fused with PTB1-RRM2-3-4 (panels 2, 7, and 12), PTB4-RRM1-2 (panels 3, 8, and 13), PTB4-RRM1-2 lacking NLS (panels 4, 9, and 14), and PTB4-RRM3-4 (panels 5, 10, 15). GFP fluorescence was visualized with an FITC filter. DNA was stained with Hoechst 33258 and examined with a DAPI filter.
The cleavage patterns of known PTB isoforms were also investigated by using purified 3Cpro and [35S]methionine-labeled PTB1, -2, and -4 generated by in vitro translation. PTB2 and -4 were completely cleaved when incubated with 3Cpro for 1 and 3 h (Fig. 2D, lanes 5, 6, 8, and 9). The two major cleavage products of 26 and 35 kDa, as indicated by the solid arrowhead and solid arrow, and a minor protein of 16 kDa, as indicated by the open arrow, were generated by 3Cpro from PTB2 and -4. The intensity of the minor band increased as the incubation time was extended (Fig. 2D, lanes 5, 6, 8, and 9). A band of the same size was also detected when PTB1 was incubated with 3Cpro, in addition to a 41-kDa polypeptide, as indicated by the open arrowhead (Fig. 2D, lanes 2 and 3). It is probable that the 41-kDa band corresponds to the C-terminal cleavage product of PTB1 detected by the antibody (Fig. 2B, lanes 3 to 6) and that the 16-kDa band corresponds to the N-terminal end of PTB1, which is common to all PTB isoforms. These in vitro data and poliovirus infection experiments strongly suggest that PTBs are cleaved by polioviral 3Cpro and/or 3CDpro at several sites. PTB2 and -4 contain a major target site, which is highly susceptible to 3Cpro, around the central region of the proteins, and at least one minor target site, less susceptible to 3Cpro, at the N-terminal region of PTB1, -2, and -4.
Determination of 3Cpro target sites in PTB.
In order to determine the 3Cpro target sites in PTBs, purified PTB1 and -4 were incubated with 3Cpro, and the cleavage products were resolved by SDS-PAGE. The cleavage products of 41 kDa from PTB1 and 26 kDa from PTB4 were subjected to automated Edman degradation to determine their N-terminal sequences. Five residues were unambiguously assigned for each fragment, and their sequences were exactly matched by parts of PTB1 and -4. The sequence of 41-kDa fragments started from both Ala-149 and Ala-153, which are depicted as minor cleavage sites in Fig. 3A. Two amino acid peaks, corresponding to amino acid residues preceded by these two alanines, were detected at each cycle of Edman degradation, which indicates that the 41-kDa band contains two polypeptides beginning with Ala-149 and Ala-153. The 26-kDa polypeptide from PTB4 matched the amino acid sequence from Ala-322 in PTB4, which is depicted as the major cleavage site in PTB4. Note that the major cleavage site resides in exon 9 of PTB2 and PTB4, but that exon 9 is deleted in PTB1 by alternative splicing (Fig. 3A). Therefore, PTB2 and PTB4 are highly susceptible to 3Cpro, which yields two major cleavage products, as indicated by the solid arrow and solid arrowhead in Fig. 2D. However, PTB1 is only partially cleaved by 3Cpro at the minor cleavage sites, indicated by the open arrow and open arrowhead (compare lanes 5, 6, 8, and 9 with lanes 2 and 3 in Fig. 2D). These minor cleavage sites in PTB2 and PTB4 are also partially cleaved by 3Cpro with an efficiency that is similar to that of the corresponding site in PTB1, as indicated by the open arrow in Fig. 2D.
Confirmation of 3Cpro target sites by site-directed mutagenesis.
