Abstract
Human immunodeficiency virus type 1 (HIV-1) requires the incorporation of cyclophilin A (CypA) for replication. CypA is packaged by binding to the capsid (CA) region of Gag. This interaction is disrupted by cyclosporine (CsA). Preventing CypA incorporation, either by mutations in the binding region of CA or by the presence of CsA, abrogates virus infectivity. Given that CypA possesses an isomerase activity, it has been proposed that CypA acts as an uncoating factor by destabilizing the shell of CA that surrounds the viral genome. However, because the same domain of CypA is responsible for both its isomerase activity and its capacity to be packaged, it has been challenging to determine if isomerase activity is required for HIV-1 replication. To address this issue, we fused CypA to viral protein R (Vpr), creating a Vpr-CypA chimera. Because Vpr is packaged via the p6 region of Gag, this approach bypasses the interaction with CA and allows CypA incorporation even in the presence of CsA. Using this system, we found that Vpr-CypA rescues the infectivity of viruses lacking CypA, either produced in the presence of CsA or mutated in the CypA packaging signal of CA. Furthermore, a Vpr-CypA mutant which has no isomerase activity and no capacity to bind to CA also rescues HIV-1 replication. Thus, this study demonstrates that the isomerase activity of CypA is not required for HIV-1 replication and suggests that the interaction of the catalytic site of CypA with CA serves no other function than to incorporate CypA into viruses.
Cyclophilin A (CypA) is a ubiquitous cytosolic protein that was originally discovered as the major binding protein for the immunosuppressive drug cyclosporine (CsA) (20). The only known catalytic function of CypA in vitro is the cis/trans isomerization of peptide bonds between the carboxyl end of an amino acid and the amino end of a proline within a polypeptide chain. It has been demonstrated that CypA accelerates slow, proline-limited steps in the refolding of several proteins, including collagen, transferrin, and Drosophila rhodopsin (17). The hydrophobic pocket of CypA contains the catalytic domain responsible for isomerization. CsA, by binding to this cavity, inhibits the isomerase activity of CypA (21, 41).
The earliest hint for a role for CypA in human immunodeficiency virus type 1 (HIV-1) replication came from studies performed by Luban and colleagues, who showed by using the yeast two-hybrid system that CypA interacts directly with HIV-1 Gag, the polyprotein precursor of the virus structural proteins (22). Further studies demonstrated that cytosolic CypA is specifically incorporated into nascent virus by binding to a proline-rich domain of the capsid (CA) region of Gag (12). Furthermore, crystal structure analysis reveals that the catalytic domain of the isomerase interacts with a proline-rich stretch of the single exposed loop of CA (36). Corroborating this observation, the CypA-Gag interaction can be competitively disrupted by CsA. Either the addition of CsA onto virus producer cells or the introduction of mutations in the CypA-binding domain of the CA region of Gag prevents the incorporation of CypA into budding viruses (4, 12, 35). Most important, these viruses which lack CypA are less infectious than wild-type viruses (3, 5, 12, 35). This demonstrates that HIV-1 requires the incorporation of host CypA to efficiently replicate in host cells. Given that CA forms the coat which surrounds and protects the viral genome and that CypA possesses a cis/trans isomerase activity, it has been proposed that CypA acts as an uncoating factor (18, 23). In this model, CypA, by catalyzing the cis/trans isomerization of peptidyl-prolyl bonds of prolines in CA, destabilizes CA-CA interactions, triggering the breakdown of the shell of CA and permitting the delivery of the viral genome into the host cell cytosol.
In the present study, we developed a system which allows us to evaluate domains of CypA, including its catalytic domain, that may be necessary for HIV-1 infection.
MATERIALS AND METHODS
DNA constructs.
The proviral clones R9 (NL4.3 derivative) and R9 G89V were described previously (14, 31). To allow an efficient in-trans incorporation of CypA, human CypA was fused to the C terminus of viral protein R (Vpr) in the context of the pLR2P-vprRT vector (39) (a generous gift from X. Wu) after removal of the reverse transcriptase (RT) coding region. A PCR-amplified MluI-XhoI DNA with the human CypA coding region was ligated in the vector above, creating the Vpr-CypA vector. Furthermore, we introduced by PCR amplification an HIV-1 protease site between Vpr and CypA to allow liberation of CypA after its virus incorporation. The cleavage sequence from the junction between p7 and p1 (ERQAN-FLGKI) has been chosen because a previous study showed that this site fused to the C terminus of Vpr does not influence virus infectivity (33).
Transfections and infections.
All viruses used in this study were transiently expressed by calcium phosphate transfection with a mixture of 20 μg of R9 proviral DNA together with 20 μg of Vpr-CypA vector as described previously (37). Viral supernatants, harvested 72 h posttransfection, were filtered through a 0.2-μm-pore-size filter to remove cellular debris. Cell-free infections were performed as previously described (33). Briefly, virus (1 ng of p24) was added to CD4+ HeLa cells (80,000 cells). Forty-eight hours postinfection, infected cells were detected by 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gal) staining. Viral load was standardized by p24 antigen by using the enzyme-linked immunosorbent assay (NEN-Dupont). CsA (Sigma) was used at a concentration of 20 μM on virus producer cells.