Poliovirus 3Cpro requires a glutamine residue at the P1 position of the scissile bond (50, 61, 64-66, 80). To assess whether glutamine 148, 152, and 321 residues in PTB4 are essential for efficient cleavage by 3Cpro, we mutated the glutamine residues to alanine residues. Five different PTB4 mutant proteins were synthesized in micrococcal nuclease-treated RRL (Promega) in the presence of [35S]methionine. The cleavage patterns of PTB and its mutants by 3Cpro are shown in Fig. 3B. 3Cpro cleaved PTB4 into approximately 35-, 26-, and 16-kDa fragments, as indicated by the solid arrow, solid arrowhead, and open arrow for lane 2 in Fig. 3B. A mutant containing mutation of Gln-321 to Ala (Q321A) did not produce polypeptides of 35 or 26 kDa (lanes 4, 6, 8, and 12 in Fig. 3B). These findings indicate that glutamine at amino acid residue 321 is the major determinant of the 3Cpro target site. The PTB4 mutant Q321A yields two cleavage products of 41 and 16 kDa, which is similar to PTB1, but PTB1 lacks the major cleavage site, as indicated by the open arrowhead and open arrow in Fig 3B (lane 4). Double mutants PTB4(Q148/321A) and PTB4(Q152/321A) also yielded partially cleaved products of 41 and 16 kDa. The smaller fragment of PTB4(Q148/321A) was slightly bigger than that of PTB4(Q152/321A) (compare lane 6 with lane 8 in Fig. 3B). This result is probably due to the cleavage of PTB4 by 3Cpro at the remaining minor cleavage sites (Q152↓153A and Q148↓149A) in the mutants PTB4(Q148/121A) and PTB4(Q152/321A), respectively. Protein cleavage by 3Cpro was abolished when a triple mutation (Q148/152/321A) was introduced into PTB4 (lane 12 in Fig. 3B), which indicates that the minor cleavage sites in PTB4 are cleaved by 3Cpro and that the three cleavage sites function independently of each other.
Poliovirus infection redistributes PTBs to different subcellular compartments.
At the early stage of poliovirus infection, PTB2 and PTB4 are divided into two parts: RRM1-2 (∼35 kDa) and RRM3-4 (∼26 kDa) (Fig. 1A). The cleavage of PTB1 and the second cleavage of RRM1-2 of PTB2 and PTB4 occur during the late stage of poliovirus infection (Fig. 1A). In the absence of poliovirus infection, PTB is mainly localized in the nucleus (Fig. 4A, panel 1), which is attributed to the nuclear localization signal (NLS) residing with the first 55 aa of PTB (36, 72, 74). PTB is redistributed to the cytoplasm upon poliovirus infection. The majority of PTBs are detected in the cytoplasm 6 h postinfection (Fig. 4A, panel 2). Note that the nuclei of infected cells are condensed 6 h postinfection (compare panel 3 with 4 in Fig. 4A).
The contribution of transcription blockage by poliovirus infection to the redistribution of PTBs was investigated by blocking cellular mRNA transcription with actinomycin D treatment. In fact, actinomycin D treatment induced the partial redistribution of PTBs to the cytoplasm, as seen in Fig. 4B, panel 2. The majority of PTBs, however, stayed in the nucleus 6 h after actinomycin D treatment (Fig. 4B, panels 1 and 2). On the other hand, the majority of PTBs were redistributed 6 h postinfection (Fig. 4A, panel 2). This indicates that main reason for the redistribution of PTB is not transcription blockage, which, itself, is induced by poliovirus infection. The possible mechanisms for the redistribution of PTB by poliovirus infection are discussed below.
Redistribution of PTB was further confirmed by transfecting the PTB4 gene containing the GFP and the RFP genes at its N- and C-termini, respectively (Fig. 4C). Before poliovirus infection, the PTB fused with both GFP and RFP was confined in the nucleus, but excluded from the nucleolus (panel 4 in Fig. 4C), as detected by green fluorescence for GFP (panel 1 in Fig. 4C) and by red fluorescence for RFP (panel 2 in Fig. 4C). Upon poliovirus infection, PTB4 was redistributed throughout the cell in a pattern similar to that found for the PTB antibody (compare panel 2 in Fig. 4A with panels 5 and 6 in Fig. 4C), which indicates that both the N- and C-terminal parts of PTB move into the cytoplasm upon poliovirus infection and that GFP- and RFP-fused PTB4 proteins reflect the localization pattern of PTB.
The distribution patterns of PTB fragments, potentially generated by polioviral 3Cpro, were investigated by using PTB fragments fused with GFP (Fig. 4D). Full-length PTB is detected in the nucleus (panels 1 and 2 in Fig. 4C), and RRM2-3-4 of PTB1, which is the major C-terminal fragment of PTB1, was partially redistributed to the cytoplasm (panels 2 and 12 in Fig. 4D). The same was true for the RRM3-4 of PTB4, which is the major C-terminal cleavage product of PTB4, even though the redistribution pattern is more dramatic and shows punctated forms in the nucleus (panel 5 in Fig. 4D). The other major PTB4 cleavage product, PTB(1-324), resides in the nucleus (panels 3 and 13 in Fig. 4D). The extreme N-terminal end of PTB4, aa 1 to 57, is likely to be responsible for the nuclear localization of PTB(1-324), since PTB4 RRM1-2ΔN (aa 58 to 324) is distributed to both the nucleus and cytoplasm (Fig. 4D, panels 4 and 14). In conclusion, all of the PTB fragments, lacking the N-terminal end (aa 1 to 57), localize at least in part in the cytoplasm.
Effect of PTB fragments on translation of polioviral mRNA.
The effect of fragmentation of PTB by polioviral 3Cpro on the translation of polioviral mRNA was investigated, because PTB is known to facilitate the IRES-dependent translation of polioviral mRNA (28, 33, 37, 38). The effect of PTB fragments on polioviral IRES-dependent translation was investigated by cotransfecting a reporter plasmid (Fig. 5A), yielding a dicistronic mRNA composed sequentially of Renilla luciferase (RLuc), polioviral IRES, firefly luciferase (FLuc), and effector plasmids (Fig. 5B) producing variable parts of PTBs. The cap-dependent translation and the polioviral IRES-dependent translation were monitored with Renilla luciferase and firefly luciferase activities, respectively. Cotransfection of full-length PTB4 augmented the translation of polioviral IRES-dependent translation in a dose-dependent manner (bars 2 to 4 in Fig. 5C). Truncated PTB1, GFP/PTB1-RRM2-3-4, strongly inhibited the translation of polioviral mRNA in a dose-dependent manner (lanes 5 to 7 in Fig. 5C), which indicates that truncated PTB has a dominant-negative effect on polioviral mRNA translation. The effect of the major fragment of PTB4 and PTB2 was also investigated by cotransfecting GFP/PTB4-RRM3-4 or GFP/PTB4-RRM1-2 with the reporter plasmid. The C-terminal region of PTB4 partially inhibited polioviral mRNA translation (bars 14 to 16 in Fig. 5C), but on the other hand, the N-terminal domain of PTB4, expressed from plasmid GFP/PTB4-RRM1-2, did not affect the translation of polioviral mRNA (bars 8 to 10 in Fig. 5C). This is probably due to the nuclear localization of PTB4-RRM1-2 protein, which contains an NLS at the N-terminal end of the molecule. In poliovirus-infected cells, however, the N-terminal domain is redistributed in the cytoplasm, as shown by the GFP signal from PTB4 fused with GFP at the N terminus (panel 5 in Fig. 4C). This is likely to be due to the modification of the NPC by the poliovirus infection (see below and reference 29). In order to mimic the localization pattern of the N-terminal part of PTB4 in poliovirus-infected cells, the NLS of PTB4 was deliberately removed, as shown in the Fig. 5B diagram of GFP/PTB4-RRM1-2ΔN. The N-terminal part of PTB4, which contains RRM1 and -2, but lacks an NLS, inhibited the translation of polioviral mRNA (bars 11 to 13 in Fig. 5C). This indicates that the translation of polioviral mRNA is inhibited indirectly by its own product, 3Cpro and/or 3CDpro, through the proteolytic cleavage of PTBs (PTB1, PTB2, and PTB4).
FIG.5.
Effect of PTB and its derivatives on poliovirus IRES activity. (A) Schematic diagram of the reporter plasmid. This plasmid expresses a dicistronic mRNA consisting of the Renilla luciferase (RLuc) gene at the first cistron and the polioviral IRES-firefly luciferase (FLuc) gene at the second cistron. The RLuc and FLuc are directed by scanning and IRES-dependent translation, respectively. (B) Schematic diagram of effector plasmids. These plasmids produce full-length PTB4 or PTB1 and PTB4 derivatives fused with GFP. (C) Effect of PTB or its derivatives on poliovirus IRES activity. 293T cells were cotransfected with 0.75 μg of dicistronic plasmid and effector plasmids as indicated in the chart at the bottom. The total amount of effector plasmids was maintained at a constant level by adding control plasmid pEGFP-C1 when necessary. Forty-eight hours after transfection, Renilla luciferase and firefly luciferase activities were measured as described in Materials and Methods, and the relative ratio of firefly luciferase to Renilla luciferase activity in each cell lysate was calculated. The columns and bars represent the means and standard deviations of four independent transfection experiments. The numbers in the chart represent micrograms of DNA.
Here we report that a polioviral protease, 3Cpro, digests a cellular RNA-binding protein, PTB, that augments polioviral IRES-dependent translation. Cleavage of PTB is likely to inhibit the translation of polioviral mRNA by reducing the availability of intact PTB and/or by the inhibitory effect of PTB fragments on IRES-dependent translation (Fig. 5C [see below]).