Protein analyses and antibodies.
Purification and immunoblot analysis of viruses produced from 293T transfected cells were conducted as previously described (37). Anti-CA and anti-Vpr antisera were obtained through the AIDS Research and Reference Program. Rabbit anti-CypA serum was obtained by immunization with recombinant human CypA protein generated using the previously described GST-CypA plasmid (22).
RESULTS
Incorporation of Vpr-CypA fusion protein into HIV-1.
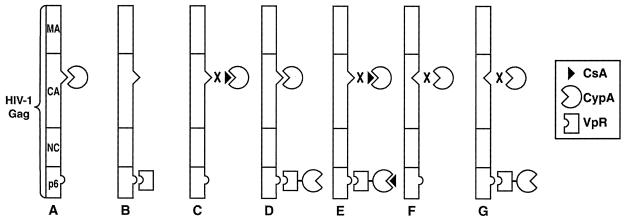
To determine whether the isomerase activity of CypA is required for HIV-1 replication, it is necessary to mutate the catalytic domain of CypA and assess the effect of this mutation on HIV-1 infection. However, as CypA is an endogenous cytosolic protein which is not encoded by the HIV-1 viral genome, analysis of CypA does not lend itself to traditional molecular analysis by mutagenesis. Overexpression in trans of mutant CypA in virus-producing cells would at best result in the incorporation of a mixture of both endogenous and mutant CypA into the virus. Thus, we sought an approach that would allow us to introduce mutant forms of CypA into HIV-1 yet block the incorporation of endogenous CypA. We exploited the fact that foreign proteins can be efficiently incorporated in trans into nascent HIV-1 particles when fused to Vpr. Importantly, Vpr is incorporated into HIV-1 via a pathway that is distinct from that used by CypA (35). Specifically, CypA is incorporated into nascent viruses by binding to the CA region of Gag (Fig. 1A), whereas Vpr is packaged into particles by binding to the p6 domain of Gag (Fig. 1B). We fused the gene encoding human CypA to the C terminus of Vpr as described previously (39), creating a Vpr-CypA chimera. In the absence of the drug CsA, we expect to observe the incorporation of both endogenous CypA and the Vpr-CypA chimera (Fig. 1D). However, in the presence of CsA, only the Vpr-CypA chimera would be incorporated into the virus but not endogenous CypA (Fig. 1E). Furthermore, we expect to observe the packaging of the Vpr-CypA chimera even in a virus mutated in the CypA-binding domain of the CA region of Gag (Fig. 1G). This approach would not only allow the incorporation of CypA independently of the CA region of HIV-1 Gag but also the incorporation of CypA mutants which lack cis/trans isomerase activity.
FIG. 1.
Model for in-trans CypA incorporation into HIV-1. (A) Under wild-type conditions, endogenous CypA is incorporated via the glycine 89-proline 90 CA region of HIV-1 Gag. (B) Vpr is incorporated via the p6 region of HIV-1 Gag. (C) CsA prevents endogenous CypA incorporation by competitively inhibiting CypA-Gag interactions. (D) In the absence of CsA, both endogenous CypA and CypA fused to Vpr are incorporated. (E) CsA prevents the incorporation of endogenous CypA but not that of the Vpr-CypA chimera. (F) Mutation of the glycine 89-proline 90 CA site inhibits endogenous CypA incorporation. (G) The Vpr-CypA chimera is incorporated despite the lesion in the glycine 89-proline 90 CA packaging signal.
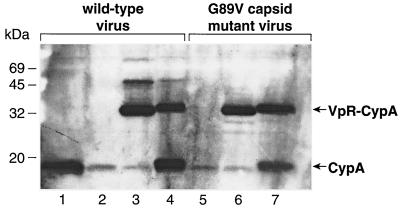
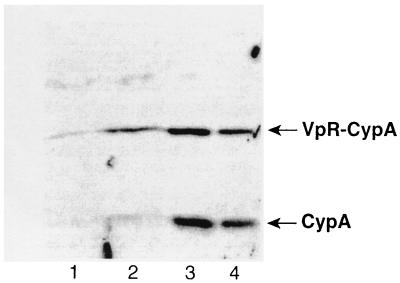
We then tested the feasibility of this approach. Specifically, 293T cells were cotransfected with the R9 proviral DNA (NL4.3 derivative) (14) and the Vpr-CypA construct as described previously (37). Twenty-four hours posttransfection, cells were grown in the presence or absence of CsA. Seventy-two hours posttransfection, supernatants of transfected cells were harvested, filtered, and centrifuged over a sucrose cushion. Amounts of pelleted viruses were quantified by p24 enzyme-linked immunosorbent assay. Purified viruses were analyzed by immunoblotting using anti-CypA antibodies. As expected, CsA strongly decreased the incorporation of endogenous CypA (18 kDa) compared to untreated virus (Fig. 2, lanes 1 and 2). In sharp contrast, CsA does not inhibit the incorporation of the Vpr-CypA fusion protein (32 kDa) (Fig. 2, lane 3). This demonstrates that our system permits the virus incorporation of CypA independently of the CA region of Gag. As the fused Vpr protein might interfere with the activity of CypA within the Vpr chimera, we also introduced an HIV-1 protease cleavage site between Vpr and CypA (called Vpr-Pr-CypA) permitting the release of CypA from Vpr after viral incorporation. We found that, like the Vpr-CypA chimera, the Vpr-Pr-CypA fusion protein is efficiently incorporated into particles despite the presence of CsA (Fig. 2, lane 4). The increase in size of uncleaved Vpr-Pr-CypA compared to the Vpr-CypA is due to the presence of the protease site. Importantly, we found that approximately half of the Vpr-Pr-CypA molecules are cleaved. Note that the slight increase in size of the cleaved CypA (18 kDa) is due to the presence of a few residues from the protease cleavage site. This confirms that this population of CypA arises exclusively from the cleavage of the Vpr-Pr-CypA chimera.