DISCUSSION
At the early stage of poliovirus infection, cap-dependent translation of host cellular mRNAs, which occurs through scanning, is inhibited by the cleavage of eIF4G mediated by the virus-encoded protease 2Apro (6, 19, 21, 51). The cleavage of eIF4G by 2Apro separates the eIF4E-binding site residing at the N terminus of the protein from eIF3- and eIF4A-binding sites that are essential for the recruitment of ribosome to mRNA (53). This cleavage results in the inhibition of cap-dependent translation, since cap-dependent translation requires eIF4G. On the other hand, translation of polioviral mRNAs, which is directed by an IRES element, continues until the late stage of virus infection, since IRES-dependent translation does not require the N-terminal part of eIF4G. However, translation of polioviral mRNAs eventually decreases at the late stage of virus infection (40).
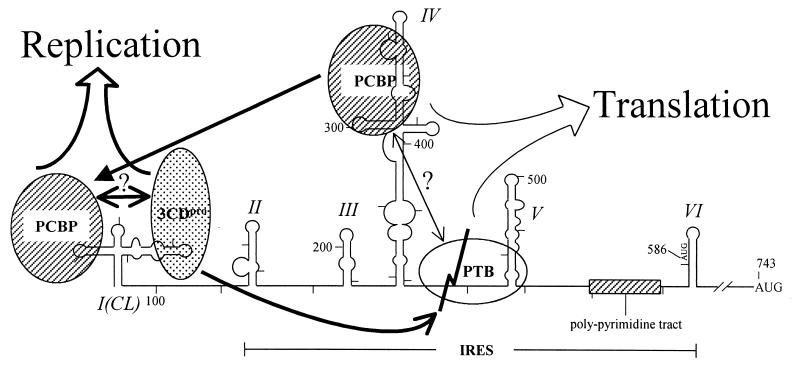
The translational machinery on the positive-sense RNA moves in the opposite direction against the replicational machinery synthesizing negative-strand RNA. When two machineries collide, translation continues, but replication stops (8, 24). Therefore, the blockage of translation is required for the molecular switching of viral RNAs from translation to replication. PCBP2, an RNA-binding protein, was suggested to play a key role in the mechanism of such a molecular switching of poliovirus (22, 24). According to the model, PCBP2, interacting with domain IV of poliovirus IRES, drives the translation of poliovirus mRNA at the early stage of virus infection (11, 12). Upon production of poliovirus proteins, one of the translation products, named 3CDpro, interacts with the CL RNA and thereby increases the affinity between PCBP2 and the CL structure at the 5" end of polioviral RNA. In this process, PCBP2 previously bound to domain IV of poliovirus IRES is transferred to the CL structure, and, consequently, the translation of poliovirus mRNA decreases, and replication of poliovirus RNA commences. The authors showed that 3CDpro dramatically increases the affinity of PCBP2 for CL RNA (from Kd values of ∼95 nM to ∼1 nM), but its affinity for domain IV remains the same (∼15 nM) (22). In this model, however, one enigma remains unsolved. As the authors pointed out, the overall cytoplasmic concentration of PCBP in the HeLa cell is about 100 nM, which is sufficient for PCBP to bind to both the CL structure and domain IV of the polioviral RNA, and the period of inhibition of translation of polioviral mRNA should be long enough not to block replication for a whole round of RNA synthesis. The simple transference of PCBP from domain IV to the CL structure is not likely to be sufficient to continuously inhibit translation.
Here we report that polioviral 3Cpro and/or 3CDpro, which exerts protease activity (50, 52, 54, 65-67, 80), cleaves a cellular protein, PTB1, and its isoforms, PTB2 and PTB4, which are required for the IRES-dependent translation of polioviral mRNAs. We also show that the PTB fragments generated by protease 3Cpro are mobilized from the nucleus to the cytoplasm (Fig. 4) and inhibit translation of polioviral mRNA (Fig. 5C). Accordingly, we propose that the cleavage of PTB by polioviral 3Cpro contributes at least in part to the molecular switch from translation to replication. The molecular switch mechanisms by the cleavage of PTBs and by the transference of PCBP from domain IV to the CL structure are not mutually exclusive. In fact, both of these activities are mediated by 3CDpro or its variant, 3Cpro, of poliovirus, and may contribute to molecular switching (Fig. 6). It is possible that PCBP and PTB bind to the polioviral IRES as a complex, since PCBP interacts with PTB through its N-terminal half (48) and PTB binds to several sites (nt 70 to 288 and 443 to 539 [domain V shown in Fig. 6], as well as nt 630 to 730) in the poliovirus IRES (35).
FIG. 6.
Model of molecular switching from translation to replication of poliovirus RNA. Proteins identified as interacting with the 5"NTR of poliovirus are depicted by ovals. Different proteins are shaded differently. For simplicity of the model, only one PTB binding is drawn, even though several PTB-binding sites are present in polioviral IRES. The events required for translation at the early stage of viral infection are depicted with thin lines. The effect of the interaction between PCBP and PTB on translation is hypothetical, even though the protein-protein interaction between these proteins was demonstrated experimentally. Changes after the production of 3CDpro are depicted by thick lines. PTB is cleaved by 3CDpro as indicated by the folded line on PTB. The C-terminal PTB may stay on the IRES element blocking translation from this mRNA. At the same time, the removal of the N-terminal end of PTB prevents its interaction with other proteins, such as PCBP, hnRNP K, hnRNP L, and PTB itself (48). The PCBP, previously bound to the IRES element, may be transferred to the CL element in the presence of 3CDpro. With these changes, replication will commence.
Knowing these properties of PTB, PCBP, and poliovirus IRES, we can envision the molecular switch process by revising the molecular switching mechanism proposed by Gamarnik and Andino (24). At the early stage of poliovirus infection, PTB and PCBP, together with other factors, facilitate the translation of poliovirus mRNA. PTB may stabilize the binding of PCBP to the IRES element and vice versa, since PTB interacts with both PCBP (48) and poliovirus IRES (33, 35). When 3CDpro accumulates during the late stage of viral infection, the PTBs are cleaved by 3Cpro and/or 3CDpro at the major and the minor cleavage sites (Fig. 3A). Consequently, the number of intact PTBs required for translation decreases, and the number of cleaved PTBs inhibiting translation increases in the cytoplasm. This results in an overall inhibition of poliovirus IRES-dependent translation and allows the replication of poliovirus mRNA to commence (8, 24).
How do the cleavage products of PTBs block the translation of poliovirus mRNA? The fragments of PTBs may interfere with the binding of intact PTBs to poliovirus IRES. The RNA-binding domain of PTB resides in RRM3 and RRM4 (63, 72). Therefore, the C-terminal parts of PTBs (RRM2-3-4 of PTB1 and RRM3-4 of PTB2 and PTB4) are likely to stay on the poliovirus IRES and by so doing block the binding of intact PTBs to the IRES element. The cleaved PTBs, however, cannot interact with PCBP, since both RRM1 and RRM2 are required for protein-protein interaction between PTB and PCBP (48). The disconnection between PCBP and PTB may facilitate liberation of PCBP from the RNA-protein complex, especially when PCBP is associated with 3CDpro bound to the CL, which results in an inhibition of the translation of poliovirus mRNA.
Alternatively, the cleaved PTBs may no longer function as translational activators that facilitate the recruitment of translational machinery to the IRES element regardless of the presence of PCBP. In this respect, the C-terminal parts of PTBs (RRM2-3-4 of PTB1 and RRM3-4 of PTB2 and PTB4) containing RNA-binding activity may block translation by preventing the binding of the intact PTBs to the IRES element. The N-terminal part of PTBs (RRM1-2 of PTB2 and PTB4) inhibits translation (Fig. 5C, bars 11 to 13), possibly by sequestrating RNA-binding proteins such as PCBP, PTB, hnRNP K, and hnRNP L, which are known to interact with the N-terminal part of PTB (32, 48). Another possible explanation for the translational inhibition by the PTB fragments is an indirect effect of the disturbance of nuclear function of PTB, such as modulation of alternative splicing of cellular mRNAs. According to this assumption, the PTB fragments should have a dominant-negative effect on the nuclear function of full-length PTB, resulting in generation of a putative translational factor(s) required for poliovirus IRES-dependent translation. We consider this model to be less plausible, even though we cannot rule out the possibility, since all of the inhibitory fragments of PTBs are localized in the cytoplasm (lanes 5 to 7 and 11 to 16 in Fig. 5C) and the PTB fragment localized in the nucleus did not inhibit translation of poliovirus RNA (lanes 8 to 10 in Fig. 5C).
The conservation of the glutamine-glycine pairs is one of the hallmarks of processing of poliovirus polyprotein by 3Cpro (50, 52, 54, 61, 64-67, 80). However, it has been known that the 3Cpro target sites in the picornaviruses are not strictly conserved. The glycine moiety at the P1" position of the cleavage sites can be replaced by a serine, threonine, alanine, valine, or methionine residue in in vitro cleavage assays (4, 68). Moreover, a systematic mutagenesis study of the poliovirus 3C/3D junction revealed that changes of the glycine at P1" to alanine or serine were tolerable in terms of virus production (45). This indicates that Gln-Ala and Gln-Ser pairs, as well as the Gln-Gly pair, can be cleaved by 3Cpro in vivo. Here, we report that a cellular protein, PTB, is cleaved by 3Cpro at Gln-Ala pairs. The three cleavage sites of 3Cpro in PTB are at Gln-Ala pairs instead of the Gln-Gly pair. By using site-directed mutagenesis, we showed that the glutamine at aa 148, 152, and 321 of PTB4 is essential for cleavage by 3Cpro (Fig. 3B).