FIG. 2.
Incorporation of Vpr-CypA fusion protein into HIV-1. 293T cells were transfected with proviral DNA encoding wild-type R9 (lanes 1 to 4) or R9 G89V CA mutant (lanes 5 to 7) and were cotransfected with Vpr-CypA (lanes 3 and 6) or Vpr-Pr-CypA (lanes 4 and 7) in the presence (lanes 2 to 4) or the absence (lanes 1, 5, 6, and 7) of CsA. Purified viruses (3 μg of p24 per lane) were analyzed for their CypA content by immunoblotting using anti-CypA antibodies. CsA (20 μM) was added to 293T cells 24 h posttransfection.
Previous studies showed that the introduction of mutations in the CypA-binding domain of CA prevents the incorporation of endogenous CypA. Specifically, mutations of glycine 89 or proline 90 in the proline-rich stretch of the exposed loop of HIV-1 CA obstruct host CypA incorporation (12, 14, 36, 40). Thus, we analyzed the incorporation of Vpr-CypA and Vpr-Pr-CypA into these CA mutants. Specifically, 293T cells were cotransfected with the R9 G89V proviral clone (31) and Vpr-CypA or Vpr-Pr-CypA construct. As expected, the CA G89V mutation drastically inhibits the packaging of endogenous CypA (Fig. 2, lane 5). Importantly, we observed an efficient incorporation of both Vpr-CypA (Fig. 2, lane 6) and Vpr-Pr-CypA (Fig. 2, lane 7) fusion proteins into R9 G89V mutant virus, similar to that observed in wild-type R9 virus. These results further demonstrate that this Vpr fusion system permits the incorporation of CypA independently of the CA region of Gag.
Incorporation of Vpr-CypA fusion protein into CypA-deficient viruses restores their infectivity.
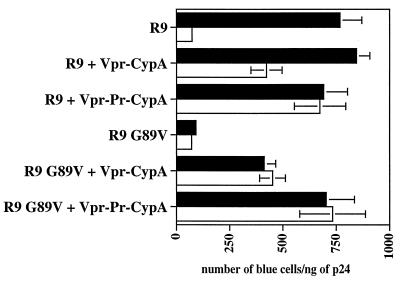
After showing that the addition of CsA to virus producer cells or the introduction of mutations in the CypA-binding site of CA (G89V) inhibits the incorporation of endogenous CypA (Fig. 2), we verified that these CypA-deficient viruses are not infectious. CD4+ HeLa cells containing an integrated LTR-beta-galactosidase gene were used as targets for single-round infection assays as previously reported (37). As expected, we found that both CsA-produced and CA G89V mutant viruses exhibit a 10-fold decrease in infectivity compared to untreated wild-type R9 virus (Fig. 3). This confirms that HIV-1 requires the incorporation of CypA for efficient replication.
FIG. 3.
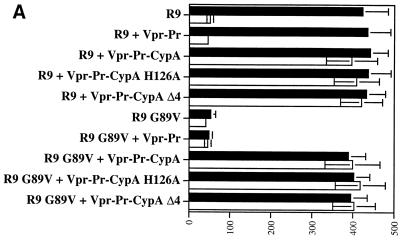
The Vpr-CypA chimera rescues the infectivity of CypA-deficient viruses. 293T cells were transfected with proviral DNA encoding wild-type R9 or R9 G89V CA mutant and were cotransfected with Vpr-CypA or Vpr-Pr-CypA in the presence □ or the absence ▪ of CsA. Viruses (1 ng of p24) were tested for their capacity to infect CD4+ HeLa cells. Infectivity was monitored by X-Gal staining. Results (triplicates) are expressed as number of blue cells per nanogram of p24. Results are representative of two independent experiments.