When we compared the efficiencies of cleavages of Gln-Ala pairs in PTB, it was found that the kinetics of their cleavage differ widely. Cleavage after Gln 321 occurs efficiently, but cleavages after Gln148 and Gln152 occur slowly. No cleavage of other Gln-Ala pairs in PTB was detected. The 321Gln-Ala322 site in an Ala-X-Pro-Gln/Ala (where X is any amino acid) context was cleaved more easily than the 148Gln-Ala149 and 152Gln-Ala153 sites in an Ala-X-Y-Gln/Ala (Y is any amino acid except proline) context. This is consistent with the results obtained from the cleavage kinetics of the TATA-binding transcription factor, TBP, which contains Ala-X-Pro-Gln/Gly. Those results suggested that both alanine at P4 and proline at P2 affect cleavage of TBP by 3Cpro (18). The cleavage pattern of PTB4, shown in Fig. 2A, also suggests that the proline residue at P2 contributes to determine the efficiency of cleavage by 3Cpro.
PTB is a shuttling protein that migrates between the nucleus and the cytoplasm (58), even though most of the protein is detected in the nucleus (36, 72, 74). Upon poliovirus infection, PTB protein is evenly distributed to the nucleus and the cytoplasm in most cells (Fig. 4A, panel 2). By using the GFP/PTB4/RFP construct, which contains GFP and RFP at the N terminus and C terminus of PTB4, respectively, we demonstrated that both the N-terminal (RRM1-2 and RRM1) and C-terminal (RRM3-4 and RRM2-3-4) fragments of PTB were redistributed to the cytoplasm after poliovirus infection (Fig. 4C, panels 5, 6, and 8). It is intriguing that not only the C-terminal parts of PTB4 (RRM3-4) lacking NLS, but also the N-terminal parts of PTB4 (RRM1-2) containing an RRM are redistributed to the cytoplasm. We suspect that this is due to the leakage of proteins from the nucleus to the cytoplasm or to impairment of the nuclear import machinery induced by poliovirus infection. Recently, Gustin and Sarnow reported that poliovirus infection induces degradation of Nup153 and p62, components of the NPC (29). The modification of the NPC results in a redistribution of hnRNP A1, K, and C from the nucleus to the cytoplasm. Therefore, it is plausible that the modification of the NPC affects the distribution pattern of PTB. Moreover, inhibition of host cell transcription by poliovirus infection (15, 18, 85, 86) is likely to contribute partially to the relocalization of PTB, since transcriptional inhibition of host cell transcription by actinomycin D induces the partial redistribution of PTB to the cytoplasm (compare panel 1 with panel 2 in Fig. 4B).
The cleavage of PTB is likely to influence cellular physiology other than the translational inhibition of poliovirus mRNA. First, the translation of cellular mRNAs containing IRES elements modulated by PTB, such as Apaf-1 (59) and BiP (49), is likely to be affected by the cleavage of PTB. Second, the splicing of pre-mRNAs, which is modulated by PTB (55, 70, 72, 79), is also likely to be changed. The effect of PTB cleavage on cellular functions remains to be elucidated.
Acknowledgments
We thank Thomas Pfister and Eckard Wimmer (State University of New York at Stony Brook) for their gifts of anti-PTB monoclonal antibody, pHRV-14, pET3Chc, H1-HeLa cells, and 293T cells. We thank members of I. Hwang's laboratory (Pohang University of Science and Technology) for help with image acquisition.
This work was supported in part by the G7, NRL, and the Molecular Medicine Research Group Programs of MOST and by a grant from the PNRC of KOSEF.
REFERENCES
- 1.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 2.Andino, R., N. Böddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. [DOI] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5" end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, E., M. Luo, G. Vriend, M. G. Rossmann, A. C. Palmenberg, G. D. Parks, M. J. Nicklin, and E. Wimmer. 1987. Implications of the picornavirus capsid structure for polyprotein processing. Proc. Natl. Acad. Sci. USA 84:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltimore, D. 1968. Structure of the poliovirus replicative intermediate RNA. J. Mol. Biol. 32:359-368. [DOI] [PubMed] [Google Scholar]
- 6.Barco, A., E. Feduchi, and L. Carrasco. 2000. A stable HeLa cell line that inducibly expresses poliovirus 2Apro: effects on cellular and viral gene expression. J. Virol. 74:2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5" cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsham, G. J., N. Sonenberg, and Y. V. Svitkin. 1995. The role of the La autoantigen in internal initiation. Curr. Top. Microbiol. Immunol. 203:85-98. [DOI] [PubMed] [Google Scholar]
- 10.Belsham, G. J., and N. Sonenberg. 2000. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 8:330-335. [DOI] [PubMed] [Google Scholar]
- 11.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5" noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5" nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, K. H., E. A. Brown, and S. M. Lemon. 1993. Cell type-specific proteins which interact with the 5" nontranslated region of hepatitis A virus RNA. J. Virol. 67:6716-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark, M. E., P. M. Lieberman, A. J. Berk, and A. Dasgupta. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conte, M. R., T. Grune, J. Ghuman, G. Kelly, A. Ladas, S. Matthews, and S. Curry. 2000. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19:3132-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig, A. W. B., Y. V. Svitkin, H. S. Lee, G. J. Belsham, and N. Sonenberg. 1997. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol. Cell. Biol. 17:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, S., and A. Dasgupta. 1993. Identification of the cleavage site and determinants required for poliovirus 3CPro-catalyzed cleavage of human TATA-binding transcription factor TBP. J. Virol. 67:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrenfeld, E. 1996. Initiation of translation by picornavirus RNAs, p. 549-573. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Ehrenfeld, E., and B. L. Semler. 1995. Anatomy of the poliovirus internal ribosome entry site. Curr. Top. Microbiol. Immunol. 203:65-83. [DOI] [PubMed] [Google Scholar]
- 21.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 22.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5" untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamarnik, A. V., and R. Andino. 1996. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 15:5988-5998. [PMC free article] [PubMed] [Google Scholar]
- 24.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5" noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 26.Ghetti, A., S. Pinol-Roma, W. M. Michael, C. Morandi, and G. Dreyfuss. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20:3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gil, A., P. A. Sharp, S. F. Jamison, and M. A. Garcia-Blanco. 1991. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 5:1224-1236. [DOI] [PubMed] [Google Scholar]
- 28.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez-Escolano, L., and R. M. del Angel. 1996. Nuclear proteins bind to poliovirus 5" untranslated region. Arch. Med. Res. 27:413-419. [PubMed] [Google Scholar]
- 31.Hahm, B., S. H. Back, T. G. Lee, E. Wimmer, and S. K. Jang. 1996. Generation of a novel poliovirus with a requirement of hepatitis C virus protease NS3 activity. Virology 226:318-326. [DOI] [PubMed] [Google Scholar]
- 32.Hahm, B., O. H. Cho, J. E. Kim, Y. K. Kim, J. H. Kim, Y. L. Oh, and S. K. Jang. 1998. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 425:401-406. [DOI] [PubMed] [Google Scholar]
- 33.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellen, C. U. T., C.-K. Lee, and E. Wimmer. 1992. Determinants of substrate recognition by poliovirus 2A proteinase. J. Virol. 66:3330-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellen, C. U. T., T. V. Pestova, M. Litterst, and E. Wimmer. 1994. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5" nontranslated region. J. Virol. 68:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, S., T. J. Deerinck, M. H. Ellisman, and D. L. Spector. 1997. The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 137:965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunt, S. L., and R. J. Jackson. 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5:344-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang, S. K., and E. Wimmer. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4:1560-1572. [DOI] [PubMed] [Google Scholar]
- 40.Jen, G., B. M. Detjen, and R. E. Thach. 1980. Shutoff of HeLa cell protein synthesis by encephalomyocarditis virus and poliovirus: a comparative study. J. Virol. 35:150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joachims, M., K. S. Harris, and D. Etchison. 1995. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology 211:451-461. [DOI] [PubMed] [Google Scholar]
- 42.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaminski, A., and R. J. Jackson. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. Jackson. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 45.Kean, K. M., N. Teterina, and M. Girard. 1990. Cleavage specificity of the poliovirus 3C protease is not restricted to Gln-Gly at the 3C/3D junction. J. Gen. Virol. 71:2553-2563. [DOI] [PubMed] [Google Scholar]