We next examined if the incorporation of the Vpr-CypA chimera can rescue the infectivity of these CypA-deficient viruses. Importantly, we found that the packaging of the Vpr-CypA chimera restores R9 infectivity despite the presence of CsA (CsA-produced R9) (Fig. 3). Similarly, the Vpr-CypA fusion protein rescues the infectivity of the G89V CA mutant virus (Fig. 3). Note that we observed a partial rescue with the Vpr-CypA chimera which does not contain the internal protease site (around 50% of rescue), and almost complete rescue with the Vpr-Pr-CypA chimera containing the cleavage site (around 90% of rescue). This is likely due to the presence of Vpr still fused to CypA, which partially interferes with the activity of CypA. Together, these results further demonstrate a requirement for CypA in HIV-1 infectivity and that the Vpr-CypA chimera can functionally replace endogenous CypA.
The isomerase activity of CypA is not required for HIV-1 infectivity.
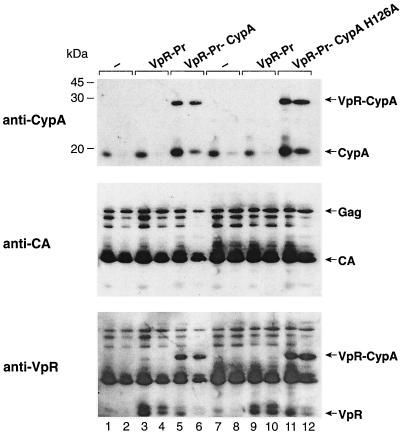
After demonstrating that the in-trans incorporation of Vpr-CypA is suitable for identifying domains of CypA necessary for HIV-1 replication, we asked whether the isomerase activity of CypA is specifically required for HIV-1 infectivity. It is crucial that the rescue of infectivity of the Vpr-CypA chimera occurs even in the presence of CsA, which is well known to inhibit the isomerase activity of CypA. Thus, our data above would seem to already suggest that the isomerase activity of CypA is not necessary for HIV-1 infection. Therefore, to further examine whether the isomerase activity of CypA is required for viral infectivity, we fused Vpr to a mutant CypA known to lack isomerase activity (41). This mutant CypA contains a lesion in the catalytic site (H126A) that abolishes both isomerase activity and the capacity to bind to the HIV-1 Gag (5, 10) or to CsA (41). First, we examined the incorporation of the Vpr-Pr-CypA H126A chimera into HIV-1. As above, 293T cells were cotransfected with the R9 proviral DNA and the Vpr-Pr-CypA H126A construct in the presence or absence of CsA. As controls, R9 was also cotransfected with the Vpr-Pr-wild-type CypA or with the Vpr-Pr empty plasmid (not fused to CypA but containing the cleavage site). Purified viruses were analyzed by immunoblotting using anti-CypA, anti-CA, and anti-Vpr antibodies. As shown above (Fig. 2), CsA prevents the incorporation of endogenous CypA into wild-type R9 virus (Fig. 4, lane 2) compared to untreated virus (Fig. 4, lane 1). In this experiment, we also examined the in-trans incorporation of Vpr not fused to CypA called Vpr-Pr. The immunoblot analysis using anti-Vpr antibodies revealed that Vpr-Pr is efficiently incorporated into R9 viruses in the presence or absence of CsA (Fig. 4, lanes 3 and 4). Note that the amount of Vpr incorporated into wild-type R9 virus is relatively low (Fig. 4, lanes 1 and 2). However, the in-trans packaging of Vpr-Pr strongly promotes the total Vpr levels in these particles (Fig. 4, lanes 3 and 4). The two specific bands detected by the anti-Vpr antibodies correspond to uncleaved Vpr-Pr and cleaved Vpr. As shown in Fig. 2, in the absence of CsA, both endogenous CypA and Vpr-Pr-CypA are incorporated into R9 (Fig. 4, lane 5). In contrast, in the presence of CsA, Vpr-Pr-CypA but not endogenous CypA is incorporated into R9 (Fig. 4, lane 6). As shown previously, approximately 50% of Vpr-Pr-CypA fusion proteins are cleaved by HIV-1 protease (Fig. 4, lane 6). Note that the summation of incorporation of endogenous CypA and cleaved CypA (from the chimera) explains the strong intensity of the CypA band (18 kDa) (Fig. 4, lane 5). As shown in Fig. 2, a slight increase in size of the cleaved CypA due to the presence of a few residues from the protease cleavage site can be observed. This confirms that this population of CypA arises exclusively from the cleavage of the Vpr-Pr-CypA chimera. It is important that the incorporation of the Vpr-Pr-CypA fusion protein does not influence either Gag packaging or Gag processing (Fig. 4, anti-CA immunoblot). Also significant, we found that the incorporation of the Vpr-Pr-CypA H126A chimera (Fig. 4, lanes 11 and 12) is similar to that of Vpr-Pr-wild-type CypA in the absence or presence of CsA. Furthermore, we found that the Vpr-Pr-CypA H126A is efficiently cleaved by the HIV-1 protease (Fig. 4, lanes 11 and 12).
FIG. 4.