- 46.Kenan, D. J., C. C. Query, and J. D. Keene. 1991. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16:214-220. [DOI] [PubMed] [Google Scholar]
- 47.Kerekatte, V., B. D. Keiper, C. Badorff, A. Cai, K. U. Knowlton, and R. E. Rhoads. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395-405. [DOI] [PubMed] [Google Scholar]
- 49.Kim, Y. K., B. Hahm, and S. K. Jang. 2000. Polypyrimidine tract-binding protein inhibits translation of BiP mRNA. J. Mol. Biol. 304:119-133. [DOI] [PubMed] [Google Scholar]
- 49a.Kim, Y. K., S. H. Back, J. Rho, S. H. Lee, and S. K. Jang. 2001. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 29:5009-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 51.Kräusslich, H. G., M. J. H. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krausslich, H. G., and E. Wimmer. 1988. Viral proteinase. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 53.Lampher, B. J., R. Kirchweager, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis factor 4G (eIF4G) with picornaviral proteases—implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 268:21975-21983. [DOI] [PubMed] [Google Scholar]
- 54.Lawson, M. A., and B. L. Semler. 1990. Picornavirus protein processing—enzymes, substrates, and genetic regulation. Curr. Top. Microbiol. Immunol. 161:49-80. [PubMed] [Google Scholar]
- 55.Lin, C. H., and J. G. Patton. 1995. Regulation of alternative 3" splice site selection by constitutive splicing factors. RNA 1:234-245. [PMC free article] [PubMed] [Google Scholar]
- 56.Luz, N., and E. Beck. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 65:6486-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michael, W. M., H. Siomi, M. Choi, S. Pinol-Roma, S. Nakielny, Q. Liu, and G. Dreyfuss. 1995. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harbor Symp. Quant. Biol. 60:663-668. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell, S. A., E. C. Brown, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 21:3364-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicklin, M. J., H. G. Krausslich, H. Toyoda, J. J. Dunn, and E. Wimmer. 1987. Poliovirus polypeptide precursors: expression in vitro and processing by exogenous 3C and 2A proteinases. Proc. Natl. Acad. Sci. USA 84:4002-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicklin, M. J. H., K. S. Harris, P. V. Pallai, and E. Wimmer. 1988. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J. Virol. 62:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh, Y. L., B. Hahm, Y. K. Kim, H. K. Lee, J. W. Lee, O. Song, K. Tsukiyama-Kohara, M. Kohara, A. Nomoto, and S. K. Jang. 1998. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pallai, P. V., F. Burkhardt, M. Skoog, K. Schreiner, P. Bax, K. A. Cohen, G. Hansen, D. E. Palladino, K. S. Harris, M. J. Nicklin et al. 1989. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 264:9738-9741. [PubMed] [Google Scholar]
- 65.Pallansch, M. A., O. M. Kew, B. L. Semler, D. R. Omilianowski, C. W. Anderson, E. Wimmer, and R. R. Rueckert. 1984. Protein processing map of poliovirus. J. Virol. 49:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmenberg, A. C. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603-623. [DOI] [PubMed] [Google Scholar]
- 67.Palmenberg, A. C., and R. R. Rueckert. 1982. Evidence for intramolecular self-cleavage of picornaviral replicase precursors. J. Virol. 41:244-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parks, G. D., J. C. Baker, and A. C. Palmenberg. 1989. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J. Virol. 63:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5"-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 70.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 71.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 72.Perez, I., J. G. McAfee, and J. G. Patton. 1997. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36:11881-11890. [DOI] [PubMed] [Google Scholar]
- 73.Pogue, G. P., C. C. Huntley, and T. C. Hall. 1994. Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses. Arch. Virol. Suppl. 9:181-194. [DOI] [PubMed] [Google Scholar]
- 74.Romanelli, M. G., F. Weighardt, G. Biamonti, S. Riva, and C. Morandi. 1997. Sequence determinants for hnRNP I protein nuclear localization. Exp. Cell Res. 235:300-304. [DOI] [PubMed] [Google Scholar]
- 75.Ruecker, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 76.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 77.Shiroki, K., T. Isoyama, S. Kuge, T. Ishii, S. Ohmi, S. Hata, K. Suzuki, Y. Takasaki, and A. Nomoto. 1999. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silvera, D., A. V. Gamarnik, and R. Andino. 1999. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5"-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 274:38163-38170. [DOI] [PubMed] [Google Scholar]
- 79.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 80.Skern. T., and H.-D. Liebig. 1994. Picornins 2A and 3C. Methods Enzmol. 244:583-595. [DOI] [PubMed] [Google Scholar]
- 81.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761-770. [DOI] [PubMed] [Google Scholar]
- 82.Trono, D., J. Pelletier, N. Sonenberg, and D. Baltimore. 1988. Translation in mammalian cells of a gene linked to the poliovirus 5" noncoding region. Science 241:445-448. [DOI] [PubMed] [Google Scholar]
- 83.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5"-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yalamanchili, P., K. Weidman, and A. Dasgupta. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176-185. [DOI] [PubMed] [Google Scholar]