In-trans incorporation of CypA mutant which lacks isomerase activity. 293T cells were transfected with a proviral DNA encoding wild-type R9 (lanes 1 to 12) or cotransfected with a construct encoding Vpr-Pr alone (lanes 3, 4, 9, and 10), Vpr-Pr-CypA (lanes 5 and 6), or Vpr-Pr-CypA H126A (lanes 11 and 12) in the presence (lanes 2, 4, 6, 8, 10, and 12) or absence (lanes 1, 3, 5, 7, 9, and 11) of 20 μM CsA. Produced viruses were analyzed for CypA, CA, and Vpr content by immunoblotting using specific antibodies. −, control.
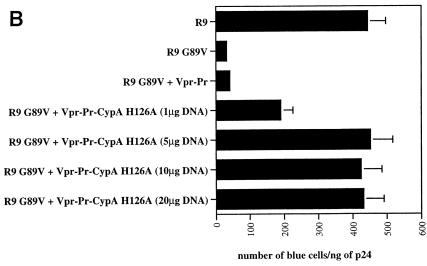
After demonstrating that the Vpr-Pr-CypA H126A chimera is efficiently incorporated in HIV-1, we examined whether this mutant CypA, which lacks isomerase activity and is unable to bind to Gag, could rescue HIV-1 replication. To explore this issue, viruses, which have incorporated the Vpr-Pr-CypA H126A chimera, were tested for their capacity to infect CD4+ HeLa cells as shown above (Fig. 3). We found that CypA-deficient viruses, such as CsA-produced R9 virus or the R9 G89V CA mutant, exhibit a 10-fold decrease in infectivity compared to untreated wild-type R9 virus. Note that the incorporation of Vpr-Pr into R9 or R9 G89V viruses does not influence their infectivity. As shown above, the in-trans incorporation of Vpr-Pr-wild-type CypA restores the infectivity of CypA-deficient viruses (CsA-produced R9 or R9 G89V CA viruses). Most importantly, the Vpr-Pr-CypA H126A chimera fully rescued R9 infectivity even in the presence of CsA (Fig. 5A). It is crucial that the CypA H126A mutant cannot bind CsA, obviating the possibility that the observed rescue of replication is due to CypA titrating away excess CsA. Corroborating this observation, we found that infectivity of the R9G89V mutant virus can also be restored by the in-trans incorporation of the Vpr-Pr-CypA H126A chimera. Altogether, these data would seem to argue against a role for CypA as an uncoating factor, at least via its isomerase activity. To rule out the possibility that the rescue does not arise from overincorporation of CypA-H126A, we performed similar experiments using decreasing amounts of Vpr-CypA-H126A. Specifically, CypA-deficient viruses were cotransfected with a range of complementary DNA encoding Vpr-CypA-H126A. Resulting viruses were then analyzed by Western blotting for CypA content and were tested for infection (Fig. 5B). Importantly, we found that even a low amount of Vpr-CypA-H126A (i.e., 5 μg of DNA; Fig. 5B, lane 2) still rescues the infectivity of CypA-deficient viruses. It is important that the rescue of CypA-deficient viruses by the Vpr-CypA H126A mutant is observed even in the presence of CsA (Fig. 4, lane 12). Thus, even if mutant CypA H126A retains a tiny, residual catalytic activity, it would be blocked by a large excess of CsA present in the system. This strongly indicates that the rescue does not arise from overincorporation of CypA-H126A.
FIG.5.
The isomerase activity of CypA is dispensable for HIV-1 infectivity. (A) 293T cells were transfected with proviral DNA encoding wild-type R9 or R9 G89V CA mutant and were cotransfected with Vpr-Pr, Vpr-Pr-CypA, Vpr-Pr-CypA H126A, or Vpr-Pr-CypA Δ4 (RKKK148/151/154/155AAAA) in the presence (□) or the absence (▪) of CsA. Viruses (1 ng of p24) were tested for their capacity to infect CD4+ HeLa cells. Infectivity was monitored by X-Gal staining. Results (duplicates) are expressed in number of blue cells per nanogram of p24. Results are representative of two independent experiments. (B) 293T cells were transfected with proviral DNA (20 μg) encoding R9 G89V CA mutant and were cotransfected with increasing concentrations of Vpr-Pr-CypA H126A (1, 5, 10, and 20 μg of DNA; lanes 1, 2, 3, and 4, respectively) in the presence of CsA. After p24 standardization, produced viruses were analyzed for CypA content by immunoblotting using anti-CypA antibodies and for infectivity using CD4+ HeLa cells. Infectivity was monitored by X-Gal staining. Results (duplicates) are expressed in number of blue cells per nanogram of p24. Results are representative of two independent experiments.
We previously showed that CypA-deficient viruses attach to target cells less efficiently than wild-type viruses (31). We also reported that recombinant CypA possesses the capacity to bind to cell surface heparan sulfates (HS) via four basic residues located within its C terminus (31). Based on these observations, we postulated that these residues might play a role in HIV-1 attachment to target cells via HS. To address this possibility using a more physiological system, we fused a mutant CypA lacking these four residues to Vpr and tested the resulting chimera for its ability to rescue HIV-1 replication in the presence of CsA (Fig. 5A). We found that this mutant also rescued HIV-1 infectivity, suggesting that these residues are not required or not sufficient for HIV-1 attachment.
DISCUSSION
HIV-1 requires the incorporation of the host protein CypA for efficient replication. CypA is packaged into HIV-1 by specifically binding to the CA region of the Gag precursor. Given that CA comprises the shell that surrounds the viral genome and that CypA possesses a cis/trans isomerase activity, it has been proposed for years that CypA acts as an uncoating factor. In this model, CypA destabilizes CA-CA interactions, triggering the rupture of the CA coat, allowing the delivery of the viral genome in the cytosol of host cells. However, because the same domain of CypA is responsible for both its isomerase activity and its capacity to be packaged in the virus, it was impossible to date to determine if the isomerase activity of CypA was required for HIV-1 replication.
In the present study, we have addressed this issue by taking advantage of a Vpr-CypA fusion protein that bypasses the interaction with the glycine 89-proline 90 packaging signal of CA in Gag and allows the incorporation of CypA into viruses even in the presence of CsA. Using this system, we found that Vpr-CypA rescued HIV-1 replication despite the presence of CsA. We then fused to Vpr a CypA mutant which has no cis/trans prolyl isomerase activity and no capacity to bind to the CA region of Gag or CsA. Importantly, this CypA mutant also rescued HIV-1 replication in the presence of CsA. Altogether, our findings demonstrate that the isomerase activity of CypA is not required for HIV-1 replication and suggest that the interaction of the catalytic site of CypA with glycine 89-proline 90 motif of CA serves no other function than to incorporate CypA into viruses.
Our present data seem to argue against a role for CypA in HIV-1 CA uncoating. However, we cannot exclude the possibility that CypA acts on loci of CA other than the glycine 89-proline 90 packaging signal. We can imagine that, during virus maturation, the processing of the Gag precursor induces major structural changes in cleaved CA that may expose new high-affinity binding sites for CypA. Postassembly actions of CypA, such as uncoating, may be mediated via these new high-affinity binding sites of mature CA proteins. Supporting this hypothesis, several studies suggest that affinities of CypA for Gag and for CA are strikingly different (6, 9, 11). Specifically, Endrich and colleagues, using recombinant proteins, showed that the processing of immature CA, yielding mature CA, elicited conformational changes in its C-terminal domain. They showed that CypA binds with a higher affinity to mature CA than to immature CA. They identified two new high-affinity binding interaction sites between CypA and mature CA in the C-terminal domain of CA around glycine 156-proline 157 and glycine 223-proline 224 (11). They proposed that these new binding sites represent loci upon which CypA acts postassembly. However, these sites are located in the C-terminal domain of CA, which is essential for CA oligomerization and CA core formation (7, 8, 15, 16, 19, 38). Given that Colgan and colleagues demonstrated that Gag multimerization is required for CypA packaging (8), we cannot exclude the possibility that these sites (glycine 156-proline 157 and glycine 223-proline 224) play only an indirect role in CypA-CA interactions. To clearly demonstrate that these sites represent authentic loci for CypA action, these sites will have to be mutated in the context of whole virus, instead of recombinant proteins, and the resulting mutant viruses will have to be examined for their capacities to assemble, to form regular cores, and to enter and to infect target cells. Corroborating the hypothesis for CA refolding upon maturation, Dietrich and colleagues showed, using recombinant proteins, that CypA binds to cleaved CA in a manner different from its binding to unprocessed Gag (9). Specifically, they showed that the mutant CypA W121F binds efficiently to Gag but fails to bind to mature CA. Altogether, these observations suggest the occurrence of maturation-dependent conformational changes in CA that may potentiate the influence of CypA on CA uncoating.
Our present findings, however, seem to argue against the hypothesis that CypA-CA contacts are critical for HIV-1 infection at a postassembly step. Specifically, we showed that CsA does not inhibit the infectivity of a virus which has packaged in trans the Vpr-CypA chimera. Since it has been shown that CsA prevents the binding of CypA to CA (2), our data suggest that the catalytic site of CypA is not necessary for CypA function in HIV-1 replication after its packaging. Although we cannot rule out the possibility that CypA acts on CA via regions other than its catalytic domain, our present observations strongly suggest that the requirement for CypA in HIV-1 replication does not depend on an action of CypA on CA. Several in vitro studies clearly demonstrated that CsA prevents CypA-CA/Gag interactions (2, 13, 22, 36); however, one report suggested that a subpopulation of CA molecules may bind CypA even in the presence of CsA (1). This discrepancy may arise from the different provenance of CA utilized. For example, some studies generated recombinant CA as glutathione S-transferase fusion proteins (2, 13, 22, 36), whereas another generated CA from the processing of recombinant Gag by the HIV-1 protease (1). It is possible that the method used to generate CA may influence its binding properties to CypA. Furthermore, we cannot exclude the possibility that the use of recombinant proteins may not reflect in vivo conditions.
If CypA acts at a postassembly step other than uncoating, what is the function of CypA in the HIV-1 life cycle? Recent reports suggest that incorporated CypA may play an important role in HIV-1 entry into target cells. Specifically, Pushkarsky and colleagues recently showed that HIV-1 entry depends on interactions between virus-associated CypA and cell surface CD147 (29). Importantly, antibodies directed against CD147 inhibit the entry of HIV-1 into target cells but not that of viruses which do not require CypA for replication, such as simian immunodeficiency virus (29). In another report, we showed that CypA-deficient viruses attach to target cells less efficiently than do wild-type viruses (31). Given that recombinant CypA possesses the capacity to bind to target cells via HS and that HIV-1 requires cell surface HS to attach to a large number of adherent cells (31, 32), we postulated that packaged CypA mediates HIV-1 adsorption to target cells via HS. Corroborating this hypothesis, we and others showed that affinity-purified anti-CypA antibodies or an excess of recombinant CypA decreasees HIV-1 uptake by target cells (31, 34). Using recombinant CypA, we identified four basic residues in the C terminus of CypA responsible for binding to cell surface HS (31). We postulated that these residues might play a role in HIV-1 attachment to target cells. To address this possibility using a more physiological system, we fused a mutant CypA lacking these four residues to Vpr and tested the resulting chimera for its ability to rescue HIV-1 replication in the presence of CsA. To our surprise, this mutant also rescued HIV-1 infectivity, suggesting that these residues are not sufficient for HIV-1 attachment. Several possibilities may explain why these residues are not necessary for HIV-1 replication. One possibility is that either additional residues of CypA or residues distinct from the four residues described above are necessary for HIV-1 attachment. We originally assumed that these basic residues of CypA participate in viral attachment arose from experiments using recombinant CypA. However, one could imagine that the use of recombinant protein to demonstrate a role for CypA in HIV-1 attachment is not adequate. Interestingly, there is a close precedent in the literature supporting this hypothesis. Specifically, it has been demonstrated that recombinant gp120 binds to cell surface HS via basic residues located within its V3 loop (30), suggesting that gp120 may mediate HIV-1 attachment either via gp120-CD4 or via gp120-HS interactions. However, a growing body of evidence indicates that gp120 is not necessary for the initial attachment of HIV-1 to target cells. Indeed, several recent studies showed that viruses with a deletion of gp120 attach to human macrophages or to CD4+ HeLa cells at levels similar to those of wild-type viruses (24, 25, 27, 31, 32). Thus, these observations strongly suggest that the use of recombinant proteins such as gp120 or CypA to demonstrate their participation in HIV-1 adsorption in the context of whole virus must be regarded with caution. Another possibility to explain why the basic residues of CypA are not necessary for HIV-1 replication using our Vpr chimera system is that, although CypA is required for HIV-1 attachment, it may not mediate virus adsorption directly. One could envision that CypA, only in conjunction with an exposed virus-associated protein, mediates HIV-1 attachment. Alternately, one could also imagine that CypA facilitates the translocation of an attachment factor to the surface of the virus during viral maturation. Nevertheless, our present in-trans Vpr-CypA incorporation system will permit us to delineate the domains of CypA necessary for both CypA-CD147 and CypA-HS interactions. This will help to better define the respective roles of these interactions in HIV-1 entry.
We recently reported that the two major HIV-1 targets—CD4+ T lymphocytes and macrophages—express opposite patterns of the candidate HIV-1 attachment receptors CD4 and HS (32). Specifically, we showed that CD4+ T lymphocytes express high CD4 levels but no HS, whereas macrophages express low CD4 levels but high HS levels. We demonstrated that HS serve as the main class of attachment receptors for HIV-1 on macrophages, suggesting that CD4 alone is not sufficient to support the initial adsorption of HIV-1 to these cells. Furthermore, we found that HIV-1 attaches efficiently to CD4+ T lymphocytes despite the absence of cell surface HS (32), suggesting that high CD4 levels are sufficient to mediate the initial attachment. If CypA is only necessary for HIV-1 attachment, we would expect to observe replication of CypA-deficient viruses in CD4+ T lymphocytes. However, a previous study showed that the nonimmunosuppressive CsA analogue SDZ NIM 811 decreases CypA incorporation and replication in CD4+ T lymphocytes (26). This observation suggested to us a postentry role for CypA in HIV-1 replication. Corroborating this hypothesis, we recently found that the use of spinoculation (centrifugal inoculation), which bypasses the mechanistic requirement for the initial virus adsorption to target cells (28), does not fully rescue the infectivity of CypA-deficient viruses pseudotyped or not with the vesicular stomatitis virus envelope (Saphire et al., unpublished). These results strongly suggest that CypA acts at least at two distinct steps in the HIV-1 life cycle: entry and postentry.
In conclusion, we showed that the trans-complementation of CypA restores the infectivity of CypA-deficient viruses even in the presence of CsA. Furthermore, we found that neither the catalytic domain of CypA nor the glycine 89-proline 90 CA-binding site is essential for HIV-1 replication. These results suggest that the interaction of the catalytic site of CypA with CA serves no other function than to incorporate CypA into viruses. We are presently using our in-trans incorporation system to identify domains of CypA essential for HIV-1 replication. The identification of these regions will help to delineate the respective roles of CypA in HIV-1 entry and postentry.
Acknowledgments
We thank J. Kuhns for secretarial assistance.
This work was supported by U.S. Public Health Service grant no. AI46958 from the National Institute of Allergy and Infectious Diseases to P.A.G.
Footnotes
Publication no. 14250-IMM from the Department of Immunology, The Scripps Research Institute, La Jolla, Calif.
REFERENCES
- 1.Agresta, B. E., and C. A. Carter. 1997. Cyclophilin A-induced alterations of human immunodeficiency virus type 1 CA protein in vitro. J. Virol. 71:6921-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billich, A., F. Hammerschmid, P. Peichl, R. Wenger, G. Zenke, V. Quesniaux, and B. Rosenwirth. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 69:2451-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bristow, R., J. Byrne, J. Squirell, H. Trencher, T. Carter, B. Rodgers, E. Saman, and J. Duncan. 1999. Human cyclophilin has a significantly higher affinity for HIV-1 recombinant p55 than p24. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:334-336. [DOI] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colgan, J., H. E. H. Yuan, E. K. Franke, and J. Luban. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 70:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich, L., L. S. Ehrlich, T. J. LaGrassa, D. Ebbets-Reed, and C. Carter. 2001. Structural consequences of cyclophilin A binding on maturational refolding in human immunodeficiency virus type 1 capsid protein. J. Virol. 75:4721-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, T., A. Weimann, A. Borsetti, C. T. Walsh, and H. G. Gottlinger. 1997. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 71:7110-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endrich, M. M., P. Gehrig, and H. Gehring. 1999. Maturation-induced conformational changes of HIV-1 capsid protein and identification of two high affinity sites for cyclophilins in the C-terminal domain. J. Biol. Chem. 274:5326-5332. [DOI] [PubMed] [Google Scholar]
- 12.Franke, E. K., H. E. H. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 13.Franke, E. K., and J. Luban. 1996. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222:279-282. [DOI] [PubMed] [Google Scholar]
- 14.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 16.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 17.Gething, M.-J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 18.Grattinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Muller, and H. G. Krausslich. 1999. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 19.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H. G. Krausslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handschumacher, R. E., M. W. Harding, J. Rice, and R. J. Drugge. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 21.Ke, H., D. Mayrose, P. J. Belshaw, D. G. Alberg, S. L. Schreiber, Z. Y. Chang, F. A. Etzkorn, S. Ho, and C. T. Walsh. 1994. Crystal structures of cyclophilin A complexed with cyclosporin A and N-methyl-4-[(E)-2-butenyl]-4,4-dimethylthreonine cyclosporin A. Structure 2:33-44. [DOI] [PubMed] [Google Scholar]
- 22.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 23.Luban, J. 1996. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell 87:1157-1159. [DOI] [PubMed] [Google Scholar]
- 24.Marechal, V., F. Clavel, J. M. Heard, and O. Schwartz. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marechal, V., M. C. Prevost, C. Petit, E. Perret, J. M. Heard, and O. Schwartz. 2001. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J. Virol. 75:11166-11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlynar, E., D. Bevec, A. Billich, B. Rosenwirth, and A. Steinkasserer. 1997. The non-immunosuppressive cyclosporin A analogue SDZ NIM 811 inhibits cyclophilin A incorporation into virions and virus replication in human immunodeficiency virus type 1-infected primary and growth-arrested T cells. J. Gen. Virol. 78:825-835. [DOI] [PubMed] [Google Scholar]
- 27.Mondor, I., S. Ugolini, and Q. J. Sattentau. 1998. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 72:3623-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pushkarsky, T., G. Zybarth, L. Dubrovsky, V. Yurchenko, H. Tang, H. Guo, B. Toole, B. Sherry, and M. Bukrinsky. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 98:6360-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roderiquez, G., T. Oravecz, M. Yanagishita, D. C. Bou-Habib, H. Mostowski, and M. A. Norcross. 1995. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J. Virol. 69:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saphire, A. C. S., M. D. Bobardt, Z. Zhang, G. David, and P. A. Gallay. 2001. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 75:9187-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serio, D., T. A. Rizvi, M. Cartas, V. S. Kalyanaraman, I. T. Weber, H. Koprowski, and A. Srinivasan. 1997. Development of a novel anti-HIV-1 agent from within: effect of chimeric Vpr-containing protease cleavage site residues on virus replication. Proc. Natl. Acad. Sci. USA 94:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherry, B., G. Zybarth, M. Alfano, L. Dubrovsky, R. Mitchell, D. Rich, P. Ulrich, R. Bucala, A. Cerami, and M. Bukrinsky. 1998. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc. Natl. Acad. Sci. USA 95:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Göttlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 36.Vajdos, F. F., S. Yoo, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1997. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 6:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 41.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